 Open Access Article
Open Access ArticleApplication and prospects of nucleic acid nanomaterials in tumor therapy
Weitong Lu†
 a,
Tianyu Chen†a,
Dexuan Xiaoa,
Xin Qina,
Yang Chen*b and
Sirong Shi*a
a,
Tianyu Chen†a,
Dexuan Xiaoa,
Xin Qina,
Yang Chen*b and
Sirong Shi*a
aState Key Laboratory of Oral Diseases & National Center for Stomatology & National Clinical Research Center for Oral Diseases, West China Hospital of Stomatology, Sichuan University, Chengdu 610041, Sichuan, China. E-mail: sirongshi@scu.edu.cn
bDepartment of Pediatric Surgery, Department of Liver Surgery & Liver Transplantation Center, West China Hospital of Sichuan University, Chengdu, Sichuan 610041, China. E-mail: chenyangwz1116@163.com
First published on 4th September 2023
Abstract
Cancer poses a great threat to human life, and current cancer treatments, such as radiotherapy, chemotherapy, and surgery, have significant side effects and limitations that hinder their application. Nucleic acid nanomaterials have specific spatial configurations and can be used as nanocarriers to deliver different therapeutic drugs, thereby enabling various biomedical applications, such as biosensors and cancer therapy. In recent decades, a variety of DNA nanostructures have been synthesized, and they have demonstrated remarkable potential in cancer therapy related applications, such as DNA origami structures, tetrahedral framework nucleic acids, and dynamic DNA nanostructures. Importantly, more attention is also being paid to RNA nanostructures, which play an important role in gene therapy. Therefore, this review introduces the developmental history of nucleic acid nanotechnology, summarizes the applications of DNA and RNA nanostructures for tumor treatment, and discusses the development opportunities for nucleic acid nanomaterials in the future.
1. Introduction
Cancer has become a significant threat to human health because of its extremely high fatality rate. According to a report by the International Agency for Research on Cancer, approximately 10 million people succumbed to cancer in 2020, and the number of cancer patients will increase by 60% globally in the next 20 years.1–3 Current clinical treatments for cancer mainly include radiotherapy, chemotherapy, and surgery; however, these treatments have shortcomings. Surgical treatment is only suitable for the early stages of cancer and the recurrence rate of cancer after surgery remains high, whereas radiotherapy and chemotherapy have toxic side effects on normal tissues and are prone to drug resistance. Therefore, the development of new tumor treatment regimens has become an important research topic in recent years. Compared with traditional antitumor drugs, nucleic acid nanomaterials possess excellent programmability, narrow particle size distribution, low cytotoxicity, and stable spatial structures; therefore, have the potential to improve the stability, delivery efficiency, and targeting ability of traditional chemotherapy drugs.4,5DNA is an important biological material that carries genetic information. Due to the stability of complementary base pairing, DNA is regarded as a bionanomaterial with extremely wide application prospects.6,7 In recent years, a variety of nucleic acid nanomaterials, such as DNA origami structures, tetrahedral framework nucleic acids (tFNAs), and dynamic DNA nanostructures have been synthesized and widely used in the field of targeted delivery of antitumor drugs. Because of their biocompatibility and modified specific targeting characteristics, nucleic acid nanomaterials can be effectively used to transport drugs to the tumor site, thereby improving the efficacy of drug delivery systems.4 More attention has also been paid to the role of RNA in cancer treatment, with siRNA and mRNA being representative molecules in gene therapy. Some RNA-based therapies have been approved by the U. S. Food and Drug Administration and European Medicines Agency,8 but their limited stability, high cost, and complexity remain significant challenges.9 Theoretically, designed RNA nanostructures can reduce RNA degradation by RNase H and improve transfection efficiency.10
Many nucleic acid nanomaterials have shown outstanding results in tumor chemotherapy, photodynamic therapy, gene therapy, and immunotherapy, and they are considered promising potential treatment methods.11–13 In this review, we outline the application and prospects of nucleic acid nanomaterials in tumor therapy and demonstrate their advantages and disadvantages by introducing their functions and properties.
2. Development of nucleic acid nanomaterials
Nucleic acid nanotechnology was first proposed by Nadrian Seeman in 1982, who first used DNA fragments to build structures, thereby creating a new field of research.14 Over the past 40 years, nucleic acid nanotechnology has developed rapidly, producing various spatial nanostructures from one to three dimensions. In 2006, Paul W. K. Rothemund first reported DNA origami technology, which allows for precise regulation of DNA nanostructures and assembly of nucleic acid nanomaterials of various shapes.15 Over the past ten years, advances in technology have led to the development of software to assist in the design of DNA nanostructures, such as caDNAno, oxDNA, and CanDo, thereby making it possible to use DNA origami technology to design more complex and delicate structures.16–18 For example, Hendrik et al. designed a 3D DNA nanostructure with complex curvatures by tuning the specific positions and patterns of the crossovers between adjacent DNA double helices.19 In 2018, Zhang et al. used DNA origami blocks to construct a “tensegrity triangle” with the assistance of caDNAno and further assembled it into a 3D rhombohedral crystalline structure.20In addition to its structural diversity, DNA origami technology can be used to precisely modify DNA nanostructures based on its ability to locate each base of the nanostructure. In 2015, Qiao enhanced the antitumor effect of DNA origami nanomaterials by modifying the surface of gold nanorods, thereby providing a promising candidate material for cancer diagnosis and treatment.11
In 2018, Fan Chunhai et al. first proposed the concept of framework nucleic acids (FNAs), which represented a new type of nucleic acid nanomaterial with unique physicochemical and biological properties.21 Among the various framework nucleic acid structures, tFNAs are widely used due to their simple synthesis, high yield, and good economic performance. tFNA is a nucleic acid nanomaterial with excellent anti-inflammatory and antioxidant capacities, which plays an important role in various disease models.22–24 For example, Qi discovered that tFNAs can exhibit protective effects by inhibiting apoptosis and alleviating oxidative stress, thereby making it suitable for treating acute kidney injury.25 Additionally, Zhu et al. reported that tFNAs can alleviate cellular inflammation by activating the AKT signaling pathway.26 As a drug delivery vehicle, tFNAs can enhance the efficacy of chemotherapy drugs; Fig. 1 shows several tFNAs drug delivery systems for cancer treatment.27–31
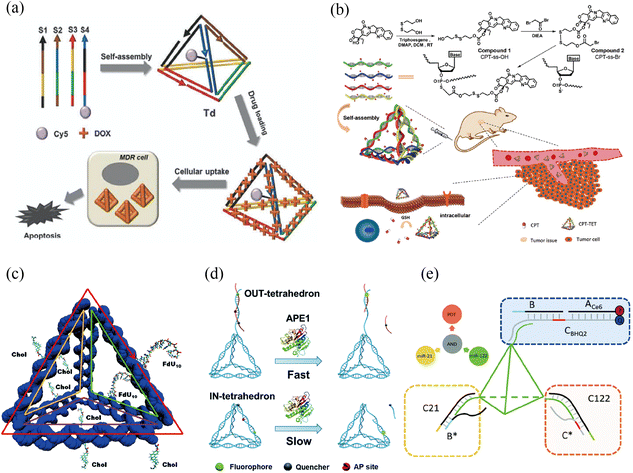 | ||
| Fig. 1 tFNA nanostructure functionalization for tumor therapy. (a) Self-assembled DNA tetrahedron for the treatment of multidrug-resistant cancer cells. Adapted with permission from ref. 27. Copyright 2013, Royal Society of Chemistry. (b) Camptothecin-grafted DNA tetrahedron for the inhibition of tumor growth. Adapted with permission from ref. 28. Copyright 2019, Wiley-VCH. (c) Cell targeting was achieved by locating multi-functional components via intercalation or insertion at one end of the tFNA. Adapted with permission from ref. 29. Copyright 2018, Royal Society of Chemistry. (d) Through intracellular ape1 binding, an apurinic/apyrimidinic endonuclease (ape1)-site-modified tFNA was employed to interfere with enzymatic catalysis, increasing tumor cell sensitivity to chemotherapeutic treatments. Adapted under the terms of the CC-BY Creative Commons Attribution 3.0 Unported license (https://creativecommons.org/licenses/by/3.0) from ref. 30. Copyright 2019, Royal Society of Chemistry. (e) A DNA-tetrahedron-based logic device was utilized to recognize microRNA for specific cell selection, after which a photosensitizer was activated for synergetic chemotherapy and photodynamic effects in the treatment of solid tumor tissue and metastasis. Adapted with permission from (ref. 31). Copyright 2021, Royal Society of Chemistry. | ||
In recent years, dynamic DNA nanostructures have been the focus of nucleic acid nanomaterial development. For example, Juul et al. designed a special three-dimensional DNA nanostructure that can realize reversible encapsulation and release of enzyme cargo without any form of covalent or noncovalent linkage for heat-sensitive drug delivery.32 In 2021, Tian Taoran designed a melittin-loaded DNA nanostructure for targeted therapy, which undergoes a conformational change after binding to the target protein, thereby releasing the drug. All of the aforementioned aspects highlight the broad potential of dynamic DNA structures in biomedicine.33
As DNA nanotechnology is rapidly developing, some researchers have also turned their attention to RNA, which shares many similarities with DNA.8 Early research on RNA nanotechnology began with the bacteriophage phi29,34 which can assemble inactive mutant pRNA into dimers, trimers, and hexamers, such as RNA dendritic polymers, of which RNA trimers represent a suitable carrier for drug delivery. In the early 2000s,35 tectoRNA and RNA units were intensively studied, and the generation of specific RNA nanoassemblies was made possible by computational design.36–39 Inspired by DNA origami, Geary et al.40 proposed a method of designing single-stranded RNA structures (RNA origami) using thermal annealing and co-transcriptional folding. In summary, canonical Watson-Crick base pairing, noncanonical base pairing, base stacking, and networks of tertiary contacts have laid the foundation for creating diverse RNA structures.40 The versatile structure and suitable pharmacokinetic and pharmacodynamic profiles of RNA nanotechnology showcase their potential in cancer therapy. These structures play an irreplaceable role in expanding the application of nucleic acid nanomaterials in cancer treatment. In the next section, we will focus on the applications of DNA and RNA structures in cancer therapy.
