Open Access Article
Open Access ArticleCopper nanoclusters: designed synthesis, structural diversity, and multiplatform applications
Ani
Baghdasaryan
 and
Thomas
Bürgi
and
Thomas
Bürgi
 *
*
Department of Physical Chemistry, University of Geneva, 30 Quai Ernest-Ansermet, 1211 Geneva 4, Switzerland. E-mail: thomas.buergi@unige.ch
First published on 26th February 2021
Abstract
Atomically precise metal nanoclusters (MNCs) have gained tremendous research interest in recent years due to their extraordinary properties. The molecular-like properties that originate from the quantized electronic states provide novel opportunities for the construction of unique nanomaterials possessing rich molecular-like absorption, luminescence, and magnetic properties. The field of monolayer-protected metal nanoclusters, especially copper, with well-defined molecular structures and compositions, is relatively new, about two to three decades old. Nevertheless, the massive progress in the field illustrates the importance of such nanoobjects as promising materials for various applications. In this respect, nanocluster-based catalysts have become very popular, showing high efficiencies and activities for the catalytic conversion of chemical compounds. Biomedical applications of clusters are an active research field aimed at finding better fluorescent contrast agents, therapeutic pharmaceuticals for the treatment and prevention of diseases, the early diagnosis of cancers and other potent diseases, especially at early stages. A huge library of structures and the compositions of copper nanoclusters (CuNCs) with atomic precisions have already been discovered during last few decades; however, there are many concerns to be addressed and questions to be answered. Hopefully, in future, with the combined efforts of material scientists, inorganic chemists, and computational scientists, a thorough understanding of the unique molecular-like properties of metal nanoclusters will be achieved. This, on the other hand, will allow the interdisciplinary researchers to design novel catalysts, biosensors, or therapeutic agents using highly structured, atomically precise, and stable CuNCs. Thus, we hope this review will guide the reader through the field of CuNCs, while discussing the main achievements and improvements, along with challenges and drawbacks that one needs to face and overcome.
1. Introduction
Metal nanoclusters with sizes comparable to the Fermi wavelength of electrons, i.e., smaller than 2 nm, show pronounced molecular-like and size-dependent optical and luminescence properties, intrinsic magnetism, strong chiral responses, etc. The ultrasmall size regime breaks down the continuum of energy bands into discrete energy levels and the collective oscillations of conduction band electrons typical for plasmonic particles upon interacting with an electromagnetic field are no longer dominant. Quantized and discrete states result in rich molecular-like features in their optical absorption spectrum. Besides the above-mentioned properties, noble metal clusters are also highly stable nanoobjects. The stability, however, for atomically precise copper nanoclusters (CuNCs) is of major concern due to the high susceptibility of the metal towards oxidation. Over last few decades, by designing synthetic protocols and applying mild and inert conditions, choosing appropriate ligands with unique structures and high metal binding affinities, the stability issue is resolved to an extent, leading to the (selective) formation, crystallization and finally the determination of tens of fascinating structures and geometries.1–3 Some structures turned out to be extraordinary not only due to the unique binding block configurations and construction moieties but also for geometries they pick up that were initially considered impossible. As for noble metals, their extraordinary stability has been investigated using several models such as the “superatom theory”4,5 and “jellium model”.6 According to the “superatom theory” and “jellium model”, valence electrons are delocalized and can be subjected to an external potential. Thus, clusters are considered superatoms and their angular momentum subshells can be filled according to the Aufbau principle.7 The superatom theory gives a series of electron shell closing numbers8 known as “magic” numbers. The electron count for any cluster with a formula of Mn(R)qm can be determined according to the following equation:| ne = n − m − q | (1) |
This rule has been widely used for spherical gold clusters, such as the very-well known Au25(SR)18,9 (negatively charged cluster, 8e− system: 1S2 1P6) and Au102(SR)44,10 (neutral cluster, 58e− system: 1S2 1P6 1D10 2S2 1F14 2P6 1G18), and also applies to CuNCs. Particularly stable atomically precise spherical CuNCs are therefore expected with electron counts of 2, 8, 18, etc., according to eqn (1) (to be discussed in section 2.2.4). For atomically precise gold nanoclusters, all magic numbers have been discovered, whereas for CuNCs only smallest 2e− systems are known and higher systems are yet to be discovered.
Keeping in mind the differences and common properties of atomically precise gold nanoclusters (AuNCs), we tried to summarize the main achievements in the field of CuNCs and illustrate the challenges that material scientists and inorganic chemists face on a daily basis. Thus, the purpose of this review is to explore the ever-growing field of copper nanoclusters, including chalcogenide, hydrido and pure thiolate-protected clusters. The review article focuses on the following topics: (i) the synthesis (top-down/bottom-up) of hydrophilic and hydrophobic clusters, and the differences and challenges when considering suitable ligands; (ii) size-dependent optical and luminescence properties of clusters by paying close attention to the effect of the core size and the nature of the protective ligand. In this sense, a special discussion will be dedicated to a novel aggregation-induced emission enhancement (AIE) phenomenon; (iii) structural analyses of atomically precise clusters by focusing mainly on the geometry of the metallic core and the arrangements of the ligands around the core; and (iv) exploration of the many possibilities for the application of CuNCs in biomedical, catalytic and optoelectronic fields. A key goal of this review is to present the great expansion of water-soluble CuNCs from the early 2000s and organic phase soluble and atomically precise CuNCs from 2012 until August 2020.
2. Synthesis of CuNCs
The synthetic procedures are of utmost importance in order to obtain high-quality nanomaterials. The great interest in synthesizing nanomaterials comes from their unique and extraordinary applications in the different areas of modern science. Therefore, various chemical and physical methods have been developed and applied for the large-scale synthesis of metal nanoclusters with precise atomic compositions. The breakthrough in the field of nanomaterials science, starting in the early 90s, made possible the synthesis of particles down to sub-nanometre sizes. Over the last few decades, tremendous progress has been made for mainly noble metals Au and Ag protected by organic and water-soluble ligands.11,12 Structure determination helped the understanding of the formation mechanisms and the development of new, efficient, and effective synthetic protocols for the synthesis of unique functional building blocks. However, despite the extensive research progress in the field of gold and silver nanoclusters, the studies focusing on the preparation and functionalization of other metals like earth-abundant copper are still in their infancy.13 The problem lies in the difficulty of preparing tiny and stable copper nanoparticles and nanoclusters. Copper, being in the same group as gold and silver, shares similar properties and can be taken as a viable alternative to expensive and rare metals. Therefore, the synthetic procedures applied for the preparation of gold and silver clusters were directly applied for the synthesis of copper nanoclusters. Modifications and slight changes made in the synthetic methods and reaction conditions helped to successfully overcome the difficulty of preparing atomically precise and stable CuNCs.14–162.1. Synthesis of water-soluble CuNCs
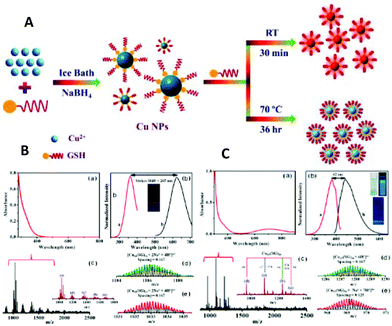 | ||
| Fig. 1 (A) Schematic illustration of the top-down synthesis of luminescent CuNCs. (B) (a) UV–vis absorption spectrum and (b) photoexcitation and photoluminescence spectra of red-emitting CuNCs in aqueous solution. (c) ESI-MS spectrum of CuNCs. (d) Isotopic patterns fitting to the formulas [Cu34(SG)16 + 2Na+ + 4H+]6+ and [Cu34(SG)13 + 2Na+ + 4H+]6+, respectively. (C) (a) UV–vis absorption spectrum and (b) photoexcitation and photoluminescence spectra of blue-emitting CuNCs in aqueous solution. (c) ESI-MS spectrum of CuNCs obtained at 70 °C. (d) Isotopic pattern fitting to the formulas [Cu25(SG)20 + 6H+]6+ and [Cu25(SG)20 + Na+ + 7H+]8+, respectively. Reprinted with permission from ref. 17. Copyright 2019 American Chemical Society. | ||
Interestingly, the bigger clusters were weakly emitting in the red, whereas the smaller ones were highly emissive in the blue spectral range. On the other hand, this showed the size-dependence of the photoluminescence properties of CuNCs (to be discussed later).
Another reported example of the top-down method involves the utilization of ammonia as an etchant to convert non-fluorescent CuNPs into small, green, fluorescent CuNCs.19 The ammonia-assisted etching of the CuNCs with an average size of 3.7 nm resulted in the formation of smaller (1.2 nm) and spherical CuNCs with an absolute quantum yield of 6.6%.
2.1.2.1. Template-mediated synthesis. The template-based method became a cutting-edge technology for the effective synthesis of various nanomaterials with designed structures, sizes, morphologies and properties.20–24 The main concept of this technique is the formation of uniform nanomaterials within the pores, channels, or cavities of a nano-matrix. Thus, the target's morphology and size can be tailored by simply changing the shape of the matrix, i.e. cylinder, rod, sphere. In general, any porous material, including naturally occurring minerals and biological substrates, can be considered as a template. Polymer networks, hydrogels, supramolecular assemblies, biomolecules such as DNA, proteins, viruses, and other microorganisms can be used as templates. Biological substances are extensively used for nanostructure synthesis owing to their naturally occurring complex structures.
Polymers: Several polymers with various structures were used as templates for the synthesis of CuNCs. However, the first report dates back almost two decades when Zhao et al. used fourth-generation (G4) poly(amidoamine) (PAMAM) dendrimers with an ethylenediamine core (G4-OH) as templates to prepare a mixture of CuNCs consisting of 4 to 64 atoms in their compositions with an average diameter of less than 1.8 nm.25 Highly stable and fluorescent (QY of 3.8% in ethanol) CuNCs were synthesized from the reduction of aqueous solutions of CuSO4 and polyethyleneimine (PEI) with hydrazine hydrate upon heating at 95 °C for 19 h (Fig. 2A).26–30 When functionalized lipoic acid (LA) attached to a tunable length of polyethylene glycol (PEG) segment was used as a template, the reduction with NaBH4 resulted in the formation of good-quality and highly fluorescent CuNCs (QY of 3.6% in water) with an average size of 2.5 nm.31 The formed clusters showed long-term stability when exposed to daylight and UV irradiation. However, when a similar reaction was carried out using both dihydrolipoic acid (DHLA) and poly(vinylpyrrolidone) (PVP)32 or just PVP alone33,34 (Fig. 2B) as capping agents and ascorbic acid as a reductant, the synthesis resulted in the formation of CuNCs with an average size of 2 nm. Polystyrene sulfonate (PSS)35 (Fig. 2C) and other multidentate polymers36 as templating scaffolds were also applied in the synthesis of CuNCs.
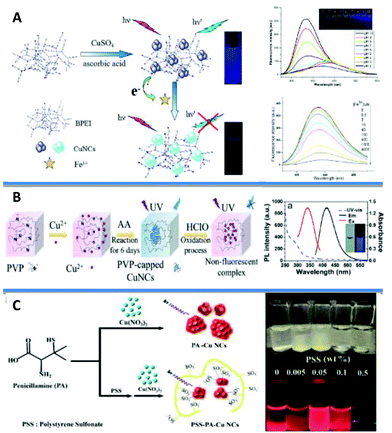 | ||
| Fig. 2 (A) Synthesis strategy for the BPEI-CuNCs, their stability under varying pH and the mechanism of the branched polyethyleneimine (BPEI)-CuNCs probe for Fe3+ sensing. Reprinted with permission from ref. 30. Copyright 2015 Elsevier. (B) Schematic diagram of PVP-capped CuNCs (on the left) and UV–vis absorption, emission, and excitation fluorescence spectra of the probe CuNCs at pH = 6.2 in 50 mM phosphate buffer (PB) (on the right). Inset: Photograph of CuNCs probe in buffer under daylight (left) and UV-light (λ = 365 nm, right), respectively. Reprinted with permission from ref. 34. Copyright 2020 American Chemical Society. (C) A scheme depicting a plausible mechanism for the synthesis of PSS-stabilized penicillamine (PA)-CuNCs and the photographs of the PSS-PA-CuNCs synthesized under different concentrations of PSS, from 0.005 to 0.5 wt%. Adapted with permission from ref. 35. Copyright 2016 Springer Nature. | ||
In 2012 Hui Zhang et al. reported the photoreductive synthesis of highly fluorescent metal nanoclusters of Cu, Ag, and Au (QYs of 2.2, 6.8, and 5.3%, respectively) in the presence of poly(methacrylic acid) functionalized with pentaerythritol tetrakis 3-mercaptopropionate (PMAA-PTMP) polymer upon UV-irradiation.37 It was found that AuNCs were relatively stable as compared to copper and silver in the presence of foreign metal ions.
