Open Access Article
Open Access ArticleIdentifying the specific-targeted marine cerebrosides against SARS-CoV-2: an integrated computational approach†
Eman Maher Zahrana,
Ahmed M. Sayedbg,
Miada F. Abdelwahab c,
Amgad Albohy
c,
Amgad Albohy d,
Basma S. Abdulrazikd,
Ayman M. Ibrahime,
Gerhard Bringmann
d,
Basma S. Abdulrazikd,
Ayman M. Ibrahime,
Gerhard Bringmann *f and
Usama Ramadan Abdelmohsen
*f and
Usama Ramadan Abdelmohsen *ac
*ac
aDepartment of Pharmacognosy, Faculty of Pharmacy, Deraya University, 61111 New Minia, Egypt
bDepartment of Pharmacognosy, Faculty of Pharmacy, Nahda University, 62513, Beni-Suef, Egypt
cDepartment of Pharmacognosy, Faculty of Pharmacy, Minia University, 61519 Minia, Egypt. E-mail: Usama.ramadan@mu.edu.eg; Fax: +20-086-2369075; Tel: +20-086-2347759
dDepartment of Pharmaceutical Chemistry, The Faculty of Pharmacy, The British University in Egypt (BUE), Cairo 11837, Egypt
eDepartment of Pharmaceutical Chemistry, Faculty of Pharmacy, Deraya University, 61111 New Minia, Egypt
fInstitute of Organic Chemistry, University of Würzburg, Am Hubland, 97074 Würzburg, Germany. E-mail: bringman@chemie.uni-wuerzburg.de; Fax: +49-931-3184755; Tel: +49-931-3185323
gDepartment of Pharmacognosy, Faculty of Pharmacy, AlMaaqal University, 61014 BasraIraq
First published on 18th November 2021
Abstract
Cerebrosides are a group of metabolites belonging to the glycosphingolipids class of natural products. So far, 167 cerebrosides, compounds 1–167, have been isolated from diverse marine organisms or microorganisms. The as yet smaller number of compounds that have been studied more in depth proves a potential against challenging diseases, such as cancer, a range of viral and bacterial diseases, as well as inflammation. This review provides a comprehensive summary on this so far under-explored class of compounds, their chemical structures, bioactivities, and their marine sources, with a full coverage to the end of 2020. Today, the global pandemic concern, COVID-19, has claimed millions of death cases around the world, making the development of anti-SARS-CoV-2 drugs urgently needed for such a battle. Accordingly, selected examples from all subclasses of cerebrosides were virtually screened for potential inhibition of SARS-CoV-2 proteins that are crucially involved in the viral–host interaction, viral replication, or in disease progression. The results highlight five cerebrosides that could preferentially bind to the hACE2 protein, with binding scores between −7.1 and −7.6 kcal mol−1 and with the docking poses determined underneath the first α1-helix of the protein. Moreover, the molecular interaction determined by molecular dynamic (MD) simulation revealed that renieroside C1 (60) is more conveniently involved in key hydrophobic interactions with the best stability, least deviation, least ΔG (−6.9 kcal mol−1) and an RMSD value of 3.6 Å. Thus, the structural insights assure better binding affinity and favorable molecular interaction of renieroside C1 (60) towards the hACE2 protein, which plays a crucial role in the biology and pathogenesis of SARS-CoV-2.
1. Introduction
Cerebrosides, a large group of glycosphingolipids, are essential components of cell membranes in eukaryotic organisms and in a few bacteria.1,2 Studies have demonstrated that cerebrosides have a prominent role in cell–cell interactions and are involved in cell regulation and signal transduction.3,4 Chemically, these biomolecules have amphiphilic properties, consisting of a ceramide moiety and one sugar unit. The hydrophobic ceramide part includes a sphingoid base (also known as sphingosine or sphingol), which is a long-chain amino-alcohol, as well as an amide-linked fatty acyl chain.5,6 The sugar residue in these monoglycosylceramides is either glucose or galactose resulting therefore in two main types, glucocerebrosides (glucosylceramides) and galactocerebrosides (galactosylceramides).7,8 Glucocerebrosides occur in fungi, plants, and animals, while galactocerebrosides are only found in fungi and animals.2 There are usually various functional groups connected to the ceramide backbone, comprising methyl branches, hydroxy groups, cis or trans desaturation, or cyclopropane rings.8Most genes responsible for the critical steps in the biosynthetic pathway of cerebrosides have been cloned from different organisms based on phylogenetic profiling. Moreover, the biological importance of most functional groups have been characterized through reverse genetic, molecular and cell biological techniques.2,8 Sphingolipid biosynthesis starts with the condensation of serine and palmitoyl-CoA to produce a long-chain base, 3-ketodihydrosphingosine, which is then reduced to yield a dihydrosphingosine. After this, a fatty acid is linked by an amide bond to the dihydrosphingosine to give rise to the corresponding dihydroceramide, which is then hydroxylated at C-4 to produce the respective ceramide. Glucose, or galactose, is added at C-1 of the ceramide to yield the corresponding glucosylceramide or galactosylceramide.4 The analysis of the geographical distribution clearly illustrates that the majority of the marine organisms investigated for the presence of cerebrosides were collected from the Pacific Ocean near the southeastern coasts of Asia, while the smallest portion was collected near the northeastern part of Africa. Several reports documented that secondary metabolites (especially those containing lipids) detected in marine sponges give them the ability to deter predators,9,10 inhibit pathogenic microbes,11 and demonstrate competitive dominance12 towards other sessile benthic organisms.
A crucial factor included in the competitive success of sponges is their ability to combat microbial attacks. Many sponges protect their surface from colonization through fouling organisms and potential pathogenic bacteria by producing secondary metabolites possessing anti-infective properties.13 Marine organisms are constantly exposed to potentially harmful bacteria, in the Pacific Ocean with bacterial abundances ranging from 6 × 104 to 2.5 × 106 cells per mL in the surrounding seawater.14 Noteworthy, the Pacific Ocean, as mentioned above, is the main area from which most of the investigated marine organisms were collected. Sponges are additionally exposed to large quantities of microbes passing through their tissues due to their filter feeding activities.15 Given the exposure of sponges to such a high number of microbes in the marine environment and the relatively low incidence of infection with diseases, production of secondary metabolites in sponges is crucial in providing effective defenses against the invasion of pathogenic microbes especially after damage or injury.16,17
Cerebrosides have been isolated from different marine micro- and macro-organisms, including sea stars,18 sponges,19,20 soft corals,21 sea cucumbers,22 bryozoa,23 ascidians,6 sea urchins,24 marine protists,25 and marine-derived fungi.26 There is an increasing interest in these natural products because they have been shown to exhibit significant biological functions such as cytotoxic,27,28 antifungal,29 immunosuppressive,19 anti-inflammatory,21 and antibacterial activities.21,30 In this comprehensive review, we report, for the first time, on the different cerebrosides isolated from various marine species, their chemical structures, biological activities, and on the site of collection for each mentioned organism.
Recently, the viral infection with coronavirus (CoV-19), a highly infectious and contagious virus of the Coronaviridae family, has caused a global outbreak of coronavirus illness (SARS-COV-2),31 which requires a continuous development of prophylaxis and treatment approaches.32 Coronaviruses are categorized into four subfamilies: alpha, beta, gamma, and delta coronaviruses, among them SARS-CoV-2, which belongs to the beta category.33 The structural organization of the viral RNA encodes for several proteins involving the E (envelope), M (membrane), N (nucleocapside) and S (spike) proteins as well as 16 other putative non-structural proteins.34 The S (spike) proteins with their binding sites are considered critical as they help viral attachment to the host cells mainly via the cellular receptor (ACE2) (a key receptor for viral entry)35 located on the epithelium of the respiratory tract of the host, which promotes its clustering and pathogenesis. Although a number of vaccines have already been developed, finding compounds with efficacy for the viral molecular targets remains a preferable approach against such highly mutable pathogens, especially those from a natural origin to avoid serious side effects. Fortunately, the recent advances in computational techniques have proven efficacy for identifying potential drug candidates from diverse natural sources.31,36,37 Since marine cerebrosides proved efficacy as antimicrobial agents, supported by their ecological roles protecting the tissues of marine invertebrates against bacterial invasion, we docked the reported marine cerebrosides against the known targets of SARS-CoV-2. The targets mainly include: viral main protease (Mpro), viral RNA-dependent RNA polymerase (nsp12), viral methyl transferase (nsp16), viral spike protein (S) as well as human angiotensin-converting enzyme 2 (hACE2), the specific viral recognition protein included in the pathogenesis pathway.38 Moreover, a molecular dynamic (MD) simulation has been carried out to validate the binding efficacy of the less fluctuating cerebroside.