3. Properties and applications of nucleic acid nanomaterials
3.1 DNA nanotechnology
In addition, tFNAs can be used for gene therapy due to their ability to link oligonucleotides to regulate tumor-related genes. The connection between tFNAs and oligonucleotides is relatively conservative. Oligonucleotides can be hydrogen-bonded with sticky ends extending from the apex of tFNAs or directly covalently bonded to one end of the single strand as an extended cantilever. In addition to the aptamers described above, oligonucleotides have been studied extensively and include microRNAs (miRNAs) and small interfering RNAs (siRNAs). miRNA is a small nucleotide with a length of approximately 20–25 bp, that is mainly produced by cells to regulate various physiological activities. siRNA is a synthetic double-stranded RNA that can interfere with the transcription of mRNA to regulate the physiological processes of cells. It is difficult for both types of oligonucleotides to cross cell membranes without carriers because of their negative charges. tFNAs can enhance the transmembrane capacity of oligonucleotides, such as miRNAs and siRNAs, and protect them from degradation. Li et al. found that tFNAs help microRNA214-3p enter A549 cells to silence the SURVIVIN gene and promote apoptosis.50 Similarly, tFNAs can also be loaded with siRNA and aptamers to target and interfere with the normal physiological processes of tumor cells to promote their apoptosis. For example, Xiao et al. linked tFNAs with AS1411 to siBraf, which interferes with Braf gene expression in melanoma; moreover, the complex showed strong cleavage of Braf mRNA and promoted apoptosis in melanoma cells.13,51
In addition to directly acting on tumor cells, tFNAs can be involved in indirect tumor therapy in various ways. Tumor immunotherapy refers to enhancing the immunogenicity of tumor cells or the killing effect of the immune system in various ways to achieve antitumor activity. Cytosine-phosphate-guanine oligodeoxynucleotides (CpG ODNs) are commonly used to stimulate dendritic cells and macrophages to secrete pro-inflammatory factors, such as tumor necrosis factor-α and interleukin-1, resulting in enhanced innate immune strength. Liu et al. co-linked antigens and CpGs on tFNAs because CpGs can help the immune system generate stronger and longer-lasting immune effects than tFNAs linked to antigens alone.52 In addition, tFNAs linked to miRNAs can achieve similar effects. Qin et al. demonstrated that tFNAs linked to microRNA-155 can promote the polarization of macrophages to a proinflammatory type and enhance innate immunity.53 In photodynamic therapy (PDT), specific wavelengths of light, photosensitizers, and oxygen are used to generate reactive oxygen species during treatment, in which tumor cells are killed through reactive oxygen species-induced apoptosis and necrosis.54 The intensity of PDT action is positively correlated with intracellular photosensitizer and oxygen concentrations. However, typical photosensitizers are hydrophobic and difficult to transport into the cells.55 Therefore, similarly to small-molecule drugs, tFNAs enhance the hydrophilicity of photosensitizers, promote their entry into cells, and increase their concentrations, thereby enhancing PDT efficacy. Kim et al. combined methylene blue with tFNAs to enhance light-induced cytotoxicity.56 Wang et al. combined IR780, a photosensitizer with photothermal and photosensitizing effects, with tFNAs and showed that tFNAs not only enabled IR780 to achieve a stronger PDT killing effect but also significantly increased the accumulation of IR780 in tumor sites; moreover, under infrared light irradiation, IR780 loaded with tFNAs exhibited stronger tumor-imaging effects than IR780 alone.57
Interestingly, tFNAs have four vertices; that is, the mixing of multiple therapeutic strategies will be the future development direction of tFNAs in the field of tumor therapy. For example, when chemotherapy drugs are combined with immunotherapy, tFNAs can be linked to doxorubicin (DOX) and CpG to achieve higher efficacy.58 Ren et al. reported tFNAs with multiple functions of targeting aptamers, e.g., DOX and DNase, which further expanded the therapeutic scope of tFNAs for tumor treatment.59 Therefore, in the future, multiple functional modifications of tFNAs will make tFNA-based treatments more comprehensive and complex, which can further improve the efficiency of treatment.
In 1986, Matsumura et al.65 reported the enhanced permeability and retention (EPR) effects of these nanostructures. Since then, the development and research of macromolecular anticancer drugs have increased, and various nanomaterials have performed well in tumor targeting based on EPR.66,67 In addition to their targeting properties, DNA origami nanostructures have good stability and biocompatibility, excellent cellular membrane penetration, and multiple modification sites for a series of biomolecules. Hence, DNA origami nanostructures are promising platforms for creating multifunctional nanomaterials for cancer therapy.68
To serve as a drug delivery system, DNA origami nanostructures must exhibit excellent stability in circulation conditions. Many studies have shown that DNA nanostructures are more stable than single- or double-stranded DNA. The complex structure of DNA origami may induce improved stability by hindering the function of nucleases.69 Moreover, encapsulation modifications have been performed to improve the stability of DNA nanostructures. Perrault et al.70 utilized a lipid bilayer to encapsulate DNA origami nanostructures, and this updated form led to longer blood circulation times compared with the plain form. Other molecules, such as PEG, polypeptides, and proteins, have been used for the same purpose.71,72
DOX is an antitumor micromolecule that can inhibit the synthesis of RNA and DNA, and has been applied a cell cycle nonspecific drug with a broad antitumor spectrum. However, DOX may produce toxic effects, such as leukopenia, thrombocytopenia, cardiotoxicity, nausea, and loss of appetite. Due to its ability to intercalate into DNA double strands, DOX serves as a classic drug that can be incorporated into DNA origami nanostructures, which are characterized by higher tumor-targeting efficiency and fewer toxic effects. Zhao et al.73 encapsulated DOX in screwy DNA nanotubes and studied their release rates, and found that DNA nanotubes could improve the cellular uptake of DOX to kill cancer cells. Jiang et al.74 also reported that DNA nanocarriers exhibit an enrichment effect and reduce drug resistance to a certain extent (Fig. 2), and also used triangular DNA nanocarriers to deliver DOX.75 The study revealed that DOX-loaded DNA origami nanostructures have improved antitumor effects and lower systemic toxicity in mouse models compared with free DOX molecules.
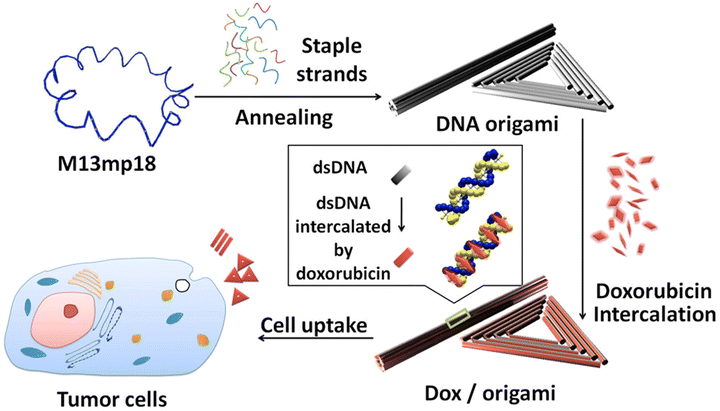 | ||
| Fig. 2 DNA origami and doxorubicin origami delivery system assembly. The long single-strand M13mp18 genomic DNA scaffold strand (blue) is folded into the triangle and tube structures through the hybridization of rationally designed staple strands. Watson–Crick base pairs in the double helices serve as docking sites for doxorubicin intercalation. After incubation with doxorubicin, the drug-loaded DNA nanostructure delivery vessels were administered to MCF 7 cells, and the effects were investigated. Adapted with permission from ref. 76. Copyright 2012, American Chemical Society. | ||
Currently, an increasing number of researchers are focusing on gene therapy, which applies exogenous nucleic acid strands, such as siRNA, microRNA, and CpG,77 to inhibit cancer cells. However, these therapeutic sequences are unstable in a complicated in vivo environment, and therefore there is an urgent need to find suitable delivery systems for these molecules. DNA origami nanostructures can be used to extend the length of usable DNA strands to ensure a connection with exogenous nucleic acid strands by protecting the nucleic acid strands and making it possible to target certain tissues. Schuller et al.78 utilized DNA origami nanostructures to deliver CpG sequences, resulting in a strong immune response and fewer side effects. Rahman et al.79 reported DNA bricks modified with siRNA that could knock down the antiapoptotic protein Bcl-2. These results indicated that DNA bricks could stabilize siRNA and effectively inhibit the growth of lung tumors.