Proteins: Proteins as structural biomolecules were widely used for the synthesis of versatile and biocompatible CuNCs (Fig. 3).38–59 Among a huge number of accessible proteins, bovine serum albumin (BSA) gained considerable attention due to its availability, high-water solubility, and ability to bind various organic and inorganic substances in a noncovalent fashion. Due to the well-defined structure, containing charged amino acids and 35 thiol groups from Cys residues, BSA offers perfect albumin-binding sites for nanocluster formation.60,61 The synthesis of CuNCs involves three steps till completion.56 In the first step, upon mixing the reactants, at neutral pH, –COOH carboxylate groups of the protein partially dissociate and immediately form a complex with Cu2+. The coordination complex resulted in the formation of a viscous paste. To enhance the solubility and further reduction with BSA, in the second reaction step, the pH of the medium needed to be increased to 12 by adding 1 M NaOH under heating at 55 °C.
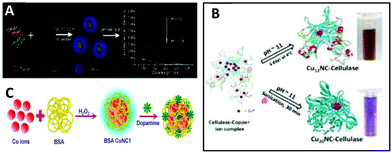 | ||
| Fig. 3 (A) Illustration of the synthesis and application of the CuNCs. Republished with permission from ref. 39. Copyright 2014 Royal Society of Chemistry. (B) Schematic representation of reaction methods employed for the synthesis of CuNCs in cellulase solution. Republished with permission from ref. 55. Copyright 2016 Royal Society of Chemistry. (C) Diagrammatic illustration of the detection of dopamine using BSA CuNC synthesized in presence of H2O2. Reprinted with permission from ref. 62. Copyright 2019 Elsevier. | ||
However, as a reducing agent, BSA is relatively weak, and to completely reduce Cu2+ to metallic copper, different reducing agents such as hydrogen peroxide49,56,62 and hydrazine hydrate (N2H4·2H2O)41,42,45 have been proposed to facilitate the last step of the reduction process. For example, the addition of a trace amount of hydrogen peroxide not only reduces the α-helix of the protein and increases the number of random and δ-structures, but also produces ˙OH radicals, which can break peptide bonds in BSA and reduce the ordered α-helix structures. Altogether, this enhances the reduction of Cu2+ by the protein and leads to faster cluster formation.56 On the other hand, while using hydrazine hydrate, with decreasing the pH, the α-helix can be transformed to δ-sheets and random coil structures, thus increasing the availability of functional groups (–OH, –NH and –COOH) that can interact with the cluster.45 Other proteins such as trypsin,39 human serum albumin (HSA),50 transferrin,53 papain,54 cellulase55 and lysozyme42,44 were also reported as effective capping agents for the preparation of fluorescent copper nanoclusters.
DNA: DNA as a naturally occurring biostructure with variable lengths and sequences has been used for the synthesis of CuNCs (Fig. 4A).63–77 Double-stranded DNA (dsDNA) with adjustable sequences and lengths has been considered as a template for the synthesis of fluorescent copper nanoparticles/nanoclusters.78–82 Moreover, it has been shown that at lower concentrations of metal precursor Cu2+ first binds to the backbone phosphate groups of DNA via a nonspecific interaction. However, with increasing the concentration, the metallization of high-affinity bases is favoured. Consequently, the reduction of the pre-formed complex with ascorbic acid leads to the formation of fluorescent CuNCs.71 The order of mixing reagents was found to be crucial for the synthesis. In general, the reaction involves the mixing of copper salt with the ligand/template in the first step followed by reduction using the reducing agent. However, in the case of dsDNA, the yield is much higher when the first copper salt is reduced with ascorbic acid and then templated inside of DNA.64 The oxygen atoms (as hard Lewis bases) of phosphodiester groups and nitrogen atoms (relatively strong Lewis bases) of the nucleobases bind to Cu2+ and therefore, the stabilization of a metal cation inhibits the metallization of the template. However, when the salt is first reduced to Cu+, it can undergo disproportionation and the formed Cu0 can occupy and cluster inside the grooves of dsDNA. Consequently, the size of the nanoparticle/nanocluster can be tuned by changing the number of DNA base pairs, i.e., much longer DNA templates resulted in the formation of nanoparticles rather than clusters. However, shorter DNA templates led to the formation of CuNCs with very low fluorescence intensity. In contrast, the PL properties can be improved by increasing the length of DNA strands to some extent.72,75 Initially it was shown that the mismatches in nucleotides in duplex do not lead to nanocluster formation, whereas complete matches resulted in the formation of fluorescent nanoclusters.64 However, it was found that DNA sequences with certain mutations can successfully template CuNCs and can be used as labels for mutation detection71 (Fig. 4B–F). Furthermore, even the deletion of entire coding regions (exons) in some genes was observed.77 Qing et al.83 and Song et al.84 demonstrated the sequence-dependent formation of CuNCs and the changes in PL properties by varying the sequence of the dsDNA template. The reader is referred to recent reviews by Yi Lv and co-workers85 and Kevin C.-W. Wu and co-workers86 on DNA-templated CuNCs and applications in label-free bioassaying.
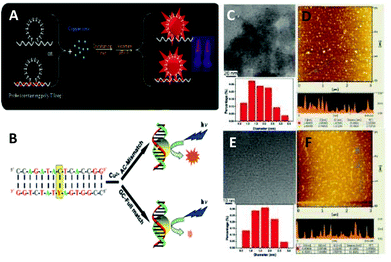 | ||
| Fig. 4 (A) Synthesis of the DNA-templated CuNCs. Reprinted with permission from ref. 67. Copyright 2017 Elsevier. (B) Schematic representation of detection strategy (Y: Single nucleotide polymorphism (SNP) site). (C) Representative TEM image and size distribution and (D) representative AFM image of Cu nanoclusters using the FULL (sequence: 5′-CCA GAT ACT CAC CGG-3′/3′-GGT CTA TGA GTG GCC-5′) duplexes as the synthetic scaffold. (E) Representative TEM image and the size distribution and (F) representative AFM image of Cu nanoclusters using the AC-mismatched (AC-MIS) duplexes (sequence: 5′-CCA GAT ACT CAC CGG-3′/3′-GGT CTA TAA GTG GCC-5′) as the synthetic scaffold. Reprinted with permission from ref. 71. Copyright 2012 American Chemical Society. | ||
The absence of grooves and base pairs in single-stranded DNA (ssDNA) excluded the formation of stable CuNCs; however, the extensive research in the field showed that highly thymine-dependent sequences of ssDNA can lead to the formation of CuNCs and the red-emissive PL properties can be improved by increasing the number of repeating thymine bases in the sequence.73,87–89 Moreover, specific selections of single-stranded DNA (ssDNA), including random ssDNA, poly(adenine) (polyA), poly(thymine) (polyT), poly(cytosine) (polyC) and poly(guanine) (polyG) have been used for the synthesis of fluorescent copper nanoparticles (CuNPs).90
2.1.2.2. Electrochemical synthesis. This method is based on the electrochemical reduction of copper salt and the deposition on the cathode in a typical electrochemical cell. The size, structure, and morphologies of the nanoparticles can be controlled by changing the applied currents91–93 and using different templates within electrolyte solutions.92 For instance, copper nanorods with mean diameter and length of 30 nm and 400 nm, respectively, can be synthesized in a two-electrode cell with a copper plate as the anode and a platinum cathode using a CTAB/CT4ABr/acetone/cyclohexane/water five-component system (where cyclohexane serves as a stabilizer).92
The synthesis of copper nanoclusters using the electrochemical method was first reported in 2010.94 The synthesis of very small CuNCs (Cun with n ≤ 14) in tetrabutylammonium nitrate (TBAN) as an electrolyte and protecting agent against aggregation and oxidation was done in a three-electrode conventional electrochemical cell, with a copper plate as the anode, a platinum sheet as the cathode and Ag/AgCl as the reference electrode. Cu ions produced from the soluble copper anode were reduced and spontaneously formed clusters. Recently, Vilar-Vidal et al. reported a selective formation of Cu13 clusters in a thermostated three-electrode electrochemical cell.95 In 2016 M. Arturo López-Quintela and co-workers reported the electrochemical synthesis of bare Cu5 clusters that showed excellent stability against UV irradiation, heating and pH variations.96 In a similar study Avelino Corma and co-workers showed that the reactivity of electrochemically prepared Cu20, Cu8 and Cu5 clusters towards O2, and thus the oxidation of the clusters, decreased with decreasing the size of the bare clusters.97 Unlike the Cu5 cluster, bigger clusters are easily oxidized and relatively less reducible in oxygen and nitrogen environments, respectively. These studies showed that very small clusters can be prepared using electrochemical synthesis methods.
2.1.2.3. Water-in-oil (w/o) microemulsion. In 1993 Lisiecki et al. reported the first synthesis of CuNPs in w/o microemulsion using NaBH4 or hydrazine as reducing agents.98 The effect of the surfactant type and content inside the microemulsion,98,99 and the effect of the amount of reducing agent100,101 on the size evolution of CuNCs was studied in terms of the α parameter, which is defined by the number of reducing agent moles used for the reaction with respect to the stoichiometric amount needed for the reduction of copper cations (Fig. 5).101
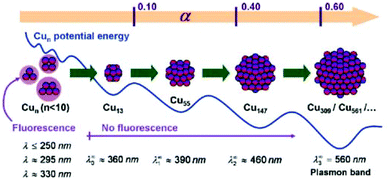 | ||
| Fig. 5 Schematic representation of copper nanocluster size evolution with increasing the molar percentage (α) of NaBH4. The clusters with n ≤ 13 are fluorescent, whereas big particles with n ≥ 309 show characteristic plasmon resonance band. Reprinted with permission from ref. 101. Copyright 2009 American Chemical Society. | ||
It was shown that with an increase in the NaBH4 concentration, bigger clusters and particles were obtained, whereas small and strongly fluorescent Cun clusters with n ≤ 13 atoms, were formed at lower α values.
2.1.2.4. Microwave-assisted synthesis. The microwave-assisted method is based on the use of microwaves as an energy source for the synthesis of copper nanomaterials.102–104 The first preparation of CuNPs with an average size of 10 nm in ethylene glycol and NaH2PO2·H2O as a reducing agent under microwave irradiation, was described in 2004 by H.-t. Zhu et al.102 A few years later, fluorescent CuNCs with an average size of 2 nm (containing Cu9 species) were successfully prepared when NaOH was introduced into the reaction mixture upon heating at high temperatures.103 Under harsh heating conditions, ethylene glycol undergoes dehydration and results in the formation of the aldehyde, which can reduce copper ions at high temperatures. It undergoes spontaneous ethoxylation and forms poly(ethylene)glycol (PEG), which later adsorbs on the surface of CuNCs. The surface passivation restricts the further agglomeration and oxidation of formed clusters. Recently, tannic acid104 and BSA62 were reported as both reducing agents and protective ligands for the preparation of clusters under MW radiation.
2.1.2.5. Modified Brust–Schiffrin technique. The Brust–Schiffrin method is a largely applied strategy to prepare atomically precise metal nanoclusters in large quantities and narrow size distributions.105,106 The method relies on the simultaneous growth of metal nanoclusters and the protection of their core with a thiol monolayer. The details of extensive mechanistic studies of the reduction process of the metal solution, in this case AuNCs containing protective ligand, as well as the structure and composition of intermediate species, can be found in the ref. 107 and 108. Similar to gold, the [TOA][CuBr2] complex was formed as an intermediate in the synthesis of copper nanoclusters (eqn (2) and (3)), the reduction of which in the presence of 2-mercapto-5-n-propylpyrimidine (MMP) resulted in the formation of CuNCs:14,107,109
| [TOA]2[CuBr4] + RSH → ½RSSR + [TOA][CuBr2] + TOABr + HBr | (2) |
| [TOA][CuBr2] + BH4− + RSH + RSSR → Cux(SR)y + … | (3) |
Li et al. showed that the choice of reducing agent is of great importance.110 Cu0 species are highly susceptible to oxidation in aqueous solution and thus NaBH4 as a strong reducing agent (E0 = −1.24 V) decomposes rapidly and consequently does not inhibit the oxidation of the metal. On the other hand, hydrazine (N2H4) being a weaker reducing agent than NaBH4 decomposes slowly but does not significantly improve the stability of the clusters. However, the authors showed that while using tetrakis(hydroxymethyl)phosphonium chloride (THPC) as a reducing agent, not only did the stability of the dihydrolipoic acid (DHLA)-stabilized cluster increase due to additional interaction with the copper core, but also the PL can be tuned from red110 to bright orange emission111 depending on the applied reaction conditions. Other ligands such as (3-mercaptopropyl)trimethoxysilane,112D-penicillamine,113–115 cysteine,116–123 mercaptobenzoic acid,124,125 peptides (in particular glutathione (GSH))17,18,126–137 and small organic molecules138–142 were successfully applied as capping agents for the synthesis of luminescent clusters in a typical Brust–Schiffrin method (Fig. 6).