2. Cerebrosides isolated from marine invertebrates
2.1. Sponges
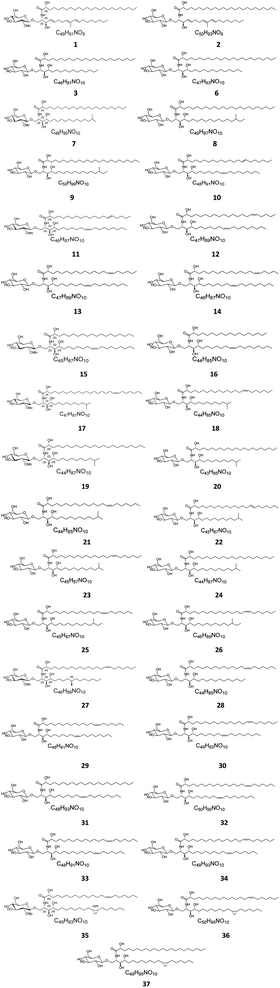
Several cerebrosides have been isolated from marine sponges (Fig. 1), where nine of them, compounds 1–9, were isolated from Agelas mauritianus, which was obtained by diving off Kume Shima in Okinawa, Japan. The nine agelasphins were obtained following antitumor and immunostimulatory bioassay-guided fractionation from the lipophilic extract of the sponge and were structurally elucidated after total synthesis, which, for most of them, delivered an α-galactosyl ceramide core.27 All compounds exhibited in vivo significant antitumor activity against B16 murine melanoma cells, with T/C ranging from 160 to 190% and a wide safety margin.27 From the glass sponge, Aulosaccus sp., collected near Kuril Islands in Russia, 28 metabolites, compounds 10–37, were obtained. These glucosyl ceramides are sphingoid bases, N-acylated with straight-chain (2R)-2-hydroxy fatty acids with no reported biological activities up to now.Two further cerebrosides with a β-D-glucosyl ceramide core, cerebroside 1 (38) and cerebroside 2 (39), were isolated from the ethanol extract of the sponge Oceanapia sp., which was collected near the Scott Reef in Australia. These compounds proved to be new modifications of N-acetylglucosamine-containing glycosphingolipids,39 which had been previously found in the sponges Amphimedon viridis29 and Halichondria cylindrata.28
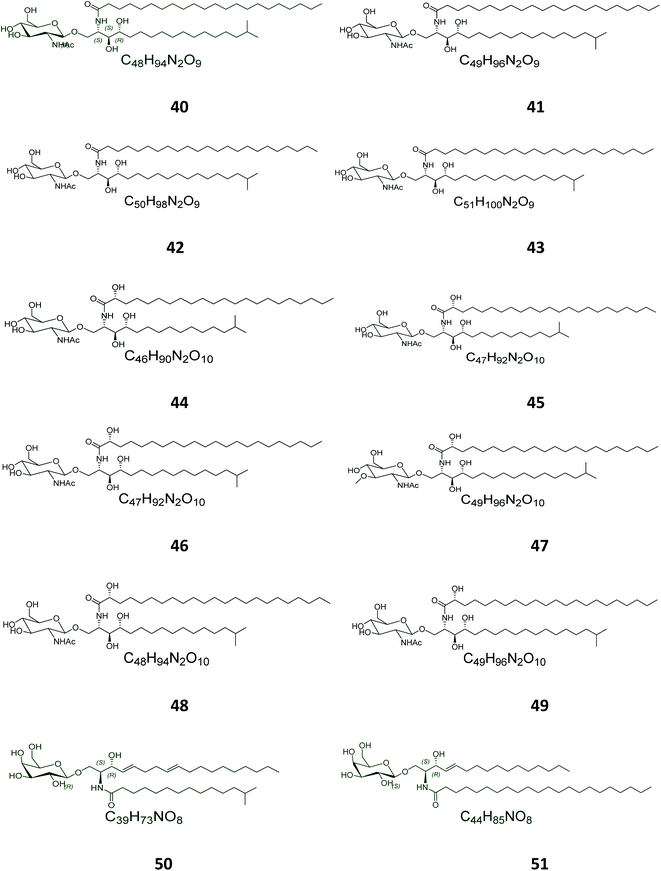
Twelve other N-acetyl-glucosaminyl ceramides, named halicylindrosides (compounds 40–51), Table S1,† were purified from the ethyl acetate fraction of the Japanese sponge Halichondria cylindrata, collected at the coast of Atami at the southwest of Tokyo. The halicylindrosides 40–49, with an N-acetyl-glucosaminyl ceramide skeleton, exhibited significant cytotoxicity against P388 murine leukemia cells, with an IC50 value of 6.8 μg mL−1, and a moderate antifungal potential against Mortierella remanniana at 250 μg per disk,28 while the halicylindrosides 50 and 51, which were also frequently isolated from the American marine sponge Halichondria panicea, found at the low intertidal rock surfaces, have a galacto-ceramide skeleton and proved inactivity in many studies (Fig. 2).40
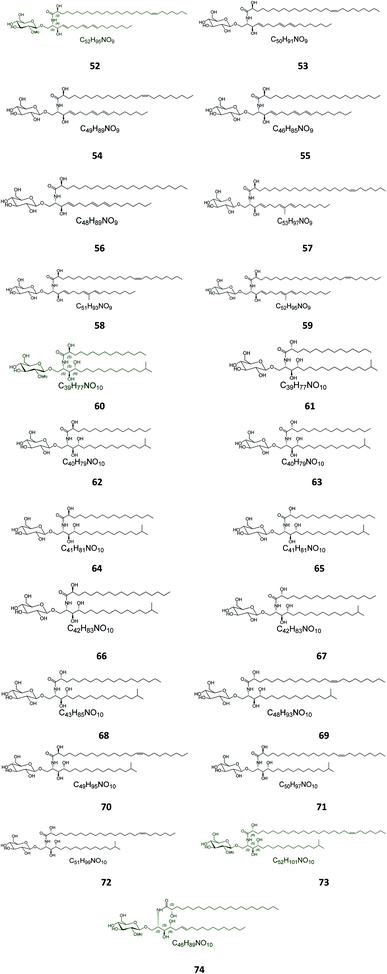
Twenty-three halicerebrosides (Fig. 3), named renierosides (compounds 52–74), were obtained from the ethyl extract of the Korean sponge Haliclona reniera, collected at deep coastal regions of Ulleung Island in Korea. Of them, eight novel compounds, renierosides A1–A5 and B1–B3 (52–59), with extraordinary amide-linked long chain fatty acid moieties, revealed no cytotoxicity against a panel of five human solid tumor cell lines (Table S1†).7
From the Red Sea at the Gulf of Eilat, near Egypt, the sponge Amphimedon viridis was collected and its extract subjected to chromatographic separation to give five glucocerebrosides named amphicerebrosides B–F (75–79). Worth mentioning, the compounds exhibited no antifungal activity against C. albicans, while their hepta-acetyl derivatives were moderately active with MIC = 12.5 μg mL−1.29 Interestingly, two galacto-cerebrosides, plakosides A (80) and B (81), which belong to a unique class of O-prenylated glycolipids, were isolated from the Italian sponge Plakortis simplex, at the coast of Half Moon Cay in Italy (Fig. 4). Plakosides showed strong immunosuppressive action on activated T cells and presented a convenient natural model for an enhanced understanding of the structural requirements for the immunomodulating potential of glycosphingolipids. They also significantly (p < 0.01) inhibited the proliferative response of the lymph node cells to Con A (0.5 μg mL−1) at the whole tested doses (0.01–10 μg mL−1).19
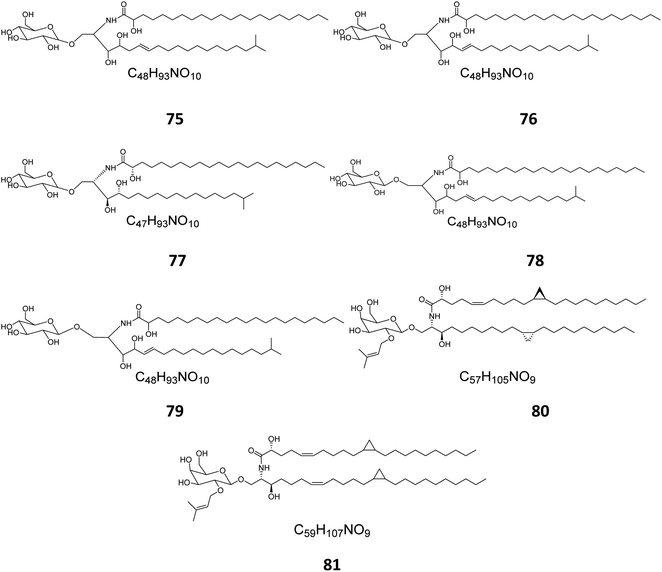 | ||
| Fig. 4 Cerebrosides isolated from sponges (compounds 75–81); the configurations at the flat-drawn stereocenters of 75, 76, 78, 79, and 81 were not presented in the literature. | ||
2.2. Star fish
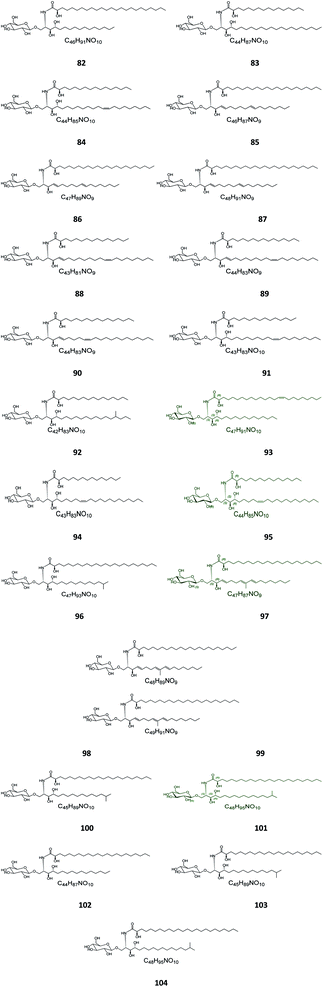
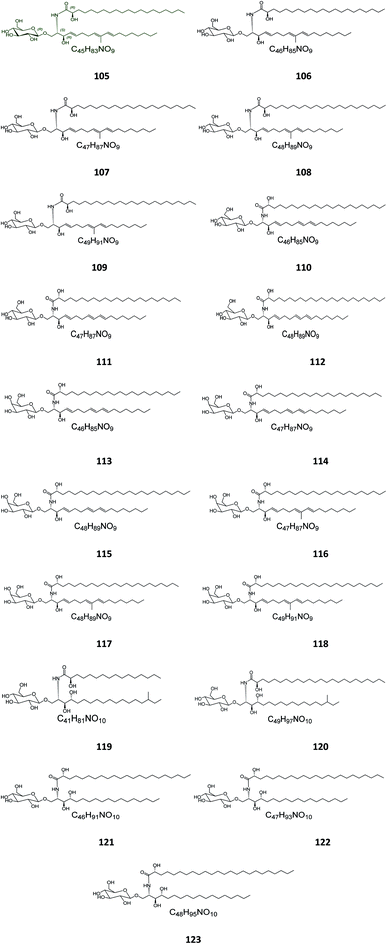
Six β-glucopyranosyl ceramides named acanthacerebrosides A–F (82–87) (Fig. 5) were isolated from the lipophilic fraction of the chloroform/methanol extract of the Japanese starfish Acanthaster planci, collected at Hakata Bay in Japan.41 Six other related β-glucopyranosyl ceramides named asteriacerebrosides A–F (88–93) were also isolated from the chloroform/methanol extract of the starfish Asterias amurensis versicolor from the same coast.42 In a similar study and by the same authors, three other related compounds named astrocerebrosides A–C (94–96) were obtained from the less polar fraction of the chloroform/methanol extract of the Japanese Astropecten latespinosus.43 Astrocerebroside B (95) was also found in the starfish Luidia maculata, which was also collected from Hakata Bay in Japan.44 From the Chinese star fish Stellaster equestris, collected from the East China Sea, Higuchi et al. isolated eight compounds, five glucosides, S-1-3–S-1-5, S-2a-3, S-2a-11 (97–101), and three galactosides, S-2b-2, S-2b-4, and S-2b-16 (102–104), discovered in the less polar fraction of its chloroform/methanol extract.45Five glucocerebrosides, named ophidiacerebrosides A–E (105–109) (Fig. 6), were isolated from the sea star Ophidiaster ophidiamus, collected at Islote Dragonera, Balearic Islands, in Spain. All of them exhibited more than 70% inhibition of L1210 murine leukemia cell growth at a concentration of 2 μg mL−1, with ophidiacerebroside A (105) being the most significant one, with 96% inhibition.18 Nine galactocerebrosides, named oreacerebrosides A–I (110–118), were obtained from the sea star Oreaster reticulatus, collected at the coast of Grand Bahama Island. When tested against rat glioma C6 cells, only oreacerebrosides A (110), D (113), and I (118) showed moderate activity, while the others were completely inactive.5 From the Japanese star fish Luidia maculata collected at Hakata Bay in Fukuoka, Japan, two glucocerebrosides, named luidiacerebrosides A (119) and B (120), were isolated from the non-polar fraction of the chloroform/methanol extract. From the Indian star fish Pentaceraster regulus, three further glucocerebrosides, named regulosides A–C (121–123), were isolated and tested for wound healing activity, but only reguloside A (121) that exhibited a moderate activity.46
2.3. Sea cucumber
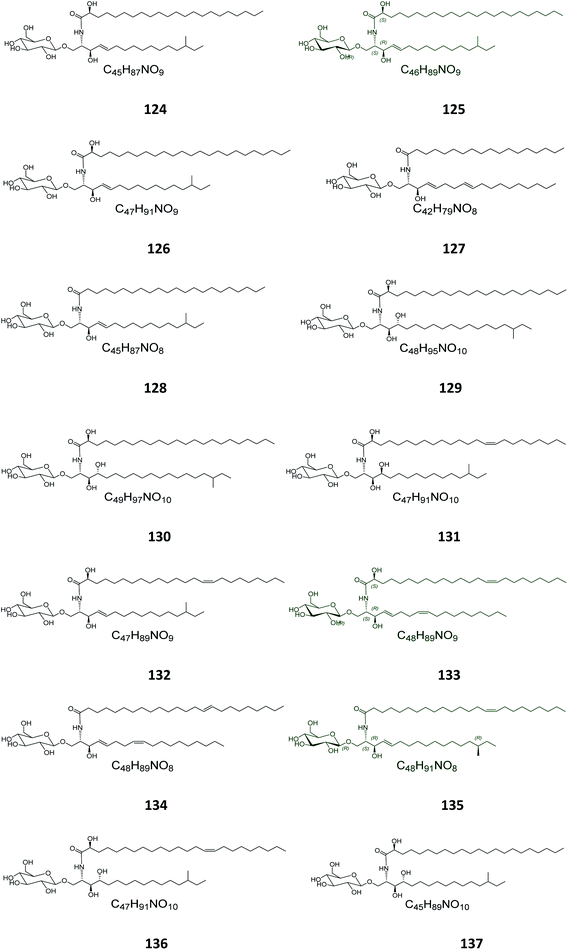
From the Japanese sea cucumber Cucumaria echinata, collected from the Ariake Sea in Japan, three glucocerebrosides were isolated and named CE-2b, CE-2c, and CE-2d (124–126) (Fig. 7),22 while four other glucosides, named PA-0-1, PA-0-5, PA-2-5, and PA-2-6 (127–130) were isolated from another Japanese sea cucumber, called Pentacta australis, which was collected from Ariake Sea in Japan.22 Back to the genus Cucumaria, from another species called C. frondosa, which was collected from deep regions of the Arctic Ocean near Russia, three further glucocerebrosides, named CF-3-1, CF-3-2, and CF-3-3 (131–133) were isolated, purified, and identified.47 From Cucumaria echinata, collected from the Sea of Genkai in Japan, four further glucocerebrosides were isolated, named CE-1-2, CE-1-3, CE-3-1, and CE-3-2 (134–137). In the brine shrimp assay, they displayed lethality at a 30 ppm concentration with a lethal rate of 22%.44,482.4. Bryozoa
Four glucocerebrosides, 138–141 (Table S1†) (Fig. 8) were obtained from the Chinese marine bryozoan Bugula neritina, which was collected at Daya Bay in the South China Sea. Only compound 140 showed significant cancer cell growth inhibition potential against murine P388 lymphocytic leukemia cells and against a panel of human cancer cell lines (GI50 0.15–2.6 μg mL−1).232.5. Ascidians
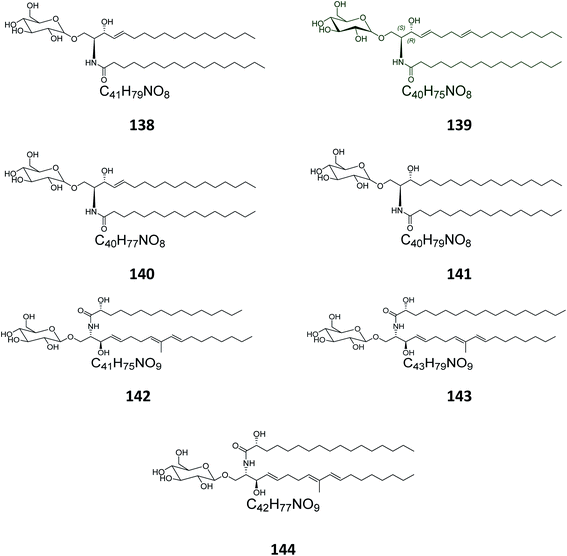
From the Spanish ascidian Phallusia fumigata, which was collected from the southern coast of Cádiz in Spain, four glucocerebrosides were isolated and identified as phallusides 1–4 (142–145).62.6. Octocorals
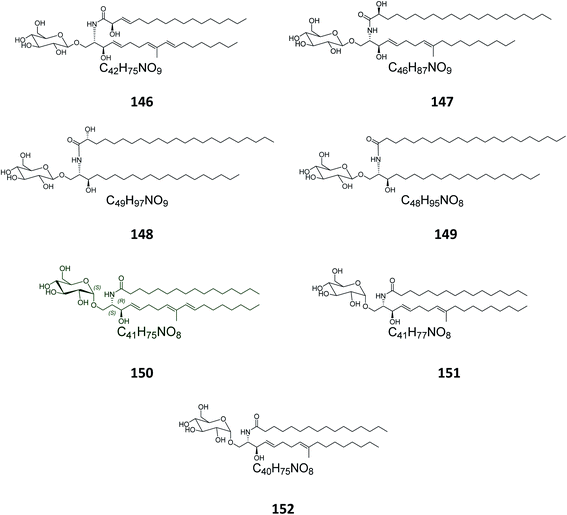
From the octocoral Sarcophyton ehrenbergi, collected at Dongsha Islands in Taiwan, two glucocerebrosides named sarcoehrenosides A (146) and B (147) (Fig. 9) were isolated. Both compounds showed relatively similar significant anti-inflammatory activity against iNOS protein expression at a concentration of 10 μM with a 47.3 ± 7.1% reduction, but were devoid of any antibacterial activity.212.7. Sea urchins
From the Indian sea urchin Temnopleurus toreumaticus, collected at the Mandapam Coast in Tamilnadu, India, two glucocerebrosides, named temnosides A and B (148, 149), were identified from the non-polar fraction.242.8. Protists
From the marine protist Thraustochytrium globosum, isolated from the surface of the tropical seagrass Thalassia testudinum collected from the Bahamas, three glucocerebrosides, named thraustochytrosides A, B, and C (150–152), were isolated.252.9. Cerebrosides isolated from marine-associated fungi
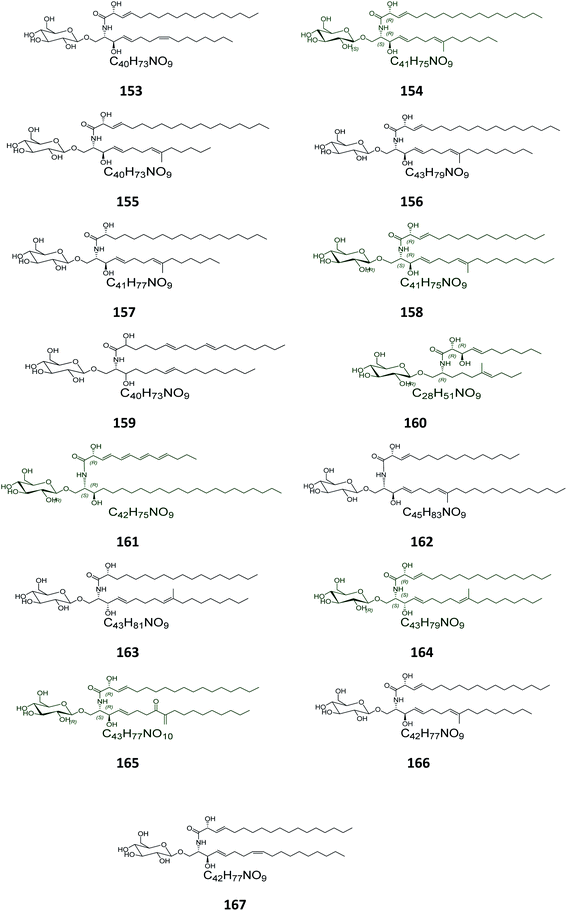
From the Chinese fungus Penicillium chrysogenum PXP-55, isolated from the roots of the mangrove plant, Rhizophora stylosa, collected in the Hainan Province in China, five glucocerebrosides were obtained, named chrysogesides A–E (153–157) (Fig. 10). Among them, only chrysogeside B (154) gave a significant antimicrobial activity against Enterobacter aerogenes, with a MIC of 1.72 μg mL−1, the other compounds were inactive against a number of bacterial and fungal strains even at higher concentrations up to 150 μg mL−1.49 From the same fungus, Penicillium chrysogenum, but collected from the Red Sea in Egypt, a glucocerebroside named LAMA-1 (158) was isolated, identified and tested for cytotoxicity against a panel of cell lines, showing a weak activity with IC50 > 100 μg mL−1. From the Red Sea Coast, now near Sharm El-Sheikh, two glucocerebrosides, named penicillosides A (159) and B (160), were isolated from a Penicillium growing on Red Sea tunicates species belonging to the genus Didemnum. Both compounds showed moderate cytotoxic activity against HeLa cells, only penicilloside A (159) exhibited antifungal potential towards Candida albicans, with an inhibition zone of 23 mm, while penicilloside B (160) proved antibacterial potential against Staphylococcus aureus and Escherichia coli, with inhibition zones of 19 and 20 mm, respectively.26Two other glucosides, named asperiamides B (161) and C (162), were obtained from the Chinese Sea water derived fungus Aspergillus niger (MF-16), which was collected from Quanzhou Gulf in Fujian Province, China. Moreover, two glucosides named flavusides A (163) and B (164) were obtained from the marine-derived fungus Aspergillus flavus grown on the edible green alga Codium fragile collected in Geomun-do Island in Yeosu, Korea. Both compounds exhibited a moderate antibacterial activity against the methicillin-resistant S. aureus and against the multidrug-resistant S. aureus, with MIC values of 15.6 μg mL−1 and 31.2 μg mL−1, respectively.50