Aptamers are short, simple-stranded DNA or RNA oligonucleotides that specifically bind to target molecules.80 Aptamers tend to fold and form stable three-dimensional structures through base pairing, electrostatic interactions, or hydrogen bonding, and can wrap around target molecules with high affinity. To obtain a high uptake efficiency of DNA origami nanostructures, aptamers can be added to certain strands of the DNA nanostructures. Song et al.81 incorporated DOX, AuNRs, and the tumor-specific aptamer MUC-1 into DNA origami nanostructures to kill drug-resistant cancer cells, which significantly enhanced the internalization of the nanostructure by cancer cells. Schaffert et al.82 synthesized a two-dimensional DNA nanostructure with the modified transport protein transferrin (TF). The results showed that transferrin improved the cellular uptake of the DNA nanostructures, and the internalization efficiency was influenced by the number of modified transferrins.
In recent years, multidrug resistance has become a major obstacle in cancer therapy. To solve this problem, researchers have combined multiple chemotherapeutic drugs within the same nano-delivery system, which is called combination therapy. Liu et al.83 designed a multifunctional DNA origami nanostructure to deliver DOX and shRNA to destroy multidrug-resistant breast cancer cells (MCF-7R), and found that the application of shRNA successfully reversed drug resistance in MCF-7R cells, which led to an enhanced therapeutic effect of DOX. Pan et al.84 established a versatile DNA origami platform to combine chemotherapy with RNA. In this study, two different antisense oligonucleotides (ASOs) were used to target P-glycoprotein (P-gp) and Bcl-2. Meanwhile, they connected the aptamer MUC-1 to the surface of the DNA platform to target cancer cells. These results demonstrated that this DNA platform increased the therapeutic effect of DOX with the assistance of ASOs and aptamers. These studies indicated that DNA origami nanostructures perform well in drug delivery and combination therapy.
In addition to drug delivery, DNA origami nanostructures are promising platforms for cancer detection.85 With the rise in cancer incidence and the development of tumor-targeting therapies, the early diagnosis of cancer has become increasingly crucial. Various DNA origami detectors have been constructed for cellular imaging by modification with fluorescent molecules. For example, Shen et al.86 incorporated biscyanine into DNA nanotubes to visualize live cells directly, and Samanta et al.87 combined DNA origami with infrared-emitting QDs for biophotonics applications.
| Year | Dynamic DNA nanostructures | Ref. |
|---|---|---|
| 1999 | Paired double crossover structure | 106 |
| 2001 | Toehold-mediated strand displacement (TMSD) based DNA tweezers | 107 |
| 2003 | pH-based DNA nanostructure | 108 |
| 2005 | DNA walker | 109 |
| 2007 | DNA box actuated by TMSD | 110 |
| 2011 | DNA beacon which detects biomolecules | 111 |
| 2012 | Logic-gated DNA box which displays molecular cargo | 112 |
| 2015 | DNA structure connected with shape complementarity | 63 |
| 2015 | DNA-based hinges, sliders, and hybrid mechanisms | 64 |
| 2016 | Rotary device made from multiple tightly-fitted components | 113 |
| 2018 | Gold nanocrystal-mediated slider | 114 |
| 2019 | Molecular algorithm that executes logic and outputs literal Arabic numerals | 115 |
| 2019 | DNA box actuated by changing pH | 116 |
| 2019 | Thermally-actuated nanovalve | 117 |
| 2019 | Self-regulating DNA nanotubes | 118 |
| 2021 | Robotic nanobee | 33 |
As single strands of DNA forming double-stranded structures through hydrogen bonds, DNA nanostructures are affected by changes in pH or ion concentration. Moreover, a wide range of ions have been reported to be suitable for insertion into DNA strands and can induce the formation of complicated structures.89,90 In addition, some DNA strands are easily affected by environmental pH. For instance, the i-motif sequence includes abundant cytosine and tends to maintain a linear strand in a neutral environment. However, as the pH decreases, the i-motif folds and forms a secondary structure.91 Meanwhile, the guanine-rich sequence, which is paired with the i-motif, can construct a G-quadruplex in the same plane.92 Therefore, the i-motif and G-quadruplex are often used as switch structures. Keum et al.93 developed a pH-sensitive DNA nanocarrier to control the release of proteins based on the i-motif sequence. Park et al.94 utilized the i-motif and G-quadruplex to deliver DOX through a triggered release. Moreover, they combined photodynamic and photothermal therapy with chemotherapy, which led to enhanced antitumor effects and fewer toxic side effects.
Aptamers bind tightly to target molecules by folding into secondary structures, such as G-quadruplexes and DNA loops, and the target molecules are mainly proteins, including cytokines, kinases, cell adhesion factors, and cell surface receptors. Through their combination with certain proteins, aptamers can perform various biological functions. Researchers believe that proteins cannot approach other biomolecules after binding with aptamers, which inhibits the usual effects of the proteins. AS1411 is a classic G-quadruplex aptamer targeting the surface receptors of cancer cells (nucleolin), and reports have indicated that AS1411 can inhibit the growth of cancer cells, especially blood cancer cells.95 Considering the multiple functions of aptamers, they have been widely applied in diverse DNA nano-delivery systems.96 In addition to promoting the target efficacy and therapeutic effect of nanomedicine, aptamers can serve as switches in dynamic DNA nanostructures. Tian et al.33 constructed a nucleolin-triggered dynamic DNA tetrahedron based on the AS1411 switch (Fig. 3). This tetrahedron remains intact in a nucleolin-free environment, and when it approaches cancer cells, the AS1411 strand separates and binds to nucleolin, resulting in structural disintegration and drug release. This design significantly reduces the damage to normal tissues caused by chemotherapy drugs.
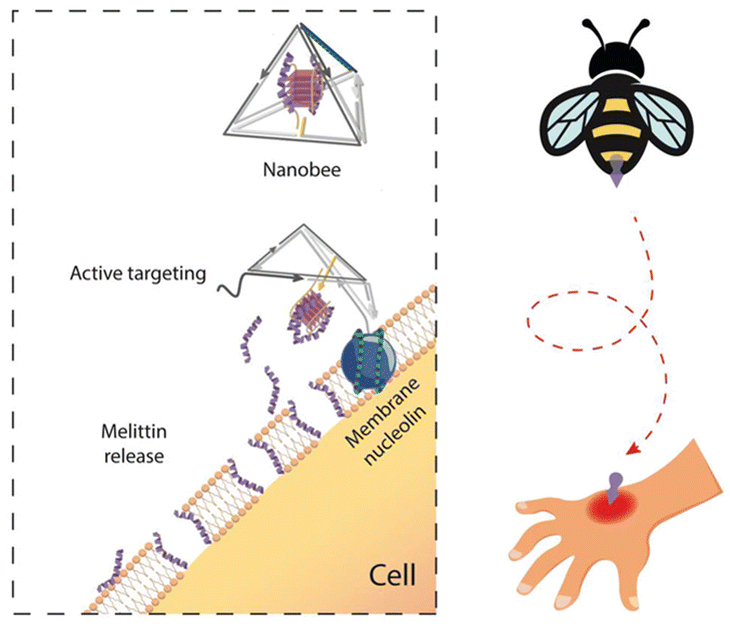 | ||
| Fig. 3 A nucleolin-triggered dynamic DNA tetrahedron based on AS1411 to reduce the damage to normal tissues caused by chemotherapy drugs. Adapted with permission from ref. 97. Copyright 2021, Wiley-VCH. | ||
The basic unit of the DNA double strand is a base pair bound by hydrogen bonds. However, by regulating the affinity of different single DNA strands, competitive binding of these DNA strands is expected, which will lead to dynamic sequence replacement. The replaced single strand may subsequently induce the replacement of other DNA strands.98 This theory is based on various DNA reactions, such as hybridization chain reaction (HCR), catalytic DNA hairpin (CDH), and catalytic DNA circuits (CDC). A hairpin-based HCR is a classic example of a DNA replacement reaction. Traditionally, hairpin-based HCRs include two DNA hairpins and an initiator. The initiator induces Hairpin-1 to release a low affiliative DNA strand, which demonstrates a high affinity to Hairpin-2 and initiates strand displacement of Hairpin-2. Subsequently, the redundant strand from Hairpin-2 further triggers the displacement reaction of Hairpin-1.99,100 Through this cascading reaction, an initial sequence can be amplified for detection, which builds a theoretical foundation for the use of DNA biosensors to detect proteins, small molecules, and nucleic acids.101 In addition, dynamic DNA nanostructures demonstrate several merits as biosensors, such as their outstanding biocompatibility and cellular internalization, high structural editability, and diverse detection targets. Based on HCR, Zhang et al.102 constructed a DNA tetrahedron modified with DNA hairpins and showed that the DNA tetrahedron could rapidly sense tumor cells by amplifying the number of receptors on the cell membrane.