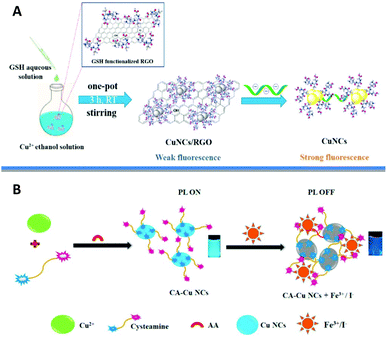 | ||
| Fig. 6 (A) Schematic illustration of the CuNCs/reduced graphene oxide (RGO) nanocomposite sensing platform for Hep sensing. Reprinted with permission from ref. 126. Copyright 2020 Elsevier. (B) Schematic representation of cysteamine (CA)-CuNCs synthesis and its sensor responses to Fe3+ and I− ions. Reprinted with permission from ref. 138. Copyright 2020 Elsevier. | ||
An alternative approach to the Brust–Shiffrin method has been proposed for the synthesis of CuNCs using hydrophilic ligands other than thiols such as tannic acid,143–145 bile acid derivatives,146 histidine,147 dopamine148 and proline.149
2.2. Synthesis of organic-phase soluble clusters
For the synthesis of atomically precise copper clusters, researchers followed similar procedures to those for Au clusters, which are well documented. However, considering the high susceptibility of copper towards oxidation, the reaction requires an inert atmosphere in most cases by employing Schlenk-line techniques connected to vacuum and inert gas lines. Moreover, the choice of the protecting ligand as well as its chemical structure and functionalities, the precursor of chalcogens plays a crucial role in the formation of certain cluster sizes. Although most of the reported structures had copper atoms in the +1 oxidation state or even in a mixed-valence state due to the presence of non-stoichiometric phases and electron deficiency, recently, a few “metallic” copper clusters have been reported (to be discussed later). A recent review by Didier Astruc summarized the synthesis of atomically precise CuNCs, the structures and their applications in galvanic reactions, hydrogen evolution, etc.150Table 1 summarizes all the reported atomically precise CuNCs published starting from 2010.| Formula | Ligand | Crystal structure | Ref. | ||
|---|---|---|---|---|---|
| Core structure | Space group | ||||
| Geometry | Composition | ||||
| Cu6(SC7H4NO)6 | C7H5NOS | Octahedron | Cu6 |
R![[3 with combining macron]](https://www.rsc.org/images/entities/char_0033_0304.gif) c c |
151 |
| [Cu6S6(SnPh)2(PPh2Et)6] | PhSnCl3/PPh2Et | Octahedron | Cu6E6 (E = S, Se, Te) |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
152 |
| [Cu6Se6(SnPh)2(PPh2Et)6] | P21/n | ||||
| [Cu6Te6(SnPh)2(PPh2Et)6] |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
||||
| [Cu7(p-S–C6H4–NMe2)7(PPh3)4] | p-Me3SiS-C6H4-NMe2/PPh3 | Trigonal prism | Cu7 | R3c | 153 |
| [Cu7(p-S–C6H4–OSiMe3)(SPh)6(PPh3)4] | p-Me3SiO-C6H4-SSiMe3/PhSH/PPh3 | ||||
| [Cu7(H){S2P(OEt)2}6] | [NH4][S2P(OEt)2] | Tetrahedron | H-Cu7 |
R![[3 with combining macron]](https://www.rsc.org/images/entities/char_0033_0304.gif) |
154 |
| [Cu7(Br){Se2P(OiPr)2}6] | [NH4][Se2P(OiPr)2] | Br-Cu7 | |||
| [Cu7H(S2COiPr)6] | KS2COiPr | Triangular pyramid | H-Cu7 |
R![[3 with combining macron]](https://www.rsc.org/images/entities/char_0033_0304.gif) |
155 |
| [Cu7H(S2COnPr)6] | KS2COnPr | ||||
| [Cu7(H){S2CR}6] 1a–c | a: R = NnPr2 | Tetrahedron | Cu7 | 2a: P![[3 with combining macron]](https://www.rsc.org/images/entities/char_0033_0304.gif) 1c 1c |
156 |
| [Cu8(H){S2CR}6](PF6) 2a–c | b: R = NEt2 | Cu8 | 2b: Pn![[3 with combining macron]](https://www.rsc.org/images/entities/char_0033_0304.gif) n n |
||
| c: R = aza-15-crown-5 | 1c: P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
||||
| [Cu8(μ-H)6(μ-dppm)5](PF6)2 | dppm = Ph2PCH2PPh2 | Octahedron | Cu6 | Pna21 | 157 |
| [Cu8S4(μ3-E)4(CAACCy)4] | CAACCy·HCl | “Butterfly”-like | Cu8E4 (E = S, Se) | C2/c | 158 |
| [Cu8Se4(μ3-E)4(CAACCy)4] | C2/c | ||||
| [Cu8(SPh)8(Ph3P)4] | PhSH | Cage | 2 × Cu4S4P2 |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
159 |
| [Cu13(SePh)13(Ph3P)4] | PhSeH | Hexagonal pyramid | Cu4Se3 + Cu9Se7 | ||
| [(CuPPh3)4(PhSn)18Cu10S31Cl2] | (PhSn)4S6 | Propellane-like Cu4S4/[Sn3S3] + [Sn2CuS3] six-membered rings | [Sn18Cu10S31] |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
160 |
| [Cu11(TBBT)9(PPh3)6](SbF6)2 | TBBT = 4-tert-butylbenzenethiol | Triangular bipyramid | Cu5 | P21/n | 161 |
| [Cu11(μ9-I)(μ3-I)3{Se2P(OiPr)2}6](OH) | NH4[Se2P(OiPr)2] | Trigonal prism | Cu11 | P21/n | 162 |
| [Cu11(μ9-I)(μ3-I)3(Se2PPh2)6](PF6) | NH4(Se2PPh2) | Pnma | |||
[Cu11H2{S2P(OiPr)2}6(C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CPh)3] CPh)3] |
HC![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CPh CPh |
Trigonal prism | Cu11H2 |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
163 |
[Cu11H2{S2P(OiPr)2}6(C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CC6H4F)3] CC6H4F)3] |
HC![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CC6H4F CC6H4F |
P21/n | |||
[Cu11H2{S2P(OiPr)2}6(C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CC6H4OMe)3] CC6H4OMe)3] |
HC![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CC6H4OMe CC6H4OMe |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
|||
| [Cu12S6(dpppt)4] | dpppt = Ph2P(CH2)5PPh2 | Octahedron | Cu12S6P8 | P42/ncm | 164 |
| [Cu12S6(dppo)4] | dppo = Ph2P(CH2)8PPh2 | Cu12S6 |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
||
| [Cu12S6(dpppt)4] | dpppt = Ph2P(CH2)5PPh2 | Octahedron | Cu12S6P8 |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
165 |
| [Cu12Se6(dppo)4] | dppo = Ph2P(CH2)8PPh2 | Octahedron | Cu12Se6P8 |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
165 |
| [Cu12S6(dppf)4] | dppf = Ph2PCpFeCpPPh2 | Octahedron | Cu12S6 |
P![[4 with combining macron]](https://www.rsc.org/images/entities/char_0034_0304.gif) 21c 21c |
166 |
| [Cu12S6(PPh2Et)8] | PPh2Et | Octahedron | Cu12S6P8 |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
167 |
| [Cu12S6(PEt3)8] | PEt3 | Octahedron | Cu12S6P8 |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
165 and 167 |
| [Cu24S12(PEt2Ph)12] | PEt2Ph | Octahedron | Dimer of Cu12S6 |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
165 |
| [Cu20S10(PPh3)8] | PPh3 | Tetragonal prism | Cu20S10 | Pbca | 167 |
| [Cu20S10(PtBu3)8] | PtBu3 | Tetragonal antiprisms | Cu20S10 | P4/nnc | 165 |
[Cu13(S2CNnBu2)6(C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CR)4](PF6) CR)4](PF6) |
R = CO2Me | Cuboctahedron | [Cu13]11+ | C2/c | 168 |
| R = 3-FC6H4 | P21/c | ||||
[Cu13(S2CNR2)6(C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CR′)4](PF6) 1a–d CR′)4](PF6) 1a–d |
a: R = nBu, R′ = CO2Me | Cuboctahedron | Cu13 | 1a: C2/c | 168 |
[Cu12(μ12-S){S2CNR2}6{C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CR′}4] 2a–c CR′}4] 2a–c |
b: R = nBu, R′ = CO2Et | S-Cu12 | 2ab: P212121 | ||
[Cu12(μ12-Cl){S2CNR2}6{C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CR′}4](PF6) 3a–e CR′}4](PF6) 3a–e |
c: R = iPr, R′ = CO2Et | Cl-Cu12 | 3d: Pnna | ||
[Cu12(μ12-Br){S2CNnBu2}6{C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CPh}4](PF6) 4e CPh}4](PF6) 4e |
d: R = nPr, R′ = 3,5-(CF3)2C6H3 | Br-Cu12 | 3e: P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
169 | |
[Cu12(μ12-Cl)(μ3-Cl){S2CNnBu2}6{C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CCO2Me}3]+ 5a CCO2Me}3]+ 5a |
e: R = nBu, R′ = Ph | Cl-Cu12 | 4e: P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
||
| 5a: C2/c | |||||
| [Cu12(SR′)6Cl2][(Cu(R′SH))6] | R′ = nBu | Cuboctahedron | [Cu12(SR′)6]6+ |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
170 |
| [Cu17(SR′′)6Cl13(THF)2(R′′SH)3][H(THF)2]2 | R′′ = CH2CH2Ph | [Cu12(SR′′)6]6+ | |||
| [Cu14H12(phen)6(PPh3)4][Cl]2 | Phen = 1,10-phenanthroline | Tetrahedron | [Cu4]4+ | P212121 | 171 |
| Cu14(C2B10H10S2)6(CH3CN)8 | 1,2-Dithiol-o-carborane | fcc | Cu14 | Fmm | 172 |
| [Cu14(R/S-DPM)8](PF6)6 | DPM = (R/S)-2-diphenyl-2-hydroxylmethylpyrrolidine-1-propyne | Cubic | Cu14 | P212121 | 173 |
| Cu16[SC(O)OC6H11]16·2C3H6O | K[SC(O)OC6H11] | “Butterfly”-like | Cu16 |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
174 |
| Cu16[SC(O)OC6H11]16·C5H8N2 |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
||||
| Cu16[SC(O)OC6H11]16·C11H9N | P2/n | ||||
| Cu16[SC(O)OC6H11]16·C10H8N2 |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
||||
| [Cu16{SC(O)OC6H11}16·C10H8N2]2 |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
||||
| [Cu18H7R10I] | R = S(C6H4)PPh2 | Tetrahedron | Cu8 |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
175 |
| [Cu19S28(SnPh)12(PEt2Ph)3][Li(THF)4] | PhSnCl3/PEt2Ph | Trigonal prism | S-Cu6 | P21/n | 176 |
| [Cu19S28(SnPh)12(PEt2Ph)3](NBu4) |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
||||
| [Cu20(CCPh)12(OAc)6] | HCCPh/Ph2SiH2 | Tetrahedron | [Cu4]2+ |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
177 |
| [Cu20H11(S2P(OiPr)2)9] | NH4[S2P(OiPr)2] | Triangular orthobicupola | Cu20 |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
178 |
| [Cu20H11(Se2P(OiPr)2)9] | NH4[Se2P(OiPr)2] | Cuboctahedron | Cu13 |
R![[3 with combining macron]](https://www.rsc.org/images/entities/char_0033_0304.gif) |
179 |
| [Cu20H11{Se2P(OiBu)2}9] | NH4[Se2P(OiBu)2] | Cuboctahedron | Cu13 |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
180 |
| [Cu20H11{S2P(CH2CH2Ph)2}9] | NH4[S2P(C2H4Ph)2] | Triangular orthobicupola | Cu20 | P21/n | |
| [Cu20H11{S2P(OnPr)2}9] | NH4[S2P(OnPr)2] | Triangular orthobicupola | Cu20 | P21/n | |
| [Cu20H11{S2P(O2Bu)2}9] | NH4[Se2P(O2Bu)2] | Triangular orthobicupola | Cu20 | C2/c | 181 |
| [Cu32H20{S2P(O2Bu)2}12] | Rhombohedra | Cu14 |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
||
| [Cu25H10(SPhCl2)18][PPh4]3 | HSPhCl2 | Cuboctahedron | Cu@Cu12 |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
182 |
| [Cu25H22(PPh3)12]Cl | PPh3/Ph2SiH2 | Icosahedron | [Cu13]11+ | Pbca | 183 |
| [Cu18H17(PPh3)10]Cl | Square antiprism | [Cu8H]7+ | P21/c | ||
| [Cu28H15(S2CNR)12]PF6 | NR = NnPr2 | Rhombicuboctahedron | Cu24 |
Pa![[3 with combining macron]](https://www.rsc.org/images/entities/char_0033_0304.gif) |
184 |
| NR = aza-15-crown-5 |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
||||
| [Cu29Cl4H22(Ph2phen)12]Cl | Ph2phen | Icosahedron | Cu13 | P31c | 185 |
| [Cu30H18 {S2P(OnPr)2}12] | NH4[S2P(OnPr)2] | Hollow icosahedron | Cu12 |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
186 |
| [Cu30H18{Se2P(OiPr)2}12] | NH4[Se2P(OiPr)2] | Hollow icosahedron | Cu12 |
R![[3 with combining macron]](https://www.rsc.org/images/entities/char_0033_0304.gif) |
|
| [Cu30H18{Se2P(OiBu)2}12] | NH4[Se2P(OiBu)2] | Hollow icosahedron | Cu12 |
R![[3 with combining macron]](https://www.rsc.org/images/entities/char_0033_0304.gif) |
|
[PdCu14H2{S2P(OnPr)2}6(C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CPh)6] CPh)6] |
HC![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CPh/Pd(PPh3)2Cl2 CPh/Pd(PPh3)2Cl2 |