Finally, the Chinese sediment-derived fungus Alternaria raphani, obtained from Hongdao sea salt in Qingdao, China, was also a source of secondary metabolites, giving three glucocerebrosides named alternarosides A, B, and C (165–167). The three compounds displayed weak antibacterial activities against Escherichia coli, Bacillus subtilis, and Candida albicans, with MIC values ranging from 70 to 400 μg mL−1.51
3. Materials and methods
3.1. Virtual screening and molecular docking
Selected cerebrosides (i.e. green-colored structures in Fig. 1–10) from the different chemical groups were investigated for their potential effects on the five main targets of SARS-COV-2, where compounds and targets were prepared as reported.52 The absolute configurations chosen for these structures are illustrated on them (Fig. 1–10). Since the exact configuration of some compounds of these selected candidates was not established in the literature, we assigned the configurations at their stereogenic centers similarly to that of their closest derivatives.The used targets included the viral main protease (6LU7), viral RNA-dependent RNA polymerase (nsp12, 7BV2), viral methyl transferase (nsp16, 6W4H), viral spike protein (6M0J), and hACE2 as a viral recognition protein (6VW1).52 Firstly, the structures of the ligands were constructed and minimized with 1000 steps of steepest-descent algorithm using Avogadro.53 Protein targets were retrieved from the protein data bank (http://www.rcsb.org/) using the codes 7BV2, 6LU7, 6W4H, 6M0J, and 6VW1 (Table 1). Proteins were prepared by deleting water molecules, adding hydrogens, merging hydrogens and then computing Gasteiger charges using AutoDock tools.54 Via the AutoDock Vina program,55 a grid box of 503 Å3 was centered on the co-crystallized ligand if available. For 6M0J and 6VW1, where no ligand was available, their grid box centered on the residues at the interacting surface including Q493 and E35, respectively. Exhaustiveness was used for all docking procedures and docking validation was done to validate the docking method and also to compare the tested compounds.56
| Ligand | 7BV2-RdRp | 6LU7-Mpro | 6W4H-methyltransferase | 6VW1-human ACE2 | 6M0J-S-protein |
|---|---|---|---|---|---|
| 1 | −5.7 | −5.2 | −6.1 | −6.3 | −4.1 |
| 7 | −4.9 | −4.9 | −5.4 | −6.3 | −4 |
| 11 | −5.2 | −4.4 | −6.1 | −6.6 | −5.8 |
| 15 | −5.5 | −5.5 | −5.7 | −6.7 | −4.1 |
| 17 | −5.4 | −4.1 | −5.9 | −6.9 | −4.5 |
| 19 | −4.8 | −4.6 | −5.6 | −6.9 | −4.5 |
| 27 | −5.6 | −5.3 | −6 | −6.5 | −4.9 |
| 35 | −5.4 | −5.2 | −5.9 | −6.7 | −4.8 |
| 40 | −5.5 | −4.3 | −6 | −6.4 | −4.5 |
| 50 | −5.5 | −5.3 | −6.5 | −6.4 | −4.8 |
| 51 | −4.9 | −4.8 | −6 | −6.0 | −4.3 |
| 52 | −5.1 | −5 | −5.8 | −7.1 | −4 |
| 60 | −5.3 | −6.1 | −6.9 | −7.1 | −5.1 |
| 73 | −4.9 | −4.9 | −5.8 | −6.9 | −4.3 |
| 74 | −5.5 | −4.9 | −5.4 | −6.4 | −4.1 |
| 93 | −5.6 | −5.1 | −5.5 | −6.3 | −4.5 |
| 95 | −5.6 | −5.9 | −5.8 | −6.8 | −4.9 |
| 97 | −5 | −5.2 | −5.8 | −6.2 | −4.1 |
| 101 | −4.9 | −4.9 | −5.7 | −6.4 | −5.1 |
| 105 | −5.5 | −5.3 | −5.8 | −6.0 | −4.9 |
| 125 | −4.9 | −5.3 | −5.7 | −6.5 | −4.7 |
| 133 | −5.6 | −5.2 | −5.1 | −6.7 | −4.7 |
| 135 | −5.5 | −5.1 | −5.9 | −6.3 | −4.1 |
| 139 | −5.1 | −5.1 | −5.9 | −6.5 | −4.6 |
| 150 | −5.7 | −5 | −5.9 | −7.0 | −4.6 |
| 154 | −5.8 | −5.2 | −5.9 | −7.0 | −4.7 |
| 158 | −5.7 | −5.6 | −6.4 | −7.6 | −5 |
| 160 | −6.4 | −6.7 | −6.3 | −7.2 | −5.2 |
| 161 | −5.3 | −5.4 | −6.7 | −7.3 | −6.2 |
| 164 | −5.7 | −5.5 | −6.4 | −6.9 | −5.1 |
| 165 | −5.3 | −4.9 | −6.3 | −6.5 | −4.3 |
| Co-crystallized ligand | −7.0 (RemdesivirTP) | −7.9 (N3) | −8.1 (SAM) | — | — |
3.2. Molecular dynamic simulation (MDS)
MD simulation was carried out in an attempt to fully explain and confirm the stability between the top scoring compounds and the active site of their corresponding target.57 MDS for the generated ligand–enzyme complexes were performed using the Nanoscale Molecular Dynamics (NAMD) 2.6 software,58 applying the CHARMM27 force field.59 Hydrogen atoms were added to the protein structures using the psfgen plugin included in the Visual Molecular Dynamic (VMD) 1.9 software. The whole generated systems were then solvated using water molecules (TIP3P) and 0.15 M NaCl. At first, the total energy of the generated systems was minimized and gradually heated to reach 300 K and equilibrated for 200 seconds. Subsequently, the MDS was continued for 50 ns, and the trajectory was stored every 0.1 ns and further analyzed with the VMD 1.9 software. The MDS output was sampled every 0.2 ns to calculate the root mean square deviation (RMSD). The parameters of the tested compounds were prepared using the online platform “Charmm-GUI (https://www.charmm-gui.org/) or the VMD force field toolkit (ffTK)60 binding free energies (ΔG) were calculated using the free energy perturbation (FEP) method. The web-based software Absolute Ligand Binder61 was used to generate the input files for NAMD software, which was performed for the simulations required for ΔGs calculations.4. Results and discussion