With the development of DNA-editing technology, DNA hydrogels have become a research hotspot in recent years. DNA hydrogels can be formed by covalent interactions between linear or polymeric DNA molecules, which is termed chemical crosslinking. Due to chemical crosslinking, DNA hydrogels tend to be irreversible and highly stable in physiological environments.103 Moreover, physical crosslinking could also promote DNA gelation through hydrogen bonding, electrostatic interactions, or DNA-metal coordination. Compared to chemical crosslinking, physically crosslinked DNA hydrogels are reversible and demonstrate better degradability, biocompatibility, and biosecurity.104 In addition, by inducing trigger-responsive units, DNA hydrogels have become smart and show prospects in drug delivery, tissue engineering, and gene editing. Ding et al.105 reported an antitumor DNA hydrogel with embedded siRNA, which displayed self-assembly by grafting DNA strands onto polycaprolactone. These results indicated that this hydrogel structure protected siRNA from RNase degradation and did not disturb the function of RNA interference.
3.2 RNA nanotechnology
Similar to DNA nanotechnology, RNA nanomaterials are attracting the attention of researchers. RNA origami involves the folding of a designed single strand into a complex structure with the participation of RNA polymerase. Since the initial framework of RNA origami presented by Geary et al.,40 there have been an increasing number of RNA origami structures being developed. Wu et al.119 designed an RNA/DNA hybrid origami nanoplatform, where target mRNA served as the scaffold and short DNA staples acted as antisense to form a tubular nanostructure. The tailored structure could be cleaved by intracellular RNase H, releasing the short DNA staples which then bind to the target mRNA in the cytoplasm and guide its degradation by RNase H, leading to gene silencing. In Ding et al.’s experiments, the tumor-associated PLK1 gene in MCF-7 was effectively silenced. Nucleic acids (NAs) are considered as natural ligands for some pattern recognition receptors (PRRs) in mammalian cells, which can be internalized into endosomes and recognized by Toll-like receptors (TLR) to enhance the mount innate responses. Therefore, Qi et al.120 developed an RNA origami structure that could stimulate NK and CD8+ T cell anti-tumor activity and alleviate tumor-mediated immune suppression environment. This structure can self-assemble in PBS solution, without the need for divalent cations to maintain its stability. Their experiments further found that this structure had excellent serum and thermal stability, and demonstrated low toxicity and inflammatory cytokines response.With the development of programmable RNA technology, there are some kinds of 2D and 3D frameworks of RNA, such as triangles, squares, pentagons, nanoprisms, tetrahedrons, and micelles, etc.85,121–125 Stewart et al.126 designed a de novo double crossover (DX) RNA tile that can self-assemble and fold into lattices for siRNA delivery and demonstrated its remarkable stability in micron-scale. In breast cancer cells (MDA-MB-1/GFP), the material can achieve an excellent silencing efficiency for GFP, and showed silencing the similar efficiency for PLK1 in human prostate cell lines (PC1). In addition, more and more attention had been attached to the biological effects of RNA that make up the framework. Hu et al.127 developed a flexible lantern RNA structure that can package Smad4 mRNA and demonstrated its mRNA delivery ability in mouse situ colorectal tumor model. The lantern structure can be recognized by ribosomes in cancer cells and translated into protein, thereby releasing the Smad4 mRNA. Due to its RNA composition, this nanostructure does not affect the functional efficacy of the mRNA compared to DNA/RNA hybrid structures. Lee et al.128 used ligased circular DNA templates, T7 promoter, and T7 RNA polymerase to amplify siRNA and self-assemble it into nucleic acid sponges composed of nanoscale pleated sheets of hairpin RNA to deliver siRNA, and validated the good performance in delivering siRNA in mouse ovarian cancer. Their designed RCT (rolling circle transcription) platform could produce a large amount of siRNA, and without deliver carrier, this structure showed lower biological toxicity Fig. 4.
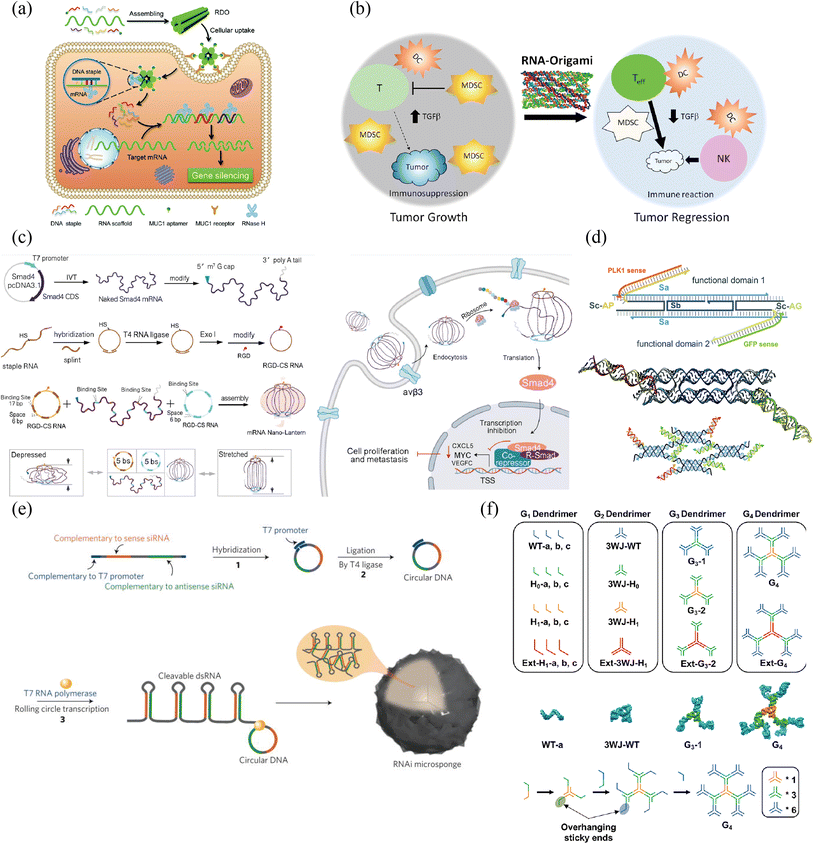 | ||
| Fig. 4 RNA nanostructure functionalization for tumor therapy. (a) RNA/DNA hybrid origami nanostructure demonstrated efficient cellular uptake. Adapted with permission from ref. 119, Copyright 2021, Royal Society of Chemistry. (b) Single-stranded RNA origami technology would not increase the level of interferons in anticancer immunotherapy. Adapted with permission from (ref. 120), Copyright 2020, American Chemical Society. (c) Lantern-shaped flexible RNA origami improved the efficiency of RNA delivery and translation. Adapted with permission from (ref. 127) Copyright 2023, Nature Publishing Group. (d) RNA lattices based on DNA tile are more resilient to nuclease degradation, and can be further functionalized with different cargos. Adapted with permission from (ref. 126), Copyright 2023, Royal Society of Chemistry. (e) The synthesis of a single RNAi-microsponge that combines carrier and cargo, provided protection for siRNA form RNase H with a high content of siRNA. Adapted with permission from (ref. 128), Copyright 2012, Nature Publishing Group. (f) Stable RNA dendrimers demonstrated its potential for hydrophobic drug delivery, shielding and controlled release. Adapted with permission from (ref. 133), Copyright 2020, Royal Society of Chemistry. | ||
Originated from bacteriophage phi29 DNA packaging motor, pRNA has the unique ability to form dimers, trimers, hexamers, and other structures through two reengineered interlocking loops, which made it a promising multivalent carrier for delivering siRNA.129 RNA is considered as a potential material in dendritic polymers due to the ultra-stable phi29 three-way junction (3WJ) motif and its derivatives serving as the central core and repetitive unit.130–132 Guo et al.133 established dendritic polymers of different generations and sizes using 2′-fluoro (2′-F) modified phosphoramidites, which can covalently bind to drugs (paclitaxel) and release them in a stepwise manner at different temperatures. Li et al.129 used T7 RNA polymerase to construct a pRNA/siRNA dimer and combined folate with pRNA to give the multimer a certain targeting ability. In their experiments, this structure had similar knockdown efficiency to other siRNA delivery vehicles in folate receptor-expressing cells such as MCF-7, but with lower cellular toxicity Table 2.