Icosahedron | PdH2@Cu14 | P21/n | |
[PdCu14H2{S2P(OnPr)2}6(C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CC6H4F)6] CC6H4F)6] |
HC![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CC6H4F/Pd(PPh3)2Cl2 CC6H4F/Pd(PPh3)2Cl2 |
Icosahedron | PdH2@Cu14 |
R![[3 with combining macron]](https://www.rsc.org/images/entities/char_0033_0304.gif) |
|
| [Cu32(H)20{S2P(OiPr)2}12] | NH4[S2P(OiPr)2] | Rhombohedra | Cu14 |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
187 and 188 |
| [Cu32(PET)24H8Cl2](PPh4)2 | PET = 2-phenylethanethiol | Bisquare antiprism | Cu14H8 |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
189 |
[Cu33(tBuC![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C)24(Mo4O16)]·BF4 C)24(Mo4O16)]·BF4 |
t
BuC![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CH CH |
Octahedron | Core/shell | P21/c | 190 |
[Cu62(tBuC![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C)34(Mo5O19)2(MoO4)2(OTf)2(OH)4]·(OTf)2 C)34(Mo5O19)2(MoO4)2(OTf)2(OH)4]·(OTf)2 |
Cubane/octahedron | Core/shell | P21/n | ||
| [Cu40Se16(S-C6H4-CN)8(dppm)8] | NC-C6H4-SH/dppm | “Disk”-like | A-B | P21/n | 191 |
| dppm = Ph2PCH2PPh2 | |||||
| [Cu43Al12](Cp*)12 | (Cp* = η5-C5Me5 | Icosahedron | Cu13 |
Im![[3 with combining macron]](https://www.rsc.org/images/entities/char_0033_0304.gif) |
192 |
| [Cu52S12(SCH2C6H4tBu)28(PPh3)8] | HSCH2C6H4tBu/PPh3 | fcc | (Cu2S)12 | C2/c | 193 |
[Cu53(RCOO)10(C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CtBu)20Cl2H18]+ CtBu)20Cl2H18]+ |
HC![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CtBu CtBu |
Icosahedral/dodecahedral | Core/shell | Pbcn | 194 |
| Cu74S15(2-PET)45 | 2-PET = 2-phenylethanethiol | Layered | A-B-A |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
195 |
| [Cu81(PhS)46(tBuNH2)10(H)32]3+ | PhSH/tBuNH2·BH3 | Planar | Cu17 | C2/c | 196 |
| [Cu93Se42(SeR)9(PPh3)18] | R = C6H4SMe | Layered | A-B-A |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
197 |
| [Cu96Se45(SeR)6(PPh3)18] | R = C6H4SMe | A-B-A |
R![[3 with combining macron]](https://www.rsc.org/images/entities/char_0033_0304.gif) c c |
||
| [Cu136S56(SR)24(dpppt)10] | R = CH2C4H3O | A-B-C | C2/c | ||
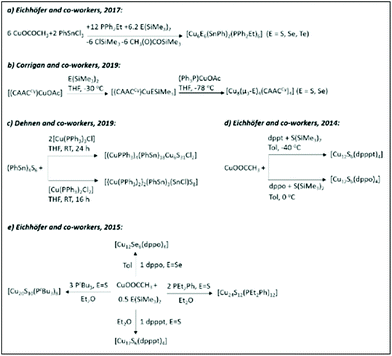 | ||
| Scheme 1 The syntheses of chalcogen-bridged CuNCs as described in ref. 152 (Reprinted with permission, Copyright 2017 American Chemical Society), ref. 158 (Reprinted with permission, Copyright 2019 American Chemical Society), ref. 160 (Adapted with permission, Copyright 2019 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim), ref. 164 and 165 (Reprinted with permission, Copyright 2015 American Chemical Society). | ||
The reaction conditions and initial concentrations of starting materials greatly influence the size distribution, composition, and crystallographic structures of the obtained clusters.
2.2.1.1. Sulphur-bridged CuNCs. The pioneering work of Prof. Fenske in the field of semiconducting chalcogenide-bridged and ligand-shielded copper nanoclusters illustrated that the cluster size influences the structure. For the majority of the reported small phosphine-protected copper clusters, a [Cu2nSn] core can be distinguished. A grand library of reported structures following the general core composition can be found in various reviews reported during the last few decades.1,2 However, this review covers the structures of CuNCs published since 2010.
John F. Corrigan and co-workers reported the synthesis (Scheme 1b) and structures of a series of small copper(I)–chalcogenide clusters [Cu4M4(μ3-E)4(CAACCy)4] (M = Cu, Au, Ag; E = S, Se) stabilized by cyclic (alkyl)(amino)carbene ligands.158 Regardless of the heteroatom or the chalcogen present in the cluster, they were isostructural and isomorphous, crystalizing in the monoclinic C2/c space group. The copper atoms in the Cu4 core have a nearly planar arrangement and form a square. Furthermore, the bridging of chalcogenide ligands with Cu4 core atoms results in the formation of an eight-membered ring resembling a “hinged” or “butterfly” geometry. Andreas Eichhöfer and co-workers synthesized [Cu12S6] clusters protected with bidentate phosphine ligands (Scheme 1d): bis(diphenylphosphino)pentane (dpppt = Ph2P(CH2)5PPh2) or bis(diphenylphosphino)octane (dppo = Ph2P(CH2)8PPh2).164
In general, the synthetic procedure involves the reaction of copper acetate with S(SiMe3)2 in toluene in the presence of bidentate phosphine ligands. The [Cu12S6(dpppt)4] cluster consists of a Cu12S6P8 core and crystalizes in the tetragonal space group P42/ncm. The structure can be viewed as an octahedron of nonbonding sulphur atoms, where the twelve edges are occupied by copper atoms. The bidentate phosphine ligands bridge copper atoms at the upper and lower square planar faces (Fig. 7A). A similar structure has been observed for the [Cu12S6(dppo)4] cluster. The only difference is that bidentate dppo ligands bridge one copper atom of the upper and one of the lower square planar face, giving rise to a helical arrangement around the Cu12S6 cluster core (Fig. 7B). Interestingly, both clusters in the solid state show bright red emission (from triplet states, phosphorescence) with quantum yields as high as 48% and 67% for dpppt and dppo protected clusters, respectively (Fig. 7C).
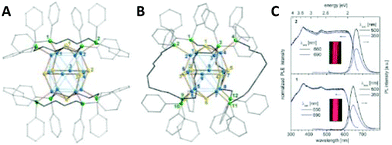 | ||
| Fig. 7 Molecular structure of (A) [Cu12S6(dpppt)4] and (B) [Cu12S6(dppo)4] clusters. Colour codes of the elements: Cu: blue, S: yellow, P: green and C: grey. H atoms are omitted for clarity. (C) Room temperature photoluminescence excitation (PLE) and emission (PL) spectra of a suspension of freshly prepared microcrystals of [Cu12S6(dpppt)4] (down) and [Cu12S6(dppo)4] (up) in toluene, measured in the integrating sphere. The inset shows the colours of the suspensions of microcrystal sunder white LED light. Republished with permission from ref. 164. Copyright 2014 Royal Society of Chemistry. | ||
The same group extended the synthesis of a series of copper chalcogenide clusters with varying core sizes and ligand functionalities (Scheme 1e and Fig. 8).165 The list included structures such as [Cu12S6(dpppt)4], [Cu12Se6(dppo)4], [Cu12S6(dppf)4],166 [Cu12S6(PPh2Et)8],167 [Cu12S6(PEt3)8],167 [Cu24S12(PEt2Ph)12], [Cu20S10(PPh3)8]167 and [Cu20S10(PtBu3)8], where dppf = Ph2PCpFeCpPPh2. Note that some of the structures from the list in ref. 165 were reported earlier. The first six compounds on the list share a similar Cu12E6 (E = S, Se) core composition (building block) and tetragonal antiprism core geometries. The [Cu24S12(PEt2Ph)12] cluster can be formally represented as a dimer of Cu12S6, whereas [Cu20S10(PtBu3)8] has a core with the composition of Cu20S10. The overall structure has an oblate shape. For the same composition with the PPh3 ligand, the cluster adopts a prolate shape. Thus, [Cu20S10(PPh3)8] and [Cu20S10(PtBu3)8] clusters can be considered as structural isomers. The crystallographic analyses of Cu–S (2.13–2.49 Å), Cu⋯Cu (2.55–3.14 Å) and S⋯S (3.64–4.62 Å) bond lengths and distances are similar in all sulphur-bridged clusters, whereas an increase was observed in the case of the selenide-bridged [Cu12Se6(dppo)4] cluster.
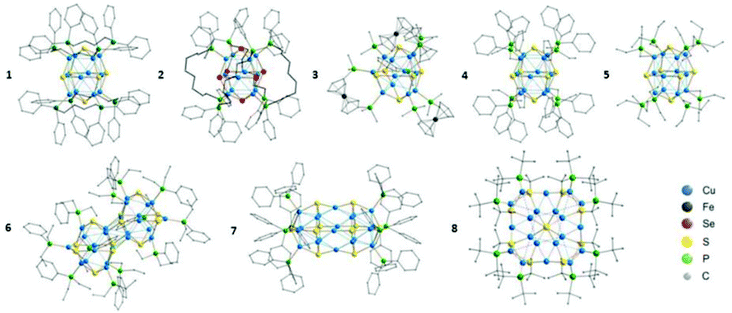 | ||
| Fig. 8 Molecular structures of [Cu12S6(dpppt)4] (1), [Cu12Se6(dppo)4] (2), [Cu12S6(dppf)4] (3), [Cu12S6(PPh2Et)8] (4), [Cu12S6(PEt3)8] (5), [Cu24S12(PEt2Ph)12] (6), [Cu20S10(PPh3)8] (7) and [Cu20S10(PtBu3)8] (8). H atoms are omitted for clarity. Reprinted with permission from ref. 165. Copyright 2015 American Chemical Society. | ||
Several groups reported the formation of non-stoichiometric clusters that display small variations in the total Cu/S ratio. The formation of such mixed valence states of the metal in the cluster has also been observed previously for selenide-bridged copper and silver nanoclusters.198,199 Andreas Eichhöfer's group reported the synthesis of ternary copper–tin–chalcogenide clusters [Cu6E6(SnPh)2(PPh2Et)6] (E = S, Se, Te, Scheme 1a)152 and [Cu19S28(SnPh)12(PEt2Ph)3]X (X = [Li(THF)4] or [NBu4], Scheme 2b)176 through the reactions of CuO(O)CCH3 or CuCl and PhSnCl3 with E(SiMe3)2 in the presence of an excess PPh2Et. In the report,152 the Cu![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chalcogenide ratio in a series is 1
chalcogenide ratio in a series is 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 regardless of the chalcogenide type. Herein, three clusters share a similar cage construction of an E6 octahedron, where two trigonal faces are capped by PhSn units. The remaining six R3PCu units bridge the free edges in μ2 and μ3 fashion for sulphide- and selenide/telluride-bridged CuNCs, respectively. In the case of [Cu19S28(SnPh)12(PEt2Ph)3]X clusters, regardless of the counter ion, the structure is composed of an inner core and an outer cluster sphere.176 The inner core is constructed from a sulphur atom coordinated with six copper atoms in a distorted trigonal prism fashion. The rest of the copper and sulphur atoms together with the remaining ligands form the outer sphere.
1 regardless of the chalcogenide type. Herein, three clusters share a similar cage construction of an E6 octahedron, where two trigonal faces are capped by PhSn units. The remaining six R3PCu units bridge the free edges in μ2 and μ3 fashion for sulphide- and selenide/telluride-bridged CuNCs, respectively. In the case of [Cu19S28(SnPh)12(PEt2Ph)3]X clusters, regardless of the counter ion, the structure is composed of an inner core and an outer cluster sphere.176 The inner core is constructed from a sulphur atom coordinated with six copper atoms in a distorted trigonal prism fashion. The rest of the copper and sulphur atoms together with the remaining ligands form the outer sphere.
 | ||
| Scheme 2 The syntheses of chalcogen-bridged CuNCs as described in ref. 166 (Reprinted with permission, Copyright 2013 American Chemical Society), ref. 176 (Republished with permission, Copyright 2012 Royal Society of Chemistry), ref. 193 (Republished with permission, Copyright 2016 Royal Society of Chemistry), ref. 197 (Adapted with permission, Copyright 2010 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim). | ||
Another example of such a phenomenon in the ternary systems with tin has been reported by the group of Stefanie Dehnen by the reaction of the binary organotin sulphide cluster [(PhSn)4S6] with [Cu(PPh3)3Cl] (Scheme 1c).160 The resulting [(CuPPh3)4(PhSn)18Cu6S31] cluster is a pseudo dimer having a [Sn18Cu10S31] core, which is separated in two identical parts connected by a single sulphur atom (Fig. 9A).