4.1 Investigation and assessment of the isolated marine cerebrosides
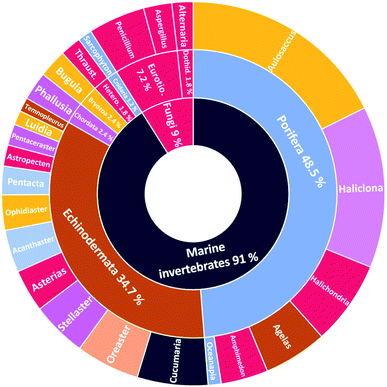
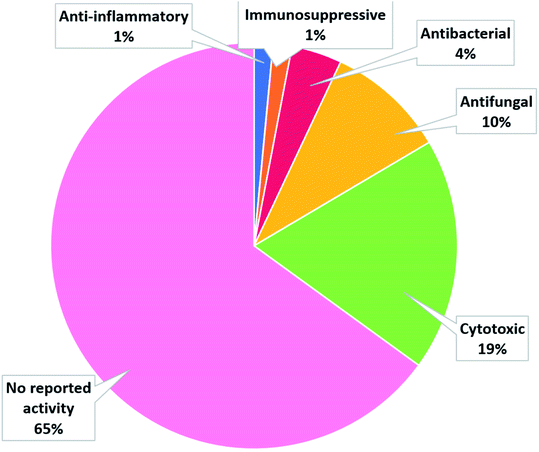
The number of cerebrosides detected in various marine invertebrates and in marine-associated fungi has gradually increased, especially during the past 30 years. A vast number of marine organisms were investigated up to date, yielding a plethora of secondary metabolites, mostly glucoside derivatives (Table S1†). The marine invertebrates released more than 90% of the identified secondary metabolites distributed between the phylum Porifera (48.5%), the phylum Echinodermata (34.7%), and other, less extensively studied Phyla. From Porifera, the major isolated compounds were obtained from the genus Aulosaccus, with 28 compounds representing about 17.7% of the total number of compounds, followed by the genus Haliclona, with 23 compounds (13.7%), while the other four genera represent smaller percentages. While from Echinodermata, the majority of compounds (13 metabolites, 6%) were obtained from sea cucumbers of the genus Cucumaria, followed by a star fish from the genus Oreaster (twelve compounds, 5.4%), with the other metabolites distributed between various organisms. Worth mentioning, the marine-derived fungi are less investigated for the production of such metabolites, here only three fungal genera were analyzed. Among these, Penicillium was the most intensely studied one, with eight isolated compounds, followed by Aspergillus with four metabolites, and finally Alternaria with three natural products (Fig. 11).The collected data in this review reveal that for most of the isolated compounds (about 65%) no biological activities have been reported, which may be attributed to the small quantities of material obtained. The majority of the biologically investigated compounds (about 19%) showed various cytotoxic potencies against a panel of cancer cell lines, while about 10% of the compounds exerted strong antifungal activities, especially against C. albicans (Fig. 12). Most of the mentioned cerebrosides (65%) were never tested for biological activities, and hence, biological investigation of this interesting class of natural products is needed.
4.2. Antiviral potential of cerebrosides against SARS CoV-2
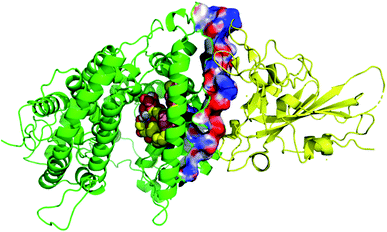
Thirty-one cerebrosides (1, 7, 11, 15, 17, 19, 27, 35, 40, 50, 51, 52, 60, 73, 74, 93, 95, 97, 101, 105, 125, 133, 135, 139, 150, 154, 158, 160, 161, 164, and 165) were selected for the docking study. They were green-colored among other compounds listed in Fig. 1–10, with a selection criterion to get representatives from each group according to their isolation source, and hence, all the reviewed structures are covered. The absolute stereochemistry of these selected structures was assigned according to the literature (shown on the green-colored compounds in Fig. 1–10. Since the exact configuration of some compounds of these selected candidates was not established in the literature, we assigned the configurations at their stereogenic centers similarly to that of their closest derivatives.Due to the large size of the investigated ligands, the docking was done in 503 Å3 centered on the co-crystallized ligand for the first three targets. The results revealed that all the tested compounds did not give a good docking score or even a clear docking pose with the targets except with hACE2 (Table 1). Among the docked structures against hACE2 target, the five top scoring structures, 52, 60, 158, 160, and 161, gave docking scores of −7.1, −7.1, −7.6, −7.2, and −7.3 kcal mol−1, respectively. It is well-known that the integrity of the SARS-CoV body is achieved via four proteins: The S (spike), E (envelope), M (membrane) and N (nucleocapsid) proteins, of which the S protein mediates the viral entry into the host cells and is recognized by the hACE2 (Naqvi et al. 2020![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 71). As a result, both S-protein and ACE2 are considered to play a crucial role in the biology and pathogenesis of SARS-CoV-2.62 Accordingly, the docking pose of the five cerebrosides was found to be underneath the first α1-helix (residues 19–45), which includes K31, E35, and D38 residues and which is known to interact with the receptor binding domain (RBD) of the S-protein with its corresponding residues L455, Q493, and S494, respectively (Fig. 13).63As a representative, the binding mode of LAMA-1 (158) is illustrated in Fig. 14.