| Nucleic acid nanostructure | Cargo or junctions | Target cell | Ref. |
|---|---|---|---|
| tFNAs | Doxorubicin | SCC7 cells | 134 |
| Taxol | Not specified | 44 | |
| Luteolin, kaempferol and apigenin | Peripheral lymphocytes | 135 | |
| Paclitaxel | A549 and A549/T cells | 47 | |
| Anti-HER2 aptamer | HER2-positive breast cancer cells | 48 | |
| 5-Fluorouracil, DNA aptamer and | MCF-7 cell line | 49 | |
| miRNA-214-3p | A549 cells | 50 | |
| Anti-braf siRNA and DNA aptamer | A375 cells | 13 | |
| siRNA, folate molecules | HeLa cells | 136 | |
| Antigen and CpG adjuvants | DC and B cell | 52 | |
| miRNA-155 | Macrophages | 53 | |
| Methylene blue | B16F10, SCC7 and MDA-MB231 cells | 137 | |
| IR-780 | MCF-7 cells | 59 | |
| Doxorubicin, aptamer and 17E DNA zyme | SMMC-7721 and HEK293T cell lines | 58 | |
| DNA origami | Doxorubicin | MDA-MB-231 cells, MDA-MB-468 cells, and MCF-7 cells | 138 |
| Doxorubicin | MCF-7 cells | 76 | |
| Doxorubicin | MDA-MB-231-GFP cells | 75 | |
| CpG | Primary splenic cells | 78 | |
| siRNA | DMS53 cell line; H1299 cell line | 79 | |
| Doxorubicin; surface-modified gold nanorods and MUC-1 aptamers | MCF-7/ADR cells | 81 | |
| Iron transport protein transferrin | KB carcinoma cell line | 139 | |
| shRNA, doxorubicin and MUC-1 aptamers | MCF-7R cells | 83 | |
| Antisense oligonucleotides and doxorubicin | Hela/adriamycin (ADR) cells | 84 | |
| Carbazole-based biscyanine | MCF-7 cells | 140 | |
| Dynamic DNA nanostructure | Au-Gi, i-motif sequence, ZnPc and doxorubicin | MDA-MB-231 cells | 94 |
| AS1411 aptamer and melittin | L929 cells and A375 cells | 33 | |
| DNA hairpin | AS49, MCF-7 and A375 cells | 141 | |
| DNA-grafted polycaprolactone and siRNA | U2OS cells | 142 | |
| RNA origami | Antisense | MCF-7 cells | 119 |
| Not specified | RAW 264.7 macrophage cell line | 120 | |
| Smad4 mRNA | SW480 and SW620 cells | 127 | |
| Self-assembled RNA | siRNA | MDA-MB-231 cells | 143 |
| siRNA | T22 cells | 128 | |
| Self-assembled RNA based on phi29 | Paclitaxel, folate | RAW 264.7 cells, human KB cells | 133 |
| siRNA, pRNA | MCF-7 cells and HeLa cells | 129 |
4. Conclusion
Nucleic acid nanomaterials have become an important field of research for tumor therapy, and various DNA and RNA structures have been considered as potential drug carriers because of their high biocompatibility, excellent editability, and easy internalization. However, the comprehensive application of nucleic acid nanomaterials for tumor treatment as a drug delivery system still faces monumental challenges, including (1) instability of synthesis, high cost, and high error rate of self-assembly, (2) various unknown biomedical effects, and (3) biosafety concerns arising from the uncertain in vivo stability, pharmacokinetic properties, and long-term cytotoxicity. Solving these challenges will greatly enhance the further application of nucleic acid nanomaterials in the field of tumor therapy.Author contributions
W. T. Lu and T. Y. Chen conceived and wrote the manuscript. D. X. Xiao, X. Qin, Y. Chen and S. R. Shi reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.Conflicts of interest
The authors report on declarations of interest.Acknowledgements
This study was supported by the National Natural Science Foundation of China (82101077), Sichuan Science and Technology Program (2023NSFSC1516), Postdoctoral Science Foundation of China (2021M692271), West China School/Hospital of Stomatology Sichuan University, no. RCDWJS2023-5, Fundamental Research Funds for the Central Universities, and Research and Develop Program, West China Hospital of Stomatology Sichuan University.References
- J. Ferlay, M. Colombet and I. Soerjomataram, et al., Cancer statistics for the year 2020: An overview, Int. J. Cancer, 2021, 149(4), 778–789 CrossRef CAS PubMed.
- R. L. Siegel, K. D. Miller and H. E. Fuchs, et al., Cancer Statistics, 2021, Ca-Cancer J. Clin., 2021, 71(1), 7–33 CrossRef PubMed.
- R. L. Siegel, K. D. Miller and H. E. Fuchs, et al., Cancer statistics, 2022, Ca-Cancer J. Clin., 2022, 72(1), 7–33 CrossRef PubMed.
- Q. Hu, H. Li and L. Wang, et al., DNA Nanotechnology-Enabled Drug Delivery Systems, Chem. Rev., 2019, 119(10), 6459–6506 CrossRef CAS PubMed.
- X. Xu, W. Ho and X. Zhang, et al., Cancer nanomedicine: from targeted delivery to combination therapy, Trends Mol. Med., 2015, 21(4), 223–232 CrossRef CAS PubMed.
- K. Xiao, J. Liu and H. Chen, et al., A label-free and high-efficient GO-based aptasensor for cancer cells based on cyclic enzymatic signal amplification, Biosens. Bioelectron., 2017, 91, 76–81 CrossRef CAS PubMed.
- M. Avci-Adali, N. Wilhelm and N. Perle, et al., Absolute Quantification of Cell-Bound DNA Aptamers During SELEX, Nucleic Acid Ther., 2013, 23(2), 125–130 CrossRef CAS PubMed.
- M. Zhao, R. Wang and K. Yang, et al., Nucleic acid nanoassembly-enhanced RNA therapeutics and diagnosis, Acta Pharm. Sin. B, 2023, 13(3), 916–941 CrossRef CAS PubMed.
- A. R. Chandrasekaran, J. A. Punnoose and L. Zhou, et al., DNA nanotechnology approaches for microRNA detection and diagnosis, Nucleic Acids Res., 2019, 47(20), 10489–10505 CrossRef CAS PubMed.
- L. Zhu, J. Ye and S. Wang, et al., Dual amplification ratiometric biosensor based on a DNA tetrahedron nanostructure and hybridization chain reaction for the ultrasensitive detection of microRNA-133a, Chem. Commun., 2019, 55(77), 11551–11554 RSC.
- Q. Jiang, Y. Shi and Q. Zhang, et al., A Self-Assembled DNA Origami-Gold Nanorod Complex for Cancer Theranostics, Small, 2015, 11(38), 5134–5141 CrossRef CAS PubMed.
- Y. Du, Q. Jiang and N. Beziere, et al., DNA-Nanostructure–Gold-Nanorod Hybrids for Enhanced In Vivo Optoacoustic Imaging and Photothermal Therapy, Adv. Mater., 2016, 28(45), 10000–10007 CrossRef CAS PubMed.
- D. Xiao, Y. Li and T. Tian, et al., Tetrahedral Framework Nucleic Acids Loaded with Aptamer AS1411 for siRNA Delivery and Gene Silencing in Malignant Melanoma, ACS Appl. Mater. Interfaces, 2021, 13(5), 6109–6118 CrossRef CAS PubMed.
- N. C. Seeman, Nucleic acid junctions and lattices, J. Theor. Biol., 1982, 99(2), 237–247 CrossRef CAS PubMed.
- P. W. K. Rothemund, Folding DNA to create nanoscale shapes and patterns, Nature, 2006, 440(7082), 297–302 CrossRef CAS PubMed.
- S. M. Douglas, A. H. Marblestone and S. Teerapittayanon, et al., Rapid prototyping of 3D DNA-origami shapes with caDNAno, Nucleic Acids Res., 2009, 37(15), 5001–5006 CrossRef CAS PubMed.
- R. V. Reshetnikov, A. V. Stolyarova and A. O. Zalevsky, et al., A coarse-grained model for DNA origami, Nucleic Acids Res., 2018, 46(3), 1102–1112 CrossRef CAS PubMed.
- V. Linko and M. A. Kostiainen, Automated design of DNA origami, Nat. Biotechnol., 2016, 34(8), 826–827 CrossRef CAS PubMed.
- H. Dietz, M. Douglas Shawn and M. Shih William, Folding DNA into Twisted and Curved Nanoscale Shapes, Science, 2009, 325(5941), 725–730 CrossRef CAS PubMed.
- T. Zhang, C. Hartl and K. Frank, et al., 3D DNA Origami Crystals, Adv. Mater., 2018, 30(28), 1800273 CrossRef PubMed.
- Z. Ge, H. Gu and Q. Li, et al., Concept and Development of Framework Nucleic Acids, J. Am. Chem. Soc., 2018, 140(51), 17808–17819 CrossRef CAS PubMed.
- G.-H. Chen, X.-L. Zhang and H. Chen, et al., miR-22 represses osteoblast viability with ESR1 presenting a direct target and indirectly inactivating p38 MAPK/JNK signaling, J. Gene Med., 2020, 22(6), e3174 CrossRef CAS PubMed.
- T. Zhang, W. Cui and T. Tian, et al., Progress in Biomedical Applications of Tetrahedral Framework Nucleic Acid-Based Functional Systems, ACS Appl. Mater. Interfaces, 2020, 12(42), 47115–47126 CrossRef CAS PubMed.
- S. Li, T. Tian and T. Zhang, et al., Advances in biological applications of self-assembled DNA tetrahedral nanostructures, Mater. Today, 2019, 24, 57–68 CrossRef CAS.
- Q. Zhang, S. Lin and L. Wang, et al., Tetrahedral framework nucleic acids act as antioxidants in acute kidney injury treatment, Chem. Eng. J., 2021, 413, 127426 CrossRef CAS.
- J. Zhu, M. Zhang and Y. Gao, et al., Tetrahedral framework nucleic acids promote scarless healing of cutaneous wounds via the AKT-signaling pathway, Signal Transduction Targeted Ther., 2020, 5(1), 120 CrossRef CAS PubMed.