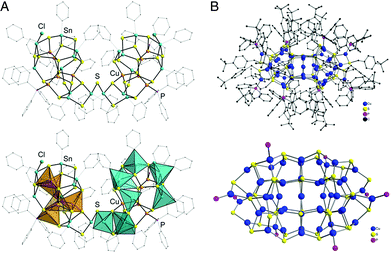 | ||
| Fig. 9 (A) Molecular structure of the [(CuPPh3)4(PhSn)18Cu6S31] cluster in ball-stick representation (top) and with coordination polyhedra (bottom) around the Sn (blue) and Cu (orange) atoms. Organic groups are shown as transparent wires. H atoms and two disordered organic substituents are omitted for clarity. Adapted with permission from ref. 160. Copyright 2019 Wiley-VCH Verlag GmbH &Co. KGaA, Weinheim. (B) Top: Molecular structure of [Cu52S12(SCH2C6H4tBu)28(PPh3)8] in the solid state. Bottom: Structure of the inorganic cluster core. Republished with permission from ref. 193. Copyright 2016 Royal Society of Chemistry. | ||
Several larger CuNCs also fall into this category of clusters with non-stoichiometric composition. Examples involve the [Cu52S12(SCH2C6H4tBu)28(PPh3)8]193 (Scheme 2c) and [Cu136S56(SCH2C4H3O)24(dpppt)10] clusters197 (Scheme 2d) by the Fenske group. The former cluster consists of a (Cu2S)12 core surrounded by eight PPh3 ligands and 28 [CuSCH2C6H4tBu] subunits, where all the copper atoms are oxidized (+1) (Fig. 9B). [Cu136S56(SCH2C4H3O)24(dpppt)10] is so far the largest reported sulphide-bridged cluster. Being at the borderline between molecular and bulk compounds, this cluster has a layered structure with a cubic closest packing (fcc; stacking A-B-C).197 In this case, some of the copper positions are partially occupied resulting in copper content deficiency. Recently, our group reported the structure of a Cu74S15(2-PET)45 (2-PET = 2-phenylethanethiol) cluster with similar Cu deficiencies.195 The reduction of a copper salt with 2-PET ligand was completed with the addition of triethylamine (TEA) as a base. The crystallization of the sample from the saturated solution resulted in the formation of dark reddish-orange crystals. The single-crystal analysis revealed that the cluster had a rod-like structure ((Fig. 10A) spliced into seven sulphur atom layers, organized in an A-B-A fashion (Fig. 10B).
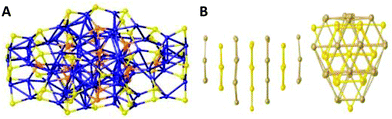 | ||
| Fig. 10 (A) The core structure of the Cu74S15(2-PET)45 cluster. Bridged-sulphur atoms are in orange, and sulphur atoms belonging to ligands are in yellow. The layers are labelled as L1 to L7 from left to right. (B) Side and top views of the layer organization of the sulphur atoms. Two types of layers (A in yellow, B in gold) organize themselves in an A-B-A pattern. Reprinted with permission from ref. 195. Copyright 2020 American Chemical Society. | ||
2.2.1.2. Selenium/tellurium-bridged CuNCs. Similar to what has been discussed in the previous section, selenium- and tellurium-bridged CuNCs with and without stoichiometric compositions have been reported in the literature (Table 1 and Scheme 1). For instance, all three types of clusters in the series [Cu6E6(SnPh)2(PPh2Et)6] (E = S, Se, Te) share the same E6 octahedron and have almost identical ligand binding motifs.152 On the same note, [Cu4M4(μ3-E)4(CAACCy)4] (M = Cu, Ag, Au; E = S, Se) clusters reported by John F. Corrigan and co-workers, have the same “butterfly”-geometries regardless of the heteroatom doping and bridging chalcogen.158 [Cu12Se6(dppo)4] cluster165 is isostructural to its sulphide-bridged analog,164 yet a slight variation in bond lengths was observed. The same group also reported Te-bridged [Cu4(μ4-Te)(μ4-η2-Te2)(μ-dppf)2] and [Cu12(μ5-Te)(μ8-η2-Te2)(μ-dppf)4] clusters containing telluride and ditelluride ligands in their CuTe architectures (Scheme 2a).166 Large Se-bridged CuNCs have been reported back in 2010 by the group of Prof. Fenske: [Cu93Se42(Se-C6H4-SMe)9(PPh3)18] (1) and [Cu96Se45(Se-C6H4-SMe)6(PPh3)18] (2) (Scheme 2d).197 Moreover, depending on the applied temperature conditions, the same reaction procedure can yield two different cluster compounds. Both clusters have similar layered arrangements in an A-B-A fashion, where all the copper atoms occupy identical positions inside the selenium lattices. Interestingly, compound (1) is a dimer in the solid-state. A similar A-B layered arrangement of the chalcogenide substructure was found in a [Cu40Se16(S-C6H4-CN)8(dppm)8] cluster, the structure of which was published in 2013 by Robert Langer et al.191 Starting from 2013, C. W. Liu together with Jean-Yves Saillard reported several selenide-CuNCs, namely [Cu7Br{Se2P(OiPr)2}6],154 [Cu11(μ9-I)(μ3-I)3{Se2P(OiPr)2}6](OH) and [Cu11(μ9-I)(μ3-I)3(Se2PPh2)6](PF6),162 [Cu20H11(Se2P(OiPr)2)9],179 [Cu20H11{Se2P(OiBu)2}9]180 and [Cu30H18{Se2P(OiPr)2}12], [Cu30H18{Se2P(OiBu)2}12],186 accompanied by extensive computational analyses of the structures. Note that in these clusters, the Se atoms are not bridged but rather are part of the ligand structure and composition (selenolates).
 | ||
| Scheme 3 The syntheses of thiolate-protected CuNCs as described in ref. 151 (Adapted with permission, Copyright 2016 Wiley-VCH Verlag GmbH &Co. KGaA, Weinheim), ref. 170 (Reprinted with permission, Copyright 2019 American Chemical Society), and ref. 174 (Adapted with permission, Copyright 2017 Wiley-VCH Verlag GmbH &Co. KGaA, Weinheim). | ||
The Cu6 core has an octahedral geometry and is protected by six SR-units through a bridging bonding motif (Fig. 11A). In 2017 the C.W. Liu group reported the synthesis and structure of a Cu16L16·2CH3COCH3 cluster (L is cyclohexylmonothiocarbonate), where the coordinated solvent molecule(s) can be easily replaced with other N-donor ligands through coordinated-solvent-replacement reactions (Scheme 3b).174 Regardless of the type of coordinated solvent molecule, all structures are similar. The Cu16 core can be seen as an assembly of three pentanuclear units arranged in a “butterfly”-like geometry (Fig. 11B). The central pentanuclear unit resembles a pyramid, whereas the other two units can be considered as butterfly motifs. The entire metallic framework is protected with cyclohexylmonothiocarbonate ligands binding in a bridging mode. Hayton and co-workers recently reported a novel synthetic strategy for the preparation of “atlas-sphere”-like [Cu12(SR′)6Cl12][(Cu(R′SH))6] (R′ = nBu) and [Cu17(SR′′)6Cl13(THF)2(R′′SH)3][H(THF)2]2 (R′′ = CH2CH2Ph) clusters (Scheme 3c), both of them having a [Cu12S6]6+ cuboctahedral core (Fig. 11C and D).170 Two other thiolate-protected copper clusters Cu7 and Cu74 were reported in 2013 by Olaf Fuhr and co-workers153 and in 2020 by Burgi and co-workers,195 respectively. Note that the former cluster also contains phosphine ligands in the ligand shell, whereas in the last case, the Cu74 core contains 15 sulphur atoms incorporated into the metallic framework, which is further protected with 45 thiolate (in this case 2-PET: 2-phenylethylthiol) ligands (Fig. 10).
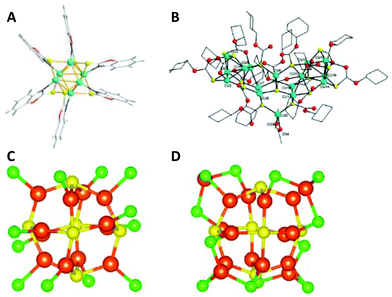 | ||
| Fig. 11 (A) The total structure of a Cu6(C7H4ONS)6 cluster. Colour legend: green sphere, Cu; yellow sphere, S; blue sphere, N; red sphere, O; light grey sphere, C; deep grey sphere, H. Adapted with permission from ref. 151. Copyright 2016 Wiley-VCH Verlag GmbH &Co. KGaA, Weinheim. (B) Solid-state structure of a Cu16[SCOOC6H11]16·2C3H6O cluster. Adapted with permission from ref. 174. Copyright 2017 Wiley-VCH Verlag GmbH &Co. KGaA, Weinheim. The core structures of [Cu12(SR′)6Cl12][(Cu(R′SH))6] (R′ = nBu) (C) and [Cu17(SR′′)6Cl13(THF)2(R′′SH)3][H(THF)2]2 (R′′ = CH2CH2Ph) clusters (D). For clarity, the organic part that is connected to the sulphur atoms and occupies one of the six square faces of the cuboctahedron has been omitted. Colour legend: orange, Cu; yellow, S; green, Cl; red, O; grey, C. Reprinted with permission from ref. 170. Copyright 2019 American Chemical Society. | ||
In 2013 T. Pradeep and co-workers reported the synthesis of a “pure” thiolate-protected cluster; no single crystal analyses were conducted, however, the molecular composition of the cluster was assigned by MALDI and ESI mass spectrometry.201
Recently, the group of Manzhou Zhu reported the synthesis of two thiolated CuNCs with the composition of [Cu8(SPh)8(Ph3P)4]159 (1) and [Cu11(TBBT)9(PPh3)6](SbF6)2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 161 (where TBBT = 4-tert-butylbenzenethiol). Note that these are not entirely “pure” thiolate-protected clusters and have phosphine ligands contributing to the ligand shell composition. Interestingly, the ligand exchange of (1) with PhSeH ligand resulted in the drastic core transformation and formation of a new [Cu13(SePh)13(Ph3P)4] cluster with different atom-packing modes (the Se atom possesses more coordination modes with Cu (μ2, μ3, μ4, μ6) than S (μ2, μ3).
161 (where TBBT = 4-tert-butylbenzenethiol). Note that these are not entirely “pure” thiolate-protected clusters and have phosphine ligands contributing to the ligand shell composition. Interestingly, the ligand exchange of (1) with PhSeH ligand resulted in the drastic core transformation and formation of a new [Cu13(SePh)13(Ph3P)4] cluster with different atom-packing modes (the Se atom possesses more coordination modes with Cu (μ2, μ3, μ4, μ6) than S (μ2, μ3).
The groups of Prof. Trevor W. Hayton and Prof. C. W. Liu revolutionized the field of copper-hydride clusters over the past few years by reporting a series of CuNCs with fascinating structures and ligand functionalities. The list extends from the small clusters to relatively large ones including those with partial Cu(0) character. Schemes 4–6 summarize some of the synthetic protocols used for the preparation of clusters.
 | ||
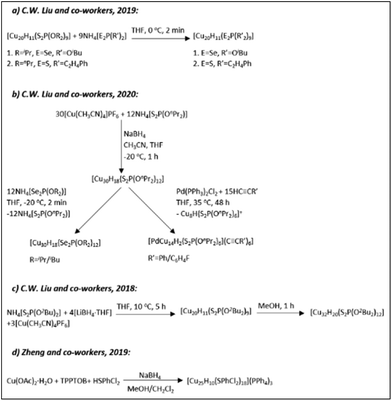
| Scheme 4 The syntheses of hydrido CuNCs as described in ref. 163 (Reprinted with permission, Copyright 2020 American Chemical Society), ref. 171 (Adapted with permission, Copyright 2015 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim), ref. 178, 179, 184 and 185 (Reprinted with permission, Copyright 2016 American Chemical Society). | ||
 | ||
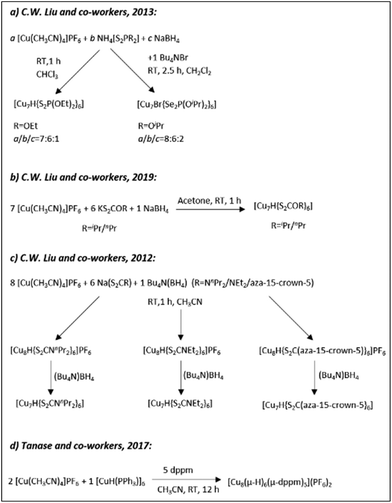
| Scheme 5 The syntheses of hydrido CuNCs as described in ref. 180 (Adapted with permission, Copyright 2019 Wiley-VCH Verlag GmbH &Co. KGaA, Weinheim), ref. 181 (Adapted with permission, Copyright 2018 Wiley-VCH Verlag GmbH &Co. KGaA, Weinheim), ref. 182 and 186 (Adapted with permission, Copyright 2020 Wiley-VCH Verlag GmbH &Co. KGaA, Weinheim). | ||
 | ||
| Scheme 6 The syntheses of hydrido CuNCs as described in ref. 154 and 155 (Reprinted with permission, Copyright 2019 Elsevier), ref. 156 and 157 (Adapted with permission, Copyright 2017 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim). | ||
In 2014 the Liu group reported the synthesis (Scheme 4a) and structure of a novel [Cu28H15{S2CNPr2}12]PF6 cluster composed of a rhombicuboctahedral framework of 24 Cu atoms.184 The Cu24 core of the clusters was further enclosed by a truncated octahedron of 24 thiolate ligands (Fig. 12A). The hydrides were found in the centre of the core (one central interstitial hydride); eight were outer-triangular-face-capping hydrides and six were face-truncating hydrides forming a bridge between the inner and outer copper atom shells.