71). As a result, both S-protein and ACE2 are considered to play a crucial role in the biology and pathogenesis of SARS-CoV-2.62 Accordingly, the docking pose of the five cerebrosides was found to be underneath the first α1-helix (residues 19–45), which includes K31, E35, and D38 residues and which is known to interact with the receptor binding domain (RBD) of the S-protein with its corresponding residues L455, Q493, and S494, respectively (Fig. 13).63As a representative, the binding mode of LAMA-1 (158) is illustrated in Fig. 14.
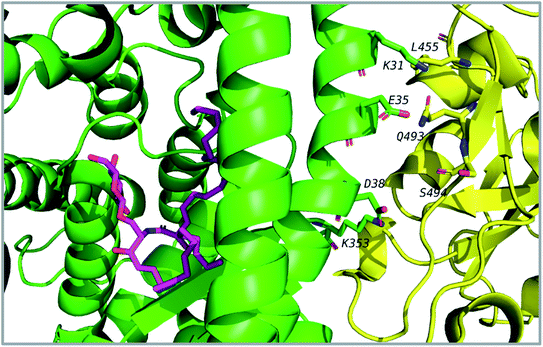 | ||
| Fig. 14 Binding mode of 158 (pink) in hACE2 (green) is underneath the α1-helix responsible for recognition of SARS-CoV-2 S-protein (yellow). Some residues at the interface are shown and labeled. | ||
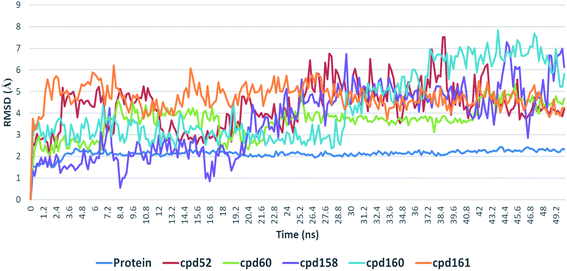
4.3. Conformational stability of protein–ligand complexes
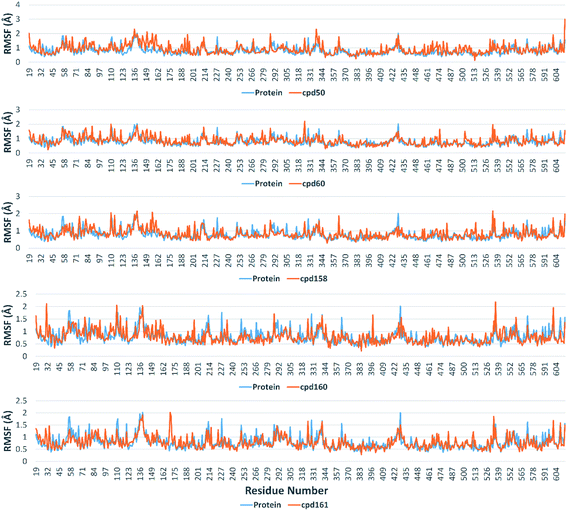
Further in silico validation was achieved through a number of MDS experiments and binding free energy (ΔG) calculations. Top scoring compounds in docking experiments (52, 60, 158, 160, and 161) showed convergent binding stability and ΔGs values over 50 ns of MDS experiments. As illustrated in Fig. 15, the RMSDs of the tested compounds over 50 ns were clustered together achieving average values ranging from 3.6 to 4.7 Å. Compound 60 was the most stable one with the least deviation and fluctuation (average RMSD = 3.6 Å), as a result, it attained the lowest ΔG value (−6.9 kcal mol−1). This indicates a good stability of the compound when combined with the hACE2. The remaining compounds (52, 158, 160, and 161) were almost identical with average RMSDs of 4.3, 3.9, 4.2, and 4.7 Å, and ΔG values of −6.1, −6.3, −6.1, and −5.7 kcal mol−1, respectively.The Root Mean Square Fluctuation (RMSF) of each protein–compound complex in comparison with that of the free protein clearly demonstrated the amino acid residues that interacted with each ligand, where they showed the least fluctuation (Fig. 16). PHE-40, MET-60, TRP-69, TRP-349, and PHE-390 were the most common amino acid residues involved in hydrophobic interactions, providing the best scoring compounds. In regard to H-bonds and water bridges, ASP-350, ASP-382, HID-401, and ARG-514 were the most common residues involved in these two types of interactions with the same compounds during the course of the simulation (Fig. 17).
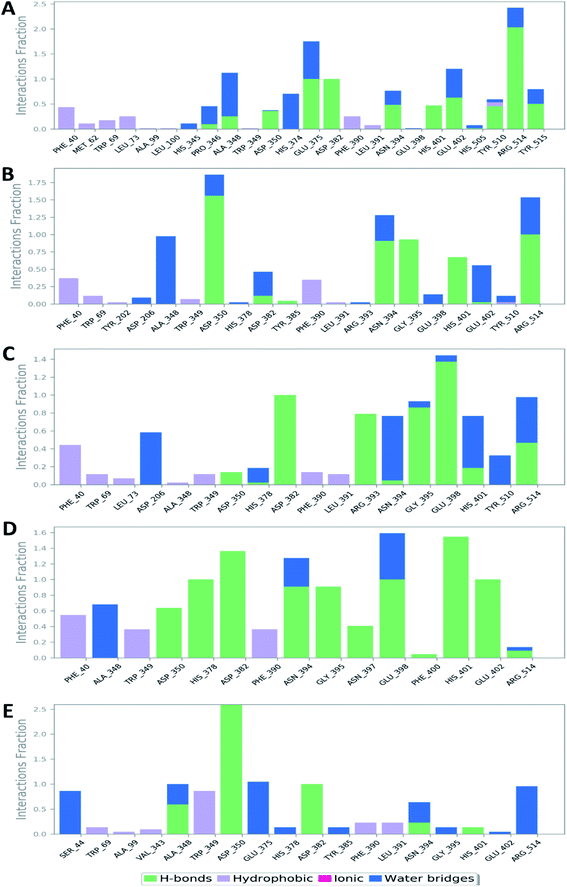 | ||
| Fig. 17 Protein–ligand contacts between the binding site of hACE-2 and compounds 52, 60, 158, 160, and 161 (A, B, C, D, and E, respectively). | ||
According to the aforementioned in silico analysis, these top-scoring cerebrosides showed stable bindings and significant interactions with the hACE2 target protein. As a result, the recognition and binding to SARS CoV-2's RBD might be affected or even the already formed hACE2–RBD complex could be destabilized. Most of the previous reports in this regard reported attenuated viral entry upon the interaction with the S-protein, where it became unable to recognize and bind to its receptor (i.e. hACE2).64–66 On the other hand, some studies also reported the way of targeting hACE2 in destabilization of its complex with the viral RBD.67,68 Further in vitro RBD–hACE2 binding assay will be needed to confirm the ability of these top-scoring cerebrosides in disrupting the binding of RBD–hACE2. In the near future, we are aiming to carry out this in vitro analysis using these cerebrosides after purification from their natural sources so that we can conduct further structural modifications to get more potent candidates. Targeting SARS CoV-2 at its entry point is considered one of the most successful fighting strategies that can enable the development of both therapeutic and prophylactic agents.68–70
5. Conclusion
Marine invertebrates and their associated microorganisms have so far been largely studied as a promising source of secondary metabolites with most diverse new chemical structures displaying a variety of interesting biological activities, yet not fully expanded. Cerebrosides present an enormous potential for the discovery of new therapeutic leads for drug expansion to fight against the current global health problems with increasing numbers over the past years. Interestingly, many cerebrosides, especially the glucosidic ones, show potent cytotoxic, antimicrobial, and anti-inflammatory activities, most of them being cytotoxic. A docking study of selected bioactive compounds was done to investigate their potential effects on SARS-CoV-2 targets. Among them, renierosides A1 (52) and C1 (60), LAMA-1 (158), penicilloside B (160), and asperiamide B (161) exhibited a potential inhibition against SARS-CoV-2 hACE2 protein suggesting a good start for the design of SARS-CoV-2 inhibitors. In addition, and based on a dynamic simulation, renieroside C1 (60) was highlighted as the most stable one with the least degree of fluctuation with an average RMSD value of 3.6 Å. Accordingly, renieroside C1 has been explored here as a potential effective therapeutic agent that might be used to control SARS-CoV-2 infection, which can be further investigated in vitro for confirmation and for determination of its exact pathway. More attention should be given for the untested cerebrosides, especially the neglected ones with the galactosyl moieties, to explore their potential against many other diseases. Moreover, scientific efforts should be given for exploring our treasure from new marine sources that can be proven to be a valuable reservoir for bioactive cerebrosides to get benefit of the rising development of oceanographic science and metabolomic screening techniques.List of abbreviations