- K. R. Kim, D. R. Kim and T. Lee, et al., Drug delivery by a self-assembled DNA tetrahedron for overcoming drug resistance in breast cancer cells, Chem. Commun., 2013, 49(20), 2010–2012 RSC.
- J. Zhang, Y. Guo and F. Ding, et al., A Camptothecin-Grafted DNA Tetrahedron as a Precise Nanomedicine to Inhibit Tumor Growth, Angew. Chem., Int. Ed., 2019, 58(39), 13794–13798 CrossRef CAS PubMed.
- A. F. Jorge, A. Aviñó and A. A. C. C. Pais, et al., DNA-based nanoscaffolds as vehicles for 5-fluoro-2′-deoxyuridine oligomers in colorectal cancer therapy, Nanoscale, 2018, 10(15), 7238–7249 RSC.
- Y. Zhang, Y. Deng and C. Wang, et al., Probing and regulating the activity of cellular enzymes by using DNA tetrahedron nanostructures, Chem. Sci., 2019, 10(23), 5959–5966 RSC.
- Q. Lin, S. Cai and B. Zhou, et al., Dual-MicroRNA-regulation of singlet oxygen generation by a DNA-tetrahedron-based molecular logic device, Chem. Commun., 2021, 57(32), 3873–3876 RSC.
- S. Juul, F. Iacovelli and M. Falconi, et al., Temperature-Controlled Encapsulation and Release of an Active Enzyme in the Cavity of a Self-Assembled DNA Nanocage, ACS Nano, 2013, 7(11), 9724–9734 CrossRef CAS PubMed.
- T. R. Tian, D. X. Xiao and T. Zhang, et al., A Framework Nucleic Acid Based Robotic Nanobee for Active Targeting Therapy, Adv. Funct. Mater., 2021, 31(5), 2007342 CrossRef CAS.
- P. Guo, C. Zhang and C. Chen, et al., Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation, Mol. Cell, 1998, 2(1), 149–155 CrossRef CAS PubMed.
- L. Jaeger, E. Westhof and N. B. Leontis, TectoRNA: modular assembly units for the construction of RNA nano-objects, Nucleic Acids Res., 2001, 29(2), 455–463 CrossRef CAS PubMed.
- E. Bindewald, C. Grunewald and B. Boyle, et al., Computational strategies for the automated design of RNA nanoscale structures from building blocks using NanoTiler, J. Mol. Graphics Modell., 2008, 27(3), 299–308 CrossRef CAS PubMed.
- F. Jossinet, T. E. Ludwig and E. Westhof, Assemble: an interactive graphical tool to analyze and build RNA architectures at the 2D and 3D levels, Bioinformatics, 2010, 26(16), 2057–2059 CrossRef CAS PubMed.
- H. M. Martinez, J. V. Maizel, Jr. and B. A. Shapiro, RNA2D3D: a program for generating, viewing, and comparing 3-dimensional models of RNA, J. Biomol. Struct. Dyn., 2008, 25(6), 669–683 CrossRef CAS PubMed.
- A. Busch and R. Backofen, INFO-RNA-a server for fast inverse RNA folding satisfying sequence constraints, Nucleic Acids Res., 2007, 35, W310–W313 CrossRef PubMed.
- C. Geary, P. W. Rothemund and E. S. Andersen, A single-stranded architecture for cotranscriptional folding of RNA nanostructures, Science, 2014, 345(6198), 799–804 CrossRef CAS PubMed.
- W. Fu, L. Ma and Y. Ju, et al., Therapeutic siCCR2 Loaded by Tetrahedral Framework DNA Nanorobotics in Therapy for Intracranial Hemorrhage, Adv. Funct. Mater., 2021, 31(33), 2101435 CrossRef CAS.
- W. Cui, X. Yang and X. Chen, et al., Treating LRRK2-Related Parkinson's Disease by Inhibiting the mTOR Signaling Pathway to Restore Autophagy, Adv. Funct. Mater., 2021, 31(38), 2105152 CrossRef CAS.
- K.-R. Kim, H. Y. Kim and Y.-D. Lee, et al., Self-assembled mirror DNA nanostructures for tumor-specific delivery of anticancer drugs, J. Controlled Release, 2016, 243, 121–131 CrossRef CAS PubMed.
- A. G. Krishna, D. V. Kumar and B. M. Khan, et al., Taxol–DNA interactions: fluorescence and CD studies of DNA groove binding properties of taxol, Biochim. Biophys. Acta – Gen. Subj., 1998, 1381(1), 104–112 CrossRef CAS PubMed.
- G. Rusak, I. Piantanida and L. Mašić, et al., Spectrophotometric analysis of flavonoid-DNA interactions and DNA damaging/protecting and cytotoxic potential of flavonoids in human peripheral blood lymphocytes, Chem.-Biol. Interact., 2010, 188(1), 181–189 CrossRef CAS PubMed.
- T. Zhang, T. Tian and Y. Lin, Functionalizing Framework Nucleic-Acid-Based Nanostructures for Biomedical Application, Adv. Mater., 2021, 2107820 Search PubMed.
- X. Xie, X. Shao and W. Ma, et al., Overcoming drug-resistant lung cancer by paclitaxel loaded tetrahedral DNA nanostructures, Nanoscale, 2018, 10(12), 5457–5465 RSC.
- W. Ma, Y. Zhan and Y. Zhang, et al., An Intelligent DNA Nanorobot with in Vitro Enhanced Protein Lysosomal Degradation of HER2, Nano Lett., 2019, 19(7), 4505–4517 CrossRef CAS PubMed.
- Y. Zhan, W. Ma and Y. Zhang, et al., DNA-Based Nanomedicine with Targeting and Enhancement of Therapeutic Efficacy of Breast Cancer Cells, ACS Appl. Mater. Interfaces, 2019, 11(17), 15354–15365 CrossRef CAS PubMed.
- S. Li, Y. Sun and T. Tian, et al., MicroRNA-214-3p modified tetrahedral framework nucleic acids target survivin to induce tumour cell apoptosis, Cell Proliferation, 2020, 53(1), e12708 CrossRef PubMed.
- H. Lee, A. K. R. Lytton-Jean and Y. Chen, et al., Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery, Nat. Nanotechnol., 2012, 7(6), 389–393 CrossRef CAS PubMed.
- X. Liu, Y. Xu and T. Yu, et al., A DNA Nanostructure Platform for Directed Assembly of Synthetic Vaccines, Nano Lett., 2012, 12(8), 4254–4259 CrossRef CAS PubMed.
- X. Qin, L. Xiao and N. Li, et al., Tetrahedral framework nucleic acids-based delivery of microRNA-155 inhibits choroidal neovascularization by regulating the polarization of macrophages, Bioact. Mater., 2022, 14, 134–144 CrossRef CAS PubMed.
- C. Fabris, G. Valduga and G. Miotto, et al., Photosensitization with zinc (II) phthalocyanine as a switch in the decision between apoptosis and necrosis, Cancer Res., 2001, 61(20), 7495–7500 CAS.
- R. V. Huis In 't Veld, J. Heuts and S. Ma, et al., Current Challenges and Opportunities of Photodynamic Therapy against Cancer, Pharmaceutics, 2023, 15(2) Search PubMed.
- K.-R. Kim, D. Bang and D.-R. Ahn, Nano-formulation of a photosensitizer using a DNA tetrahedron and its potential for in vivo photodynamic therapy, Biomater. Sci., 2016, 4(4), 605–609 RSC.
- S. Wang, Z. Liu and Y. Tong, et al., Improved cancer phototheranostic efficacy of hydrophobic IR780 via parenteral route by association with tetrahedral nanostructured DNA, J. Controlled Release, 2021, 330, 483–492 CrossRef CAS PubMed.
- J. Yan, X. Zhan and Z. Zhang, et al., Tetrahedral DNA nanostructures for effective treatment of cancer: advances and prospects, J. Nanobiotechnol., 2021, 19(1), 412 CrossRef CAS PubMed.
- T. Ren, Z. Deng and H. Liu, et al., Co-delivery of DNAzyme and a chemotherapy drug using a DNA tetrahedron for enhanced anticancer therapy through synergistic effects, New J. Chem., 2019, 43(35), 14020–14027 RSC.
- S. M. Douglas, J. J. Chou and W. M. Shih, DNA-nanotube-induced alignment of membrane proteins for NMR structure determination, Proc. Natl. Acad. Sci. U. S. A., 2007, 104(16), 6644–6648 CrossRef CAS PubMed.
- R. Iinuma, Y. Ke and R. Jungmann, et al., Polyhedra self-assembled from DNA tripods and characterized with 3D DNA-PAINT, Science, 2014, 344(6179), 65–69 CrossRef CAS PubMed.
- E. Benson, A. Mohammed and J. Gardell, et al., DNA rendering of polyhedral meshes at the nanoscale, Nature, 2015, 523(7561), 441–444 CrossRef CAS PubMed.
- T. Gerling, F. Wagenbauer Klaus and M. Neuner Andrea, et al., Dynamic DNA devices and assemblies formed by shape-complementary, non–base pairing 3D components, Science, 2015, 347(6229), 1446–1452 CrossRef CAS PubMed.