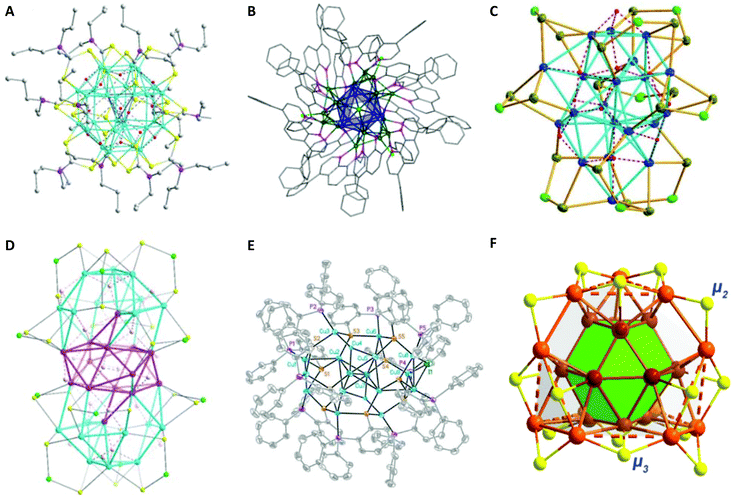 | ||
| Fig. 12 (A) The molecular structure of a [Cu28H15{S2CNPr2}12] cations. The counter ions are omitted for clarity. The following colour coding has been applied for the description: carbon: grey, copper: cyan, hydrogen: red, nitrogen: pink, sulphur: yellow. Adapted with permission from ref. 184. Copyright 2014 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. (B) Ball and stick diagrams of [Cu29Cl4H22(Ph2phen)12]Cl. The Cu13 centred-icosahedral core is highlighted in blue. The remaining Cu atoms are shown in dark green. The N atoms are purple, and the Cl atoms are lime green. All solvent molecules and outer sphere counter ions have been omitted for clarity. Reprinted with permission from ref. 185. Copyright 2016 American Chemical Society. (C) The molecular structure of the [Cu20H11{Se2P(OiPr)2}9] cluster. Isopropoxy groups have been omitted for clarity. Adapted with permission from ref. 179. Copyright 2015 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. (D) Crystal structures of a [Cu32H20{S2P(OiPr)2}12] cluster. Isopropoxy groups have been omitted for clarity. Adapted with permission from ref. 187. Copyright 2015 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. (E) Crystal structure of a [Cu18H7{(S(C6H4)PPh2)}10I] cluster. Hydrogen atoms are omitted for clarity. Adapted with permission from ref. 175. Copyright 2014 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. (F) The core structure of a [Cu25H10(SPhCl2)18]3− cluster. Metal atoms within the shell and in the two layers are bridged by μ2 and μ3 S atoms. Colour code: dark-red and light-brown, Cu; grey, C; yellow, S; bright green, Cl; purple, P. Hydrogen atoms have been omitted for clarity. Reprinted with permission from ref. 182. Copyright 2019 American Chemical Society. | ||
Hayton and co-workers in 2015 reported a novel hydride cluster compound (Scheme 4b) with a composition of [Cu14H12(phen)6(PPh3)4][X]2 (X = Cl or OTf; OTf = trifluoromethanesulfonate, phen = 1,10-phenanthroline) protected with neutral donor ligands featuring a tetrahedral [Cu4]4+ core.171 In that same year, they reported the first-ever [Cu25H22(PPh3)12]Cl (1) cluster with partial Cu(0) character (to be discussed later in section 2.2.4).183 The ligand-induced structural transformations in the latter [Cu25H22(PPh3)12]Cl (1) cluster, resulted in the formation of a new [Cu29Cl4H22(Ph2phen)12]Cl (2, Ph2phen = 4,7-diphenyl-1,10-phenanthroline) cluster in the presence of excess ligand (Scheme 4c).185 Like its precursor, the Cu29 cluster features a Cu13 icosahedral core typical to most of the known and well-characterized monolayer-protected gold nanoclusters. The core of the cluster is connected to four triangular [Cu4(Ph2phen)3(Cl)] motifs via Cu–Cu bonds in a tetrahedral arrangement (Fig. 12B).
Ligand exchange induced structural transformations and the conversion from achiral [Cu20H11{S2P(OiPr)2}9] (1) into chiral [Cu20H11{Se2P(OiPr)2}9] (2) has been observed by the group of C.W. Liu (Scheme 4d).179 Unlike the transformation from a [Cu25H22(PPh3)12]Cl cluster to a [Cu29Cl4H22(Ph2phen)12]Cl cluster,185 the ligand exchange reaction of [Cu20H11{S2P(OiPr)2}9] cluster resulted in a drastic core structure transformation while converting the achiral compound (1, Scheme 4e)178 with C3h symmetry into an intrinsically chiral compound (2) with C3 symmetry. Both enantiomers can be found in the single unit cell. The structure of the left-handed enantiomer consists of three anticlockwise copper strands of ideal C3 symmetry and features an almost ideal cuboctahedral Cu13 core (Fig. 12C). The core is further covered by a Cu6 cupola (from the top) and a capping Cu atom (from the bottom). The whole copper framework is covered by nine diselenophosphate ligands. The structure determination by neutron diffraction revealed the positions of all eleven H atoms: seven are capping μ3-H ligands and four are in interstitial μ5-H locations. Furthermore, the thermal treatment of a [Cu20H11{S2P(OiPr)2}9] cluster solution in the presence of terminal alkynes leads to subsequent core transformations and the formation of [Cu11H2{S2P(OiPr)2}6(C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CR)3] (R = Ph, C6H4F and C6H4OMe) clusters (Scheme 4f).163
CR)3] (R = Ph, C6H4F and C6H4OMe) clusters (Scheme 4f).163
Under optimized reaction conditions, ligand exchange reactions can be performed to obtain clusters with different ligand shells while preserving the composition and the structure of an initial cluster. For example, [Cu20H11{Se2P(OiBu)2}9] protected with Se-donor ligands can be obtained in good yield through the ligand exchange reaction of [Cu20H11{S2P(OiPr)2}9] with NH4[Se2P(OiBu)2] or a [Cu20H11{S2P(CH2CH2Ph)2}9] cluster can be produced from the ligand replacement reaction of [Cu20H11{S2P(OnPr)2}9] with NH4[S2P(C2H4Ph)2] (Scheme 5a).180 In a similar fashion [Cu30H18{Se2P(OiPr)2}12] and [Cu30H18{Se2P(OiBu)2}12] clusters can be obtained from the ligand exchange of [Cu30H18{S2P(OnPr)2}12] with NH4[Se2P(OiPr)2] and NH4[Se2P(OiBu)2], respectively (Scheme 5b).186 Furthermore, All three clusters possess a hollow Cu12 icosahedral core embedded inside the rhombicuboctahedral Cu18 framework. Although the ligand exchange reactions are quite straightforward and are at the centre of attention in the nanocluster community, the C. W. Liu group established a direct method for the synthesis of various polynuclear copper hydrido clusters such as [Cu20H11{S2P(O2Bu)2}9], [Cu32H20{S2P(O2Bu)2}12]181 (Scheme 5c) and [Cu32H20{S2P(OiPr)2}12]187 from the corresponding ammonium salts (as a ligand source) and copper complexes. Interestingly enough, those clusters having the same number of copper atoms in the composition adopt similar crystal structures and geometries regardless of the type of ligand. For instance, the [Cu32H20{S2P(OiPr)2}12] cluster187 is composed of a distorted hexa-capped rhombohedral Cu14 core sandwiched between two nest-like triangular cupola fragments of Cu atoms (Fig. 12D). The twenty hydrides reside inside the structure through twelve μ3-H, six μ4-H, and two μ5-H locations. Very recently, Sanghwa Lee et al. reported the structure of another [Cu32(PET)24H8Cl2](PPh4)2 cluster that possesses a rare edge-sharing bisquare anitiprismatic Cu14H8 core capped by Cu7(PET)11Cl and Cu2PET units.189
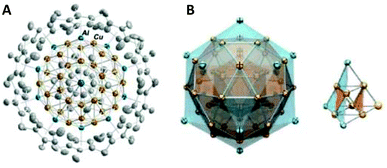
[Cu18H7L10I] (L = HS(C6H4)PPh2) and [Cu25H10(SPhCl2)18]3− hydride clusters were reported by Miguel A. Huertos et al.175 and Cunfa Sun et al.182 (Scheme 5d), respectively. The former structure consists of a Cu8 core surrounded by the remaining 10 Cu atoms that are connected to S and P atoms (Fig. 12E). The latter structure consists of a core best represented as a Cu@Cu12 centered twinned cuboctahedron (ctco).182 The remaining copper atoms form a truncated tetrahedral shell, the edges of which are bridged with 18 thiolates (Fig. 12F). All the hydrogen atoms in hydride form were located inside the metallic core (six μ6-H and four μ3-H). Very recently, the Osman M. Bakr group reported the synthesis and full structural analyses of high-nuclearity CuNCs: [Cu81(PhS)46(tBuNH2)10(H)32]3+.196 The cluster possesses an unusual Cu17 planar core, a hemispherical shell comprising a curved and a planar surface layer as well as different surface protective motifs of the ligands. This is so far the cluster with the largest number of hydrides; their presence and positions have been established by ESI-MS and DFT calculations, respectively.
Besides the relatively large hydrido copper clusters discussed above, there are also small clusters containing less than 10 copper atoms. In this context, Prof. C.W. Liu established synthetic methods for the preparation of small hydrido copper clusters, namely Cu7 and Cu8 (Scheme 6a–c).154–156 Regardless of the ligand, the clusters share a common hydrogen155,156 atom (or bromine154) located in the middle of a tetrahedron or a pyramid. Tomoaki Tanase and co-workers reported the structure of a Cu8H6 cluster, where hydrogen atoms bridge the Cu3 triangular faces of a Cu6 octahedral core (Scheme 6d).157
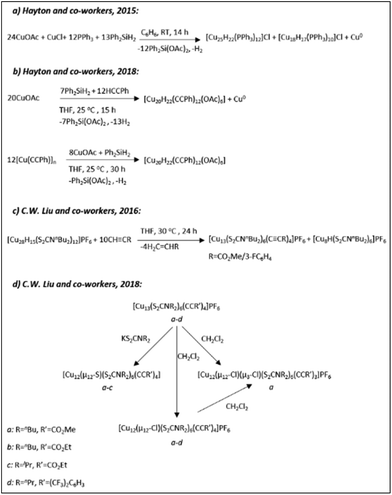 | ||
| Scheme 7 The syntheses of (hydrido) CuNCs with partial Cu(0) character as described in ref. 168 (Adapted with permission, Copyright 2016 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim), ref. 169 and 177 (Reprinted with permission, Copyright 2018 American Chemical Society), and ref. 183 (Reprinted with permission, Copyright 2015 American Chemical Society). | ||
 | ||
| Scheme 8 The syntheses of CuNCs with partial Cu(0) character as described in ref. 172 and 192 (Adapted with permission, Copyright 2018 Wiley-VCH Verlag GmbH &Co. KGaA, Weinheim), and ref. 194. | ||
In 2015 the group of Trevor. W. Hayton reported the first copper cluster, [Cu25H22(PPh3)12]Cl, having Cu(0) character and a Cu13 centred-icosahedral core (Scheme 7a and Fig. 13A).183 The core of the cluster is connected to four triangular [Cu(PPh3)]3 motifs forming a tetrahedral arrangement. What is striking about the structure is that the Cu13 core is isostructural to the known M13 core in atomically precise gold nanoclusters such as in the Au25(SR)18 cluster.207 Based on the molecular composition, to balance the charge, two out of 25 copper atoms must be in the zero oxidation state. The electron count for the cluster gives the value of 2, thus the cluster can be considered as a n* = 2 superatom with a 1S2 closed-shell configuration. A year later, the same group reported a Cu29 cluster (the structure of the cluster has been described in the previous 2.2.3. section) as a result of the ligand exchange reaction and subsequent core transformations of the Cu25 cluster in the presence of an excess ligand (Scheme 4c).185 In 2018, Hayton and co-workers reported a [Cu20(CCPh)12(OAc)6] cluster as another 2-electron superatom system (Scheme 7b).177 The structure of the latter can be depicted as a [Cu4]2+ tetrahedral core trapped inside the [Cu16(CCPh)12(OAc)6]2−shell. Earlier on, the group of C. W. Liu reported a [Cu13(S2CNnBu2(OAc)6](PF6) cluster bearing acetylide groups (OAc) in μ3 fashion capping four triangular faces of a cuboctahedral [Cu13]11+ core (Scheme 7c).168 Similar core structures were observed in a series of new Cu13 and Cu12 clusters reported by the same group in 2018 (Scheme 7d), where, in the latter, the central atom of a cuboctahedral Cu13 core was replaced with S, Cl and Br leading to the formation of inverse-coordination clusters.169
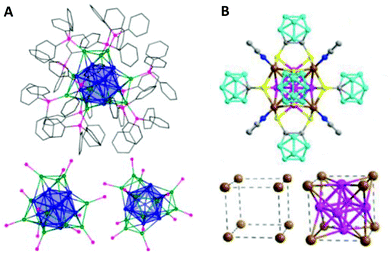 | ||
| Fig. 13 (A) Ball and stick diagrams of [Cu25H22(PPh3)12]Cl. The Cu13-centred-icosahedral core is highlighted in blue. The four [Cu(PPh3)]3 capping motifs are shown with the Cu atoms in green and P atoms in pink. The side view with carbon atoms depicted in wireframe format. All hydrogen atoms, chloride counter ions and solvent molecules have been omitted for clarity. Bottom left: Side view showing only the Cu and P atoms. Bottom right: Top view, looking down the C3 axis, showing only the Cu and P atoms. Reprinted with permission from ref. 183. Copyright 2015 American Chemical Society. (B) Molecular structure of the Cu14(C2B10H10S2)6(CH3CN)8 cluster. Bottom left: Cu88+ shell and bottom right: Cu88+ shell with Cu64+ core of the fcc-Cu14 framework. Colour codes: brown and pink = copper; yellow = sulphur; grey = carbon; blue = nitrogen; turquoise = boron. Adapted with permission from ref. 172. Copyright 2019 Wiley-VCH Verlag GmbH &Co. KGaA, Weinheim. | ||
Thomas C. W. Mak and co-workers very recently reported the synthesis of another Cu14(C2B10H10S2)6(CH3CN)8 cluster with partial Cu(0) character having a fcc core geometry (Scheme 8a).172 The six faces of the core are capped by bidentate 1,2-dithiolate-o-carborane ligands, whereas the eight vertices are connected to CH3CN ligands (Fig. 13B).