| MD | Molecular dynamics |
| ACE2 | Angiotensin converting enzyme 2 (enzyme reducing the amount of angiotensin II) |
| T/C | Treated/control |
| Con A | Concanavalin A (a T cell mitogen) |
| iNOS | Inducible nitric oxide synthase |
| hACE2 | Human angiotensin converting enzyme 2 |
| MDS | Molecular dynamic simulation |
| NAMD | Nanoscale molecular dynamic |
| VMD | Visual molecular dynamic |
| RMSD | Root mean square deviation |
| ffTK | Force field toolkit |
| FEP | Free energy perturbation |
| ΔGs | Gibbs free energy |
| RBD | Receptor binding domain |
| RMSF | Root mean square fluctuation |
Funding
This publication has received no external funding.Author contributions
E. M. Z. searched the literature and wrote the paper; M. F. A. drew the structures of the metabolites and wrote the introduction part; A. M. I. revised the drawn structures and stereoconfigurations, A. A. and B. S. A. performed the in silico study; E. M. Z., U. R. A. contributed to analyzing the collected data; E. M. Z., G. B., and U. R. A. contributed to reviewing and editing the final version of the manuscript. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors are grateful to Minia University and Deraya University for supporting this work.References
- Z. J. Wu, M. A. Ouyang, R. K. Su and Y. X. Guo, Chin. J. Chem., 2008, 26, 759–764 CrossRef CAS
.
- D. Warnecke and E. Heinz, Cell. Mol. Life Sci., 2003, 60, 919–941 CrossRef CAS PubMed
.
- T. M. Dunn, D. V. Lynch, L. V. Michaelson and J. A. Napier, Ann. Bot., 2004, 93, 483–497 CrossRef CAS PubMed
.
- G. Daum, N. D. Lees, M. Bard and R. Dickson, Yeast, 1998, 14, 1471–1510 CrossRef CAS PubMed
.
- V. Costantino, C. de Rosa, E. Fattorusso, C. Imperatore, A. Mangoni, C. Irace, C. Maffettone, D. Capasso, L. Malorni and R. Palumbo, Eur. J. Org. Chem., 2007, 2007, 5277–5283 CrossRef
.
- R. Durán, E. Zubía, M. J. Ortega, S. Naranjo and J. Salvá, Tetrahedron, 1998, 54, 14597–14602 CrossRef
.
- T. A. Mansoor, P. B. Shinde, X. Luo, J. Hong, C.-O. Lee, C. J. Sim, B. W. Son and J. H. Jung, J. Nat. Prod., 2007, 70, 1481–1486 CrossRef CAS PubMed
.
- P. Ternes, P. Sperling, S. Albrecht, S. Franke, J. M. Cregg, D. Warnecke and E. Heinz, J. Biol. Chem., 2006, 281, 5582–5592 CrossRef CAS PubMed
.
- E. Burns, I. Ifrach, S. Carmeli, J. R. Pawlik and M. Ilan, Mar. Ecol. Prog. Ser., 2003, 252, 105–114 CrossRef
.
- J. R. Pawlik, B. Chanas, R. J. Toonen and W. Fenical, Mar. Ecol. Prog. Ser., 1995, 127, 183–194 CrossRef CAS
.
- S. R. Kelly, E. Garo, P. R. Jensen, W. Fenical and J. R. Pawlik, Aquat. Microb. Ecol., 2005, 40, 191–203 CrossRef
.
- H. Madduppa, P. J. Schupp, M. R. Faisal, M. Y. Sastria and C. Thoms, Mar. Biodivers., 2017, 47, 149–151 CrossRef
.
- S. R. Kelly, P. R. Jensen, T. P. Henkel, W. Fenical and J. R. Pawlik, Aquat. Microb. Ecol., 2003, 31, 175–182 CrossRef
.
- C. J. Wiebinga, M. J. Veldhuis and H. J. De Baar, Deep Sea Res. Part I Oceanogr. Res. Pap., 1997, 44, 451–476 CrossRef CAS
.
- J.-M. Gili and R. Coma, Trends Ecol. Evol., 1998, 13, 316–321 CrossRef CAS PubMed
.
- C. Thoms, R. Ebel and P. Proksch, J. Chem. Ecol., 2006, 32, 97–123 CrossRef CAS PubMed
.
- J. C. Futch, D. W. Griffin, K. Banks and E. K. Lipp, Mar. Pollut. Bull., 2011, 62, 2308–2316 CrossRef PubMed
.
- W. Jin, K. L. Rinehart and E. A. Jares-Erijman, J. Org. Chem., 1994, 59, 144–147 CrossRef CAS
.
- V. Costantino, E. Fattorusso, A. Mangoni, M. Di Rosa and A. Ianaro, J. Am. Chem. Soc., 1997, 119, 12465–12470 CrossRef CAS
.
- E. A. Santalova, V. A. Denisenko and P. S. Dmitrenok, Lipids, 2015, 50, 1209–1218 CrossRef CAS PubMed
.
- S.-Y. Cheng, Z.-H. Wen, S.-F. Chiou, C.-W. Tsai, S.-K. Wang, C.-H. Hsu, C.-F. Dai, M. Y. Chiang, W.-H. Wang and C.-Y. Duh, J. Nat. Prod., 2009, 72, 465–468 CrossRef CAS PubMed
.
- R. Higuchi, M. Inagaki, K. Togawa, T. Miyamoto and T. Komori, Liebigs Ann. Chem., 1994, 1994, 79–81 CrossRef
.
- X.-R. Tian, H.-F. Tang, Y.-S. Li, H.-W. Lin, N. Ma, W. Zhang and M.-N. Yao, J. Asian Nat. Prod. Res., 2009, 11, 1005–1012 CrossRef CAS PubMed
.
- U. Babu, S. Bhandari and H. Garg, J. Nat. Prod., 1997, 60, 732–734 CrossRef CAS
.
- K. M. Jenkins, P. R. Jensen and W. Fenical, Tetrahedron Lett., 1999, 40, 7637–7640 CrossRef CAS
.
- S. S. Murshid, J. M. Badr and D. T. Youssef, Rev. bras. farmacogn., 2016, 26, 29–33 CrossRef CAS
.
- T. Natori, M. Morita, K. Akimoto and Y. Koezuka, Tetrahedron, 1994, 50, 2771–2784 CrossRef CAS
.
- H.-y. Li, S. Matsunaga and N. Fusetani, Tetrahedron, 1995, 51, 2273–2280 CrossRef CAS
.
- S. Hirsch and Y. Kashman, Tetrahedron, 1989, 45, 3897–3906 CrossRef CAS
.
- G. Yang, L. Sandjo, K. Yun, A. S. Leutou, G.-D. Kim, H. D. Choi, J. S. Kang, J. Hong and B. W. Son, Chem. Pharm. Bull., 2011, 59, 1174–1177 CrossRef CAS PubMed
.
- S. A. Mohamad, E. M. Zahran, M. R. A. Fadeel, A. Albohy and M. A. Safwat, Int. J. Nanomed., 2021, 16, 1789 CrossRef PubMed
.
- C. B. Mishra, P. Pandey, R. D. Sharma, M. Z. Malik, R. K. Mongre, A. M. Lynn, R. Prasad, R. Jeon and A. Prakash, Brief. Bioinform, 2021, 22, 1346–1360 CrossRef CAS PubMed
.
- Y. A. Helmy, M. Fawzy, A. Elaswad, A. Sobieh, S. P. Kenney and A. A. Shehata, J. Clin. Med., 2020, 9, 1225 CrossRef CAS PubMed
.
- Y. Han and P. Král, ACS Nano, 2020, 14, 5143–5147 CrossRef CAS PubMed
.
- D. Morniroli, M. L. Giannì, A. Consales, C. Pietrasanta and F. Mosca, Front. immunol., 2020, 11, 1480 CrossRef CAS PubMed
.
- S. S. El Hawary, A. R. Khattab, H. S. Marzouk, A. S. El Senousy, M. G. Alex, O. M. Aly, M. Teleb and U. R. Abdelmohsen, RSC Adv., 2020, 10, 43103–43108 RSC
.
- S. S. El-Hawary, M. A. Rabeh, M. A. E. Raey, E. M. A. El-Kadder, M. Sobeh, U. R. Abdelmohsen, A. Albohy, A. M. Andrianov, I. P. Bosko and M. M. Al-Sanea, J. Biomol. Struct. Dyn., 2021, 1–13 CrossRef PubMed
.
- S. S. El-Hawary, R. Mohammed, H. S. Bahr, E. Z. Attia, M. m. H. El-Katatny, N. Abelyan, M. M. Al-Sanea, A. S. Moawad and U. R. Abdelmohsen, J. Appl. Microbiol., 2021, 131(3), 1193–1211 CrossRef CAS PubMed
.
- A. Guzii, T. Makarieva, V. Denisenko, V. Svetashev, S. Rodkina, P. Dmitrenok, S. Anastyuk and V. Stonik, Russ. Chem. Bull., 2006, 55, 928–933 CrossRef CAS
.
- D. G. Nagle, W. C. McClatchey and W. H. Gerwick, J. Nat. Prod., 1992, 55, 1013–1017 CrossRef CAS PubMed
.
- Y. Kawano, R. Higuchi, R. Isobe and T. Komori, Liebigs Ann. Chem., 1988, 1988, 19–24 CrossRef
.
- R. Higuchi, J. X. JHOU, K. Inukai and T. Komori, Liebigs Ann. Chem., 1991, 745–752 CrossRef CAS
.
- R. Higuchi, M. Kagoshima and T. Komori, Liebigs Ann. Chem., 1990, 1990, 659–663 CrossRef
.
- S. Kawatake, K. Nakamura, M. Inagaki and R. Higuchi, Chem. Pharm. Bull., 2002, 50, 1091–1096 CrossRef CAS PubMed
.
- R. Higuchi, Y. Harano, M. Mitsuyuki, R. Isobe, K. Yamada, T. Miyamoto and T. Komori, Liebigs Ann., 1996,(4), 593–599 CAS
.
- U. Venkannababu, S. P. S. Bhandari and H. S. Garg, Liebigs Ann., 1997, 1997, 1245–1247 CrossRef
.
- L. Ming-Ping, S. Jun-Jie, J. Jian and Y. Yang-Hua, Chin. J. Nat. Med., 2012, 10, 105–109 CrossRef
.
- K. Yamada, E. Hara, T. Miyamoto, R. Higuchi, R. Isobe and S. Honda, Eur. J. Org. Chem., 1998, 1998, 371–378 CrossRef
.
- X. Peng, Y. Wang, K. Sun, P. Liu, X. Yin and W. Zhu, J. Nat. Prod., 2011, 74, 1298–1302 CrossRef CAS PubMed
.
- G. Yang, L. Sandjo, K. Yun, A. S. Leutou, G. D. Kim, H. D. Choi, J. S. Kang, J. Hong and B. W. Son, Chem. Pharm. Bull., 2011, 59, 1174–1177 CrossRef CAS PubMed
.
- W. Wang, Y. Wang, H. Tao, X. Peng, P. Liu and W. Zhu, J. Nat. Prod., 2009, 72, 1695–1698 CrossRef CAS PubMed
.
- E. M. Zahran, A. Albohy, A. Khalil, A. H. Ibrahim, H. A. Ahmed, E. M. El-Hossary, G. Bringmann and U. R. Abdelmohsen, Mar. Drugs, 2020, 18, 645 CrossRef CAS PubMed
.
- M. D. Hanwell, D. E. Curtis, D. C. Lonie, T. Vandermeersch, E. Zurek and G. R. Hutchison, J. Cheminformatics, 2012, 4, 1–17 CrossRef PubMed
.
- E. M. Zahran, U. R. Abdelmohsen, A. T. Ayoub, M. A. Salem, H. E. Khalil, S. Y. Desoukey, M. A. Fouad and M. S. Kamel, S. Afr. J. Bot., 2020, 131, 311–319 CrossRef CAS
.
- A. Vina, J. Comput. Chem., 2010, 31, 455–461 Search PubMed
.
- E. M. Zahran, U. R. Abdelmohsen, A. Kolkeila, M. A. Salem, H. E. Khalil, S. Y. Desoukey, M. A. Fouad and M. S. Kamel, Nat. Prod. Res., 2020, 1–5 Search PubMed
.
- E. M. Zahran, U. R. Abdelmohsen, A. S. Hussein, M. A. Salem, H. E. Khalil, S. Yehia Desoukey, M. A. Fouad and M. S. Kamel, Nat. Prod. Res., 2019, 1–5 Search PubMed
.
- J. C. Phillips, R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, E. Villa, C. Chipot, R. D. Skeel, L. Kale and K. Schulten, J. Comput. Chem., 2005, 26, 1781–1802 CrossRef CAS PubMed
.
- A. D. MacKerell Jr, D. Bashford, M. Bellott, R. L. Dunbrack
Jr, J. D. Evanseck, M. J. Field, S. Fischer, J. Gao, H. Guo and S. Ha, J. Phys. Chem. B, 1998, 102, 3586–3616 CrossRef PubMed
.
- C. G. Mayne, M. Muller and E. Tajkhorshid, Parameterizing Small Molecules Using the Force Field Toolkit (ffTK), University of Illinois at Urbana-Champaign, 2015 Search PubMed
.
- S. Jo, W. Jiang, H. S. Lee, B. Roux and W. Im, CHARMM-GUI Ligand Binder for absolute binding free energy calculations and its application, 2013 Search PubMed
.
- A. Albohy, E. M. Zahran, U. R. Abdelmohsen, M. A. Salem, T. Al-Warhi, M. M. Al-Sanea, N. Abelyan, H. E. Khalil, S. Y. Desoukey and M. A. Fouad, J. Biomol. Struct. Dyn., 2020, 1–11 CrossRef PubMed
.
- J. Shang, G. Ye, K. Shi, Y. Wan, C. Luo, H. Aihara, Q. Geng, A. Auerbach and F. Li, Nature, 2020, 581, 221–224 CrossRef CAS PubMed
.
- A. Hussain, A. Hasan, M. M. N. Babadaei, S. H. Bloukh, M. E. Chowdhury, M. Sharifi, S. Haghighat and M. Falahati, Biomed. Pharmacother., 2020, 110559 CrossRef CAS PubMed
.
- S. Xia, Q. Liu, Q. Wang, Z. Sun, S. Su, L. Du, T. Ying, L. Lu and S. Jiang, Virus Res., 2014, 194, 200–210 CrossRef CAS PubMed
.
- S. Xia, M. Liu, C. Wang, W. Xu, Q. Lan, S. Feng, F. Qi, L. Bao, L. Du and S. Liu, Cell Res., 2020, 30, 343–355 CrossRef CAS PubMed
.
- M. Razizadeh, M. Nikfar and Y. Liu, Biophys. J., 2021, 120, 2793–2804 CrossRef CAS PubMed
.
- A. E. Salih, B. Thissera, M. Yaseen, A. S. Hassane, H. R. El-Seedi, A. M. Sayed and M. E. Rateb, Mar. Drugs, 2021, 19, 406 CrossRef CAS PubMed
.
- G. Williamson and A. Kerimi, Biochem. Pharmacol., 2020, 178, 114123 CrossRef CAS PubMed
.
- S. Jiang, X. Zhang and L. Du, Expert Opin. Ther. Targets, 2021, 25, 415–421 CrossRef CAS PubMed
.
- A. A. T. Naqvi, K. Fatima, T. Mohammad, U. Fatima, I. K. Singh, A. Singh, S. M. Atif, G. Hariprasad and M. I. Hassan, Biochim. Biophys. Acta, Mol. Basis Dis., 2020, 1866(10), 165878 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available: Table S1: Cerebrosides isolated from different marine sources. See DOI: 10.1039/d1ra07103c |
| This journal is © The Royal Society of Chemistry 2021 |