- E. Marras Alexander, L. Zhou and H.-J. Su, et al., Programmable motion of DNA origami mechanisms, Proc. Natl. Acad. Sci., 2015, 112(3), 713–718 CrossRef CAS PubMed.
- Y. Matsumura and H. Maeda, A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs, Cancer Res., 1986, 46(12 Pt 1), 6387–6392 CAS.
- I. Brigger, C. Dubernet and P. Couvreur, Nanoparticles in cancer therapy and diagnosis, Adv. Drug Delivery Rev., 2002, 54(5), 631–651 CrossRef CAS PubMed.
- S. Acharya and S. K. Sahoo, PLGA nanoparticles containing various anticancer agents and tumour delivery by EPR effect, Adv. Drug Delivery Rev., 2011, 63(3), 170–183 CrossRef CAS PubMed.
- S. Mishra, Y. Feng and M. Endo, et al., Advances in DNA Origami-Cell Interfaces, ChemBioChem, 2020, 21(1–2), 33–44 CrossRef CAS PubMed.
- Q. Mei, X. Wei and F. Su, et al., Stability of DNA origami nanoarrays in cell lysate, Nano Lett., 2011, 11(4), 1477–1482 CrossRef CAS PubMed.
- S. D. Perrault and W. M. Shih, Virus-inspired membrane encapsulation of DNA nanostructures to achieve in vivo stability, ACS Nano, 2014, 8(5), 5132–5140 CrossRef CAS PubMed.
- A. Chopra, S. Krishnan and F. C. Simmel, Electrotransfection of Polyamine Folded DNA Origami Structures, Nano Lett., 2016, 16(10), 6683–6690 CrossRef CAS PubMed.
- N. Ponnuswamy, M. M. C. Bastings and B. Nathwani, et al., Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation, Nat. Commun., 2017, 8, 15654 CrossRef CAS PubMed.
- Y.-X. Zhao, A. Shaw and X. Zeng, et al., DNA origami delivery system for cancer therapy with tunable release properties, ACS Nano, 2012, 6(10), 8684–8691 CrossRef CAS PubMed.
- Q. Jiang, C. Song and J. Nangreave, et al., DNA origami as a carrier for circumvention of drug resistance, J. Am. Chem. Soc., 2012, 134(32), 13396–13403 CrossRef CAS PubMed.
- Q. Zhang, Q. Jiang and N. Li, et al., DNA origami as an in vivo drug delivery vehicle for cancer therapy, ACS Nano, 2014, 8(7), 6633–6643 CrossRef CAS PubMed.
- Q. Jiang, C. Song and J. Nangreave, et al., DNA origami as a carrier for circumvention of drug resistance, J. Am. Chem. Soc., 2012, 134(32), 13396–13403 CrossRef CAS PubMed.
- J. Vollmer and A. M. Krieg, Immunotherapeutic applications of CpG oligodeoxynucleotide TLR9 agonists, Adv. Drug Delivery Rev., 2009, 61(3), 195–204 CrossRef CAS PubMed.
- V. J. Schüller, S. Heidegger and N. Sandholzer, et al., Cellular immunostimulation by CpG-sequence-coated DNA origami structures, ACS Nano, 2011, 5(12), 9696–9702 CrossRef PubMed.
- M. A. Rahman, P. Wang and Z. Zhao, et al., Systemic Delivery of Bc12-Targeting siRNA by DNA Nanoparticles Suppresses Cancer Cell Growth, Angew. Chem., Int. Ed. Engl., 2017, 56(50), 16023–16027 CrossRef CAS PubMed.
- S. D. Jayasena, Aptamers: an emerging class of molecules that rival antibodies in diagnostics, Clin. Chem., 1999, 45(9), 1628–1650 CrossRef CAS.
- L. Song, Q. Jiang and J. Liu, et al., DNA origami/gold nanorod hybrid nanostructures for the circumvention of drug resistance, Nanoscale, 2017, 9(23), 7750–7754 RSC.
- D. H. Schaffert, A. H. Okholm and R. S. Sørensen, et al., Intracellular Delivery of a Planar DNA Origami Structure by the Transferrin-Receptor Internalization Pathway, Small, 2016, 12(19), 2634–2640 CrossRef CAS PubMed.
- J. Liu, L. Song and S. Liu, et al., A Tailored DNA Nanoplatform for Synergistic RNAi-/Chemotherapy of Multidrug-Resistant Tumors, Angew. Chem., Int. Ed. Engl., 2018, 57(47), 15486–15490 CrossRef CAS PubMed.
- Q. Pan, C. Nie and Y. Hu, et al., Aptamer-Functionalized DNA Origami for Targeted Codelivery of Antisense Oligonucleotides and Doxorubicin to Enhance Therapy in Drug-Resistant Cancer Cells, ACS Appl. Mater. Interfaces, 2020, 12(1), 400–409 CrossRef CAS PubMed.
- E. F. Khisamutdinov, H. Li and D. L. Jasinski, et al., Enhancing immunomodulation on innate immunity by shape transition among RNA triangle, square and pentagon nanovehicles, Nucleic Acids Res., 2014, 42(15), 9996–10004 CrossRef CAS PubMed.
- X. Shen, Q. Jiang and J. Wang, et al., Visualization of the intracellular location and stability of DNA origami with a label-free fluorescent probe, Chem. Commun., 2012, 48(92), 11301–11303 RSC.
- A. Samanta, Z. Deng and Y. Liu, Infrared emitting quantum dots: DNA conjugation and DNA origami directed self-assembly, Nanoscale, 2014, 6(9), 4486–4490 RSC.
- J. S. Kahn, Y. Hu and I. Willner, Stimuli-Responsive DNA-Based Hydrogels: From Basic Principles to Applications, Acc. Chem. Res., 2017, 50(4), 680–690 CrossRef CAS PubMed.
- Y. Kohwi and T. Kohwi-Shigematsu, Magnesium ion-dependent triple-helix structure formed by homopurine-homopyrimidine sequences in supercoiled plasmid DNA, Proc. Natl. Acad. Sci. U. S. A., 1988, 85(11), 3781–3785 CrossRef CAS PubMed.
- Y.-Y. Wu, Z.-L. Zhang and J.-S. Zhang, et al., Multivalent ion-mediated nucleic acid helix-helix interactions: RNA versus DNA, Nucleic Acids Res., 2015, 43(12), 6156–6165 CrossRef CAS PubMed.
- H. Abou Assi, M. Garavís and C. González, et al., i-Motif DNA: structural features and significance to cell biology, Nucleic Acids Res., 2018, 46(16), 8038–8056 CrossRef PubMed.
- B. Chu, D. Zhang and P. J. Paukstelis, A DNA G-quadruplex/i-motif hybrid, Nucleic Acids Res., 2019, 47(22), 11921–11930 CAS.
- J.-W. Keum and H. Bermudez, DNA-based delivery vehicles: pH-controlled disassembly and cargo release, Chem. Commun., 2012, 48(99), 12118–12120 RSC.
- H. Park, J. Kim and S. Jung, et al., DNA-Au Nanomachine Equipped with i-Motif and G-Quadruplex for Triple Combinatorial Anti-Tumor Therapy, Adv. Funct. Mater., 2018, 28(5) Search PubMed.
- A. D. Keefe, S. Pai and A. Ellington, Aptamers as therapeutics, Nat. Rev. Drug Discovery, 2010, 9(7), 537–550 CrossRef CAS PubMed.
- D. H. J. Bunka, O. Platonova and P. G. Stockley, Development of aptamer therapeutics, Curr. Opin. Pharmacol., 2010, 10(5), 557–562 CrossRef CAS PubMed.
- T. Tian, D. Xiao and T. Zhang, et al., A Framework Nucleic Acid Based Robotic Nanobee for Active Targeting Therapy, Adv. Funct. Mater., 2021, 31(5), 2007342 CrossRef CAS.
- L. Qian and E. Winfree, Scaling up digital circuit computation with DNA strand displacement cascades, Science, 2011, 332(6034), 1196–1201 CrossRef CAS PubMed.
- L. Liu, Q. Rong and G. Ke, et al., Efficient and Reliable MicroRNA Imaging in Living Cells via a FRET-Based Localized Hairpin-DNA Cascade Amplifier, Anal. Chem., 2019, 91(5), 3675–3680 CrossRef CAS PubMed.
- R. M. Dirks and N. A. Pierce, Triggered amplification by hybridization chain reaction, Proc. Natl. Acad. Sci. U. S. A., 2004, 101(43), 15275–15278 CrossRef CAS PubMed.
- J. Tang, Y. Lei and X. He, et al., Recognition-Driven Remodeling of Dual-Split Aptamer Triggering In Situ Hybridization Chain Reaction for Activatable and Autonomous Identification of Cancer Cells, Anal. Chem., 2020, 92(15), 10839–10846 CrossRef CAS PubMed.
- B. Zhang, T. Tian and D. Xiao, et al., Facilitating In Situ Tumor Imaging with a Tetrahedral DNA Framework-Enhanced Hybridization Chain Reaction Probe, Adv. Funct. Mater., 2022, 32, 2109728 CrossRef CAS.