The [Cu53(RCOO)10(C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CtBu)20Cl2H18]+ cluster (Scheme 8b) is another 2-electron superatom having a core–shell1–shell2–shell3 arrangement of Cu3@Cu10Cl2@Cu20@Cu20 (Fig. 14A).194 Copper atoms in the Cu3 core are in a triangular arrangement, whereas the 10 copper atoms form an icosahedron with 2 Cl atoms at the poles. The second shell of Cu20 makes a pentagonal dodecahedron, which is further trapped inside a Cu20 nanowheel with a diameter of 1.1 nm (shell 3, Fig. 14B).
CtBu)20Cl2H18]+ cluster (Scheme 8b) is another 2-electron superatom having a core–shell1–shell2–shell3 arrangement of Cu3@Cu10Cl2@Cu20@Cu20 (Fig. 14A).194 Copper atoms in the Cu3 core are in a triangular arrangement, whereas the 10 copper atoms form an icosahedron with 2 Cl atoms at the poles. The second shell of Cu20 makes a pentagonal dodecahedron, which is further trapped inside a Cu20 nanowheel with a diameter of 1.1 nm (shell 3, Fig. 14B).
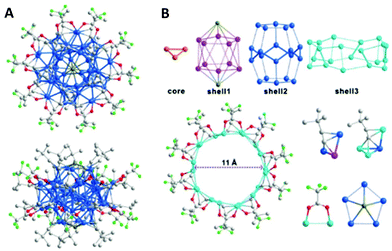 | ||
| Fig. 14 (A) Molecular structure of a Cu53 cluster: the view along an approximately five-fold axis (top), and general view (bottom). Colour codes: Cu; blue, F; light green, Cl; dark yellow, O; red, C; grey. Hydrogen atoms are omitted for clarity. (B) Shell-by-shell structural analyses of the cluster framework. Adapted with permission from ref. 194. Copyright 2019 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. | ||
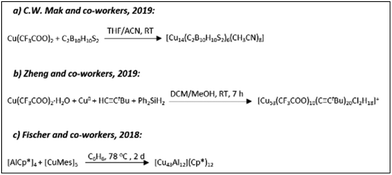
In 2018, Jana Weßing and co-workers reported the structure of a Mackay-type M55 heterometallic cluster [Cu43Al12](Cp*)12 (Scheme 8c) that possesses an ideal two-shell icosahedral structure.192 The detailed structure includes a body-centred Cu13 icosahedron protected by a heterometallic [Cu30Al12] shell (Fig. 15). The structure is completed with the binding of twelve Cp* ligands to Al centres.
 | ||
| Fig. 15 (A) The molecular structure of a [Cu43Al12](Cp*)12 cluster in the solid state, viewed along the five-fold rotational axis. H atoms and co-crystallized solvent molecules have been omitted for clarity. Colour codes: Al; blue, Cu; orange, C; grey. (B) Naked metal cores and the underlying tetrahedral M10 subunit of a cluster with highlighted M@12 M@42 M polyhedral shell geometries. The outer shell is constructed from a Cu30 icosidodecahedron embedded inside an Al12 icosahedron. Adapted with permission from ref. 192. Copyright 2018 Wiley-VCH Verlag GmbH &Co. KGaA, Weinheim. | ||
The structural variety of the above-mentioned 2-electron superatoms of CuNCs is quite big as compared to gold. Besides, high electron counts have not been observed for CuNCs, which may be related to the lower stability (oxidation) of copper compared to gold.
3. Size-dependent properties of CuNCs
3.1. Optical properties
It is well established that depending on the reaction conditions and the initial concentrations of the reacting components, the size of the obtained clusters can be drastically varied. Consequently, with the changes in size, the physicochemical properties will also be affected. Due to the quantum confinement effects, the continuum conduction and valence bands break down into discrete energy levels in clusters (with sizes smaller than 2 nm). Thus, depending on the size of the cluster core, copper nanoclusters with well-defined compositions can possess characteristic electronic transitions in the UV–vis spectrum. The UV–vis absorption spectrum is a characteristic optical fingerprint for both qualitative (identification) and quantitative (concentration) analysis of a cluster of interest. With just a glance at the spectrum, it is possible to determine the following: (i) whether the cluster is big or small by observing the presence or absence of the plasmon peak of copper particles (in this case) at 570 nm; (ii) the optical gap, by conversion of the wavelength scale into photon energy and extrapolating the absorbance to zero value. Normally, the optical gap gets bigger by decreasing the size of the cluster. DFT calculations are required to assign the bands in the UV–vis spectrum to certain electronic transitions. Interestingly, though, compared to gold clusters,208 copper clusters do not have rich multi-band UV–vis spectra, especially in the visible region. Typically, the optical spectra of CuNCs are characterized by a smooth exponential increase in their absorbance at low energies and have multiple bands and transitions at higher energies mostly governed by the absorption of the protective ligand. This phenomenon is quite elusive and not yet understood, some of the transitions have already been assigned and marked. For instance, in trinuclear copper(I) thiolate complexes and hexanuclear copper(I) selenolate [Cu6(μ-P^P)4(μ3-SePh)4](BF4)2 (P^P = dppm, (Ph2P)2NH)209 clusters the absorption bands/shoulders at ca. 250–290 nm in the UV–vis spectra were assigned as an intraligand (IL) n(S) → π* transition. For an octanuclear copper(I) methanediide cluster with a gyrobifastigium geometry, the absorption bands in the UV region were assigned as metal-centred metal–metal 3d → 4p transitions.210 Besides, the electronic transitions in the 300–500 nm region and below 260 nm were found to be ligand-to-metal charge transfer (LMCT) in tetranuclear, pentanuclear and hexanuclear Cu(I) clusters with aromatic ligands.210,211 Note that depending on the ligand shell and the size of the cluster, these transitions can be shifted either way. Eichhöfer and co-workers studied the optical properties of chalcogenide clusters differing in the size and the type of protective ligand and compared the obtained experimental results with calculated absorption spectra.165 For instance, it was found that in [Cu12S6(PPh2Et)8], [Cu20S10(PPh3)8] and [Cu24S12(PEt2Ph)12] clusters the high energy transitions (above 2.5 eV) originated from excitations of electrons from cluster core orbitals to ligand orbitals (Fig. 16A–C). Such transitions are also observable for Cu20 and Cu24 clusters at lower energies. However, in the case of Cu12, the low energy transitions also have contributions from the cluster core too, i.e., the electrons are transferred from the filled copper d-orbitals and sulphur p-orbitals to the empty p-orbitals of copper atoms. Moreover, for smaller clusters, the LUMO has contributions from the copper orbitals, whereas the HOMO mainly consists of d(Cu) and p(S) orbitals. On top of that, the low-energy excitations are very sensitive to the ligand shell and the excitation band character varies with the ligand environment even for the same core size.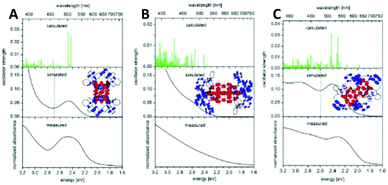 | ||
| Fig. 16 A comparison of measured electronic spectra (powdered crystals in mineral oil) of (A) [Cu12S6(PPh2Et)8], (B) [Cu20S10(PPh3)8] and (C) [Cu24S12(PEt2Ph)12] with calculated singlet excitation energies and oscillator strengths plotted as vertical green lines as well as by superimposed Gaussians of fwhm = 0.3 eV (black) to simulate the experimental spectrum. The contributions of occupied orbitals are plotted in red, and those of the unoccupied orbitals are in blue. Reprinted with permission from ref. 165. Copyright 2015 American Chemical Society. | ||
3.2. Photoluminescence
Photoluminescence (PL), as a molecular property to absorb light of a particular energy and emit it at longer wavelengths, is an intriguing phenomenon that gives rise to many applications. PL is an important technique in modern spectroscopy and biology labs to study all kinds of processes. For that purpose, intrinsic (naturally occurring, e.g. porphyrins) and extrinsic (synthesized, such as organic molecules and quantum dots) fluorophores with high quantum yields (QYs) have been applied as dyes to detect and label biological specimens and various metal ions/small molecules in contaminated environments.212 Therefore, highly fluorescent but biocompatible and nontoxic dyes are very much required. After the discovery of monolayer-protected clusters, massive research efforts have been devoted to the preparation of nanomaterial-based fluorophores. Recent progress in this field resulted in the synthesis of various cluster-based fluorophores with high QYs, which are biocompatible.24 The PL phenomenon in CuNCs is not well understood. Perhaps, the situation is relatively easier for gold clusters due to the known compositions. However, for CuNCs the situation is often more complicated due to the unresolved structural information for water-soluble CuNCs, to say the least. Several attempts were made to obtain an in-depth understanding of the PL mechanism by studying the effect of the cluster size, ligand shell and composition. The effects of the ligand concentration (cysteine) during synthesis on the size and subsequent PL properties of obtained monodisperse clusters have been explicitly investigated by Mohammad Reza Ganjali and co-workers.213 It was found that the increase in ligand concentration results in the formation of weakly emissive and bigger clusters, whereas at moderate concentrations, highly emissive clusters are formed.For CuNCs soluble in organic solvents, it was found that the quantum efficiency can be increased by applying cryogenic temperatures159,161 and modifying the ligand shell, but it also strongly depends on the crystal packing.153,165 Note that the latter affects the electronic transitions within the cluster core.
Occasionally, dual-emission is also observed in CuNCs, where the low-energy emission is assigned to the intraband HOMO–LUMO transitions within the sp-band, whereas the high-energy emission is attributed to the interband transition from excited states in the sp-band to the d-band.109,214 Thus, an in-depth understanding of PL mechanisms and corresponding photophysics can be achieved only by having a complete understanding of size-structure-composition-property relationships in metal nanoclusters.
3.3. Aggregation/assembly-induced emission (AIE)
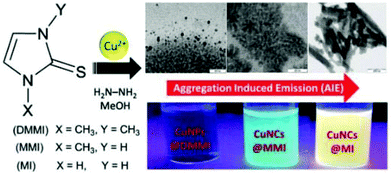 | ||
| Fig. 17 Schematic representation showing the effect of substituents on the self-assembly and aggregation-induced emission of mercaptoimidazole capped CuNCs. Republished with permission from ref. 217. Copyright 2019 Royal Society of Chemistry. | ||
Similarly, when para-substituted thiophenol (substitutional groups include –NO2, –F, –Cl, –Br, –CH3, and –OCH3) was used for capping, the self-assembly process led to the formation of ribbons with different emission properties in the red region.140 By changing the electron-donating abilities of the ligand (–F to –OCH3), the PL emission peak can be tuned from 548 to 698 nm (Fig. 18). On the contrary, 4-nitrothiophenol leads to the random aggregation of clusters into nanoparticles with no emission enhancement.