- M.-A. Shahbazi, T. Bauleth-Ramos and H. A. Santos, DNA Hydrogel Assemblies: Bridging Synthesis Principles to Biomedical Applications, Adv. Ther., 2018, 1(4), 1800042 CrossRef.
- W. K. Hu, Z. J. Wang and Y. Xiao, et al., Advances in crosslinking strategies of biomedical hydrogels, Biomater. Sci., 2019, 7(3), 843–855 RSC.
- F. Ding, Q. B. Mou and Y. Ma, et al., A Crosslinked Nucleic Acid Nanogel for Effective siRNA Delivery and Antitumor Therapy, Angew. Chem., Int. Ed., 2018, 57(12), 3064–3068 CrossRef CAS PubMed.
- C. Mao, W. Sun and Z. Shen, et al., A nanomechanical device based on the B–Z transition of DNA, Nature, 1999, 397(6715), 144–146 CrossRef CAS PubMed.
- B. Yurke, A. J. Turberfield and A. P. Mills, et al., A DNA-fuelled molecular machine made of DNA, Nature, 2000, 406(6796), 605–608 CrossRef CAS PubMed.
- D. Liu and S. Balasubramanian, A Proton-Fuelled DNA Nanomachine, Angew. Chem., Int. Ed., 2003, 42(46), 5734–5736 CrossRef CAS PubMed.
- J.-S. Shin and N. A. Pierce, A Synthetic DNA Walker for Molecular Transport, J. Am. Chem. Soc., 2004, 126(35), 10834–10835 CrossRef CAS PubMed.
- E. S. Andersen, M. Dong and M. M. Nielsen, et al., Self-assembly of a nanoscale DNA box with a controllable lid, Nature, 2009, 459(7243), 73–76 CrossRef CAS PubMed.
- A. Kuzuya, Y. Sakai and T. Yamazaki, et al., Nanomechanical DNA origami 'single-molecule beacons' directly imaged by atomic force microscopy, Nat. Commun., 2011, 2(1), 449 CrossRef PubMed.
- M. Douglas Shawn, I. Bachelet and M. Church George, A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads, Science, 2012, 335(6070), 831–834 CrossRef CAS PubMed.
- P. Ketterer, E. M. Willner and H. Dietz, Nanoscale rotary apparatus formed from tight-fitting 3D DNA components, Sci. Adv., 2016, 2(2), e1501209 CrossRef PubMed.
- M. J. Urban, S. Both and C. Zhou, et al., Gold nanocrystal-mediated sliding of doublet DNA origami filaments, Nat. Commun., 2018, 9(1), 1454 CrossRef PubMed.
- D. Woods, D. Doty and C. Myhrvold, et al., Diverse and robust molecular algorithms using reprogrammable DNA self-assembly, Nature, 2019, 567(7748), 366–372 CrossRef CAS PubMed.
- H. Ijäs, I. Hakaste and B. Shen, et al., Reconfigurable DNA Origami Nanocapsule for pH-Controlled Encapsulation and Display of Cargo, ACS Nano, 2019, 13(5), 5959–5967 CrossRef PubMed.
- P. M. Arnott and S. Howorka, A Temperature-Gated Nanovalve Self-Assembled from DNA to Control Molecular Transport across Membranes, ACS Nano, 2019, 13(3), 3334–3340 CrossRef CAS PubMed.
- L. N. Green, H. K. K. Subramanian and V. Mardanlou, et al., Autonomous dynamic control of DNA nanostructure self-assembly, Nat. Chem., 2019, 11(6), 510–520 CrossRef CAS PubMed.
- X. Wu, Q. Liu and F. Liu, et al., An RNA/DNA hybrid origami-based nanoplatform for efficient gene therapy, Nanoscale, 2021, 13(30), 12848–12853 RSC.
- X. Qi, X. Liu and L. Matiski, et al., RNA Origami Nanostructures for Potent and Safe Anticancer Immunotherapy, ACS Nano, 2020, 14(4), 4727–4740 CrossRef CAS PubMed.
- L. Zhu, J. Luo and K. Ren, Nucleic acid-based artificial nanocarriers for gene therapy, J. Mater. Chem. B, 2023, 11(2), 261–279 RSC.
- C. Geary, A. Chworos and E. Verzemnieks, et al., Composing RNA Nanostructures from a Syntax of RNA Structural Modules, Nano Lett., 2017, 17(11), 7095–7101 CrossRef CAS PubMed.
- H. Li, K. Zhang and F. Pi, et al., Controllable Self-Assembly of RNA Tetrahedrons with Precise Shape and Size for Cancer Targeting, Adv. Mater., 2016, 28(34), 7501–7507 CrossRef CAS PubMed.
- Y. Shu, H. Yin and M. Rajabi, et al., RNA-based micelles: A novel platform for paclitaxel loading and delivery, J. Controlled Release, 2018, 276, 17–29 CrossRef CAS PubMed.
- I. Severcan, C. Geary and A. Chworos, et al., A polyhedron made of tRNAs, Nat. Chem., 2010, 2(9), 772–779 CrossRef CAS PubMed.
- J. M. Stewart, M. Viard and H. K. K. Subramanian, et al., Programmable RNA microstructures for coordinated delivery of siRNAs, Nanoscale, 2016, 8(40), 17542–17550 RSC.
- M. Hu, C. Feng and Q. Yuan, et al., Lantern-shaped flexible RNA origami for Smad4 mRNA delivery and growth suppression of colorectal cancer, Nat. Commun., 2023, 14(1), 1307 CrossRef CAS PubMed.
- J. B. Lee, J. Hong and D. K. Bonner, et al., Self-assembled RNA interference microsponges for efficient siRNA delivery, Nat. Mater., 2012, 11(4), 316–322 CrossRef CAS PubMed.
- L. Li, J. Liu and Z. Diao, et al., Evaluation of specific delivery of chimeric phi29 pRNA/siRNA nanoparticles to multiple tumor cells, Mol. BioSyst., 2009, 5(11), 1361–1368 RSC.
- A. Sharma, F. Haque and F. Pi, et al., Controllable self-assembly of RNA dendrimers, Nanomedicine, 2016, 12(3), 835–844 CrossRef CAS PubMed.
- D. Shu, Y. Shu and F. Haque, et al., Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics, Nat. Nanotechnol., 2011, 6(10), 658–667 CrossRef CAS PubMed.
- H. Zhang, J. A. Endrizzi and Y. Shu, et al., Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the Phi29 hexameric motor pRNA, Rna, 2013, 19(9), 1226–1237 CrossRef CAS PubMed.
- X. Li, M. Vieweger and P. Guo, Self-assembly of four generations of RNA dendrimers for drug shielding with controllable layer-by-layer release, Nanoscale, 2020, 12(31), 16514–16525 RSC.
- K. R. Kim, H. Y. Kim and Y. D. Lee, et al., Self-assembled mirror DNA nanostructures for tumor-specific delivery of anticancer drugs, J. Controlled Release, 2016, 243, 121–131 CrossRef CAS PubMed.
- G. Rusak, I. Piantanida and L. Masić, et al., Spectrophotometric analysis of flavonoid-DNA interactions and DNA damaging/protecting and cytotoxic potential of flavonoids in human peripheral blood lymphocytes, Chem.-Biol. Interact., 2010, 188(1), 181–189 CrossRef CAS PubMed.
- H. Lee, A. K. Lytton-Jean and Y. Chen, et al., Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery, Nat. Nanotechnol., 2012, 7(6), 389–393 CrossRef CAS PubMed.
- K. R. Kim, D. Bang and D. R. Ahn, Nano-formulation of a photosensitizer using a DNA tetrahedron and its potential for in vivo photodynamic therapy, Biomater. Sci., 2016, 4(4), 605–609 RSC.
- Y. X. Zhao, A. Shaw and X. Zeng, et al., DNA origami delivery system for cancer therapy with tunable release properties, ACS Nano, 2012, 6(10), 8684–8691 CrossRef CAS PubMed.
- D. H. Schaffert, A. H. Okholm and R. S. Sørensen, et al., Intracellular Delivery of a Planar DNA Origami Structure by the Transferrin-Receptor Internalization Pathway, Small, 2016, 12(19), 2634–2640 CrossRef CAS PubMed.
- X. Shen, Q. Jiang and J. Wang, et al., Visualization of the intracellular location and stability of DNA origami with a label-free fluorescent probe, Chem. Commun., 2012, 48(92), 11301–11303 RSC.
- B. Zhang, T. Tian and D. Xiao, et al., Facilitating In Situ Tumor Imaging with a Tetrahedral DNA Framework-Enhanced Hybridization Chain Reaction Probe, Adv. Funct. Mater., 2022, 32(16), 2109728 CrossRef CAS.
- F. Ding, Q. Mou and Y. Ma, et al., A Crosslinked Nucleic Acid Nanogel for Effective siRNA Delivery and Antitumor Therapy, Angew Chem. Int. Ed. Engl., 2018, 57(12), 3064–3068 CrossRef CAS PubMed.
- J. M. Stewart, M. Viard and H. K. Subramanian, et al., Correction: Programmable RNA microstructures for coordinated delivery of siRNAs, Nanoscale, 2017, 9(15), 5019 RSC.
Footnote |
| † These authors contribute equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |