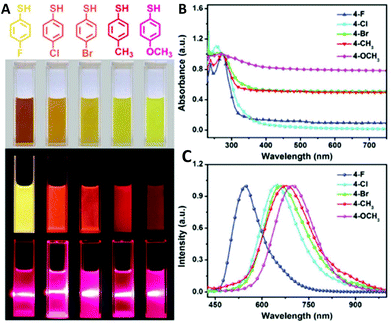 | ||
| Fig. 18 (A) Chemical structures of the capping ligands, and the corresponding images of the sample solutions. PL images with 330 nm excitation and Tyndall scattering phenomena on the irradiation (by a laser pointer) of the self-assembled architectures composed of the CuNCs capped with different ligands. Steady-state absorption (B) and PL emission spectra (C) of the self-assembled architectures composed of the CuNCs capped with different ligands. Republished with permission from ref. 140. Copyright 2017 Royal Society of Chemistry. | ||
Jinbin Liu and co-workers demonstrated the synthesis of CuNCs assemblies embedded inside the hydrophobic core of spherical micelles formed from an amphiphilic triblock copolymer template.218 The number of encapsulated CuNCs inside the core can be controlled by adjusting the block segments of the template. The clusters are assembled into uniform structures and held together through the cross-linking of a multidentate thiol ligand. It was also shown that depending on the structure and branching of the ligand, i.e. whether it is mono- or multidentate, the formed cluster assemblies displayed different PL properties. Furthermore, the brightest luminescence (absolute quantum yield of 7.3%) was observed when a hexadentate thiol ligand with a star-like structure was used.
These findings illustrated not only the effect and importance of the protective ligand in the synthesis, but also opened a new pathway for constructing highly fluorescent clusters with the AIE effect.
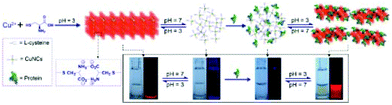 | ||
| Fig. 19 Schematic diagram of the pH-guided strategy in the fabrication of water-soluble protein/CuNCs hybrid nanostructures. The AIE behaviour of the as-prepared nanocomposite can be regulated by changing the pH of the solution. A strong emission was observed at pH 3.0 due to the aggregation of the cluster, but this further decreased significantly upon the dissolution of the composite under weakly acidic conditions (pH > 3). Reprinted with permission from ref. 219. Copyright 2017 American Chemical Society. | ||
Similarly, D-penicillamine-protected CuNCs showed changes in their emission spectra when the pH increased from 4 to 6.2.114 At pH 6.2 the emission intensity dropped 100-fold due to the dissolution of CuNCs lamellar structures/aggregates in aqueous solution and the formation of rather isolated clusters. On the contrary, Yuanyuan Huang et al. showed that the AIE enhancement of CuNCs aggregates under acidic conditions can stay unaltered when the pH is further increased to 8.220
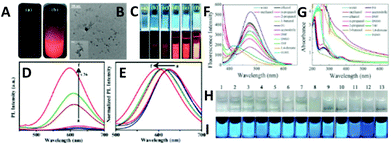 | ||
| Fig. 20 (A) Digital photographs of Cu34–32(SG)16–13 under 365 nm UV illumination (a) at ambient temperature and (b) upon freezing (temperature <0 °C). (B) TEM image of aggregates with scale bar 100 and 50 nm (inset). (C) Corresponding digital photographs with volume fraction (fv) of (a) 0%, (b) 20%, (c) 40%, (d) 60%, (e) 80%, and (f) 90% under daylight conditions and UV (365 nm) illumination. (D) PL spectra of Cu34–32(SG)16–13. (E) Shifting of the emission maxima in water–EtOH mixture with fv = 0–90%. Reprinted with permission from ref. 17. Copyright 2019 American Chemical Society. Fluorescence spectra (F) and absorption spectra (G) of the PEI-capped Cu nanoclusters dispersed in different solvents. Photographs of the PEI-capped Cu nanoclusters dispersed in different solvents under visible light (H) and UV light (I). Panels H and I show (1) water, (2) methanol, (3) ethanol, (4) n-propanol, (5) isopropanol, (6) 1-butanol, (7) ethylene glycol, (8) acetonitrile, (9) DMF, (10) DMSO, (11) THF, (12) 1,4-dioxane, and (13) EGME. Reprinted with permission from ref. 28. Copyright 2015 American Chemical Society. | ||
Similar solvent driven AIE enhancement in GSH-CuNCs was also detected by Jingcheng Hao and co-workers.221 The addition of methanol, ethanol, n-propanol, 2-propanol, DMF and DMSO resulted in the formation of milky solution under daylight conditions and strong fluorescence under UV-irradiation with the order of PL intensity being IDMF > Imethanol > IDMSO > Iethanol > I2-propanol > In-propanol. The increase in the organic solvent content leads to the reorganization of surface ligands into amphiphilic structures to minimize the contact between the organic solvent and ligand molecules at the water/organic solvent interface.
With the accumulation of amphiphilic structures at the water/organic solvent interface, highly ordered self-assemblies of CuNCs are formed (spherical particles in the size range of 100–500 nm).
Yu Ling et al. demonstrated the solvent effect in PEI-CuNCs by adding alcohols, DMF, DMSO, THF, 1,4-dioxane, EGME and acetonitrile (Fig. 20F–I).28 Except for the first two solvents, strong solvent-dependent AIE enhancement was observed. Furthermore, the highest enhancement and blue shift of the emission peak was detected upon the addition of 60% THF, which was explained by the formation of hydrogen bonds and the collapse of PEI branches because of insolubility in THF as well as conformational changes in the polymer network. Altogether, this led to the aggregation of the clusters and thus, enhancement of AIE.
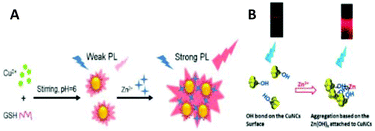 | ||
| Fig. 21 (A) Schematic illustration of the preparation of GSH-capped CuNCs and the aggregation-induced PL enhancement of CuNCs in the presence of Zn2+. Reprinted with permission from ref. 225. Copyright 2017 Elsevier. (B) Scheme for the mechanism of the aggregation-induced fluorescence enhancement for CuNCs in the presence of Zn2+. Republished with permission from ref. 110. Copyright 2016 Royal Society of Chemistry. | ||
From a mechanistic point of view, the coordination of metal cations with the ligand shell introduced instability and led to the instant formation of large aggregates.
Hao Zhang and co-workers showed that the spherical dodecanethiol (DT)-capped CuNCs with size of 1.9 nm self-assembled into ribbons (average width about 50–200 nm, length about 1–2 μm, and thickness about 13.5 nm) at 128 °C (Fig. 22A and B).139 The emission of CuNCs-based ribbons was greatly enhanced (absolute quantum yield of 6.5%, Fig. 22C). Furthermore, the emission colour can be tuned from yellow to deep blue/green just by changing the annealing temperature during the self-assembly process (thermochromic effect) (Fig. 22D).
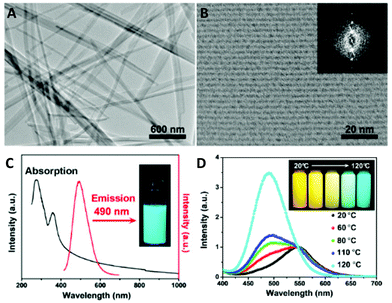 | ||
| Fig. 22 (A) TEM image of the ribbons from CuNCs self-assembly. (B) HRTEM image of the ribbons. Inset: Fourier transform image. (C) Steady-state absorption (black) and emission (red) spectra of the ribbons in chloroform. Inset: The fluorescent image with 365 nm excitation. (D) Spectra of CuNCs assemblies respectively prepared at 20, 60, 80, 110, and 120 °C. Inset: Sample excited at 365 nm at different temperatures. Reprinted with permission from ref. 139. Copyright 2015 American Chemical Society. | ||
The AIE enhancement in ribbons made of individual CuNCs is (i) due to the changes in ligand/ligand, ligand/metal, and metal/metal (cuprophilic Cu(I)⋯Cu(I)) interactions, which drastically increases the excited state relaxation dynamics. (ii) The restriction of the intramolecular vibration and rotation of capping ligands reduces the nonradiative relaxation of excited states.
The same group also studied nanosheets of clusters and showed that metal defects on the surface of highly structured assemblies influenced the PL properties (see Fig. 23).230 In short, the dangling bonds and the formation of unsaturated coordination sites on the nanosheet surface significantly altered the photophysical relaxation dynamics. When dodecanethiol (DT)-capped CuNCs assembled into a nanosheet in the presence of alcohols (ethanol and methanol), they featured pronounced AIE properties (QY 15.4%). The Cu(I)/Cu(0) ratio at the surface was found to be 1/0.32, and the presence of Cu(I) species facilitated the radiative relaxation. Moreover, at room temperature, mainly metal-defects at the surface were responsible for emission (T2 state), whereas upon lowering the temperature, the contribution of defects was negligible, and the ligand-to-metal–metal charge transfer (LMMCT)-determined triplet states (T1 state) were dominant (Fig. 23). Thus, the presence of a large number of metal defects on the surface changed the LMMCT behaviour by providing a lower energy level.
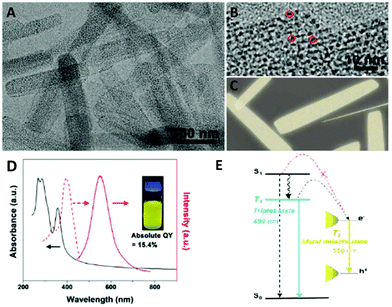 | ||
| Fig. 23 Low (A) and high (B) magnification TEM images of CuNCs self-assembled nanosheets. (C) Top- and side-view cartoon image of the nanosheets. (D) Steady-state absorption (solid), excitation (dash), and emission (dot) spectra of the nanosheets in chloroform. Inset: Sample excited at 365 nm. (E) Schematic diagram of the excited state relaxation dynamics of the nanosheets. Reprinted with permission from ref. 230. Copyright 2017 American Chemical Society. | ||
The discussions made in section 3.3 demonstrated the importance of the AIE mechanism/phenomenon for strong PL properties in CuNCs. Nonetheless, it is by far not the only important factor to consider since not all the CuNCs are explicit AIEgens and more studies on well-defined systems are needed to better understand the PL properties of clusters, which is important for applications.
3.4. Chirality in CuNCs
Chirality is a symmetry property of matter and plays an important role in biology and pharmacological sciences. Since the first discovery of optical activity in glutathione-protected gold nanoclusters by Whetten and co-workers in the 2000s,231 several groups have, both experimentally and theoretically, attempted to understand the origin of chirality in small nanoclusters containing a few tens of atoms. Several theoretical calculations were conducted to individually analyse the core, the shell and overall structure to gain insight into the origin of the strong chiral responses in metal-based transitions. To that end, several mechanisms have been proposed to explain the origin of the optical activity of various passivated clusters: an intrinsically chiral metal core, dissymmetric field model and chiral footprint.232,233 The intrinsically chiral metal core or ligand-induced chirality in metal nanoclusters, including Au38(SR)24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 234,235 and Au102(SR)44,236 has been considered as an origin of chirality, mainly due to strong Au–S bond formation and staple motif arrangements on the surface. The dissymmetric field model states that the chirality in metal nanoclusters is due to the adsorption of chiral molecules on achiral metallic cores. The chiral molecules create a dissymmetric field sensed by the metal core.237 Another model, called the “chiral footprint”, implies that when chiral molecules are adsorbed on metallic surfaces, the local environment becomes chiral.238 The induction of chirality in the cluster via chiral ligands has been considered as a relatively effective and controllable approach for chiral nanomaterial synthesis. In this regard, Shuang-Quan Zang and co-workers reported the synthesis of the atomically precise [Cu14(R/S-DPM)8](PF6)6 cluster with strong CD and CPL properties.173 In the CD spectra, the mirror-image signals in the range of 300–460 nm contained additional signals different from the pure ligand, indicating the chirality transfer from the ligand to the core. This is the first report on CPL-active atomically precise CuNCs. More intriguingly, in solution, the clusters were non-emissive, whereas they became extremely red-emissive in the crystalline or solid phase (Fig. 24) and they showed AIE enhancement.
234,235 and Au102(SR)44,236 has been considered as an origin of chirality, mainly due to strong Au–S bond formation and staple motif arrangements on the surface. The dissymmetric field model states that the chirality in metal nanoclusters is due to the adsorption of chiral molecules on achiral metallic cores. The chiral molecules create a dissymmetric field sensed by the metal core.237 Another model, called the “chiral footprint”, implies that when chiral molecules are adsorbed on metallic surfaces, the local environment becomes chiral.238 The induction of chirality in the cluster via chiral ligands has been considered as a relatively effective and controllable approach for chiral nanomaterial synthesis. In this regard, Shuang-Quan Zang and co-workers reported the synthesis of the atomically precise [Cu14(R/S-DPM)8](PF6)6 cluster with strong CD and CPL properties.173 In the CD spectra, the mirror-image signals in the range of 300–460 nm contained additional signals different from the pure ligand, indicating the chirality transfer from the ligand to the core. This is the first report on CPL-active atomically precise CuNCs. More intriguingly, in solution, the clusters were non-emissive, whereas they became extremely red-emissive in the crystalline or solid phase (Fig. 24) and they showed AIE enhancement.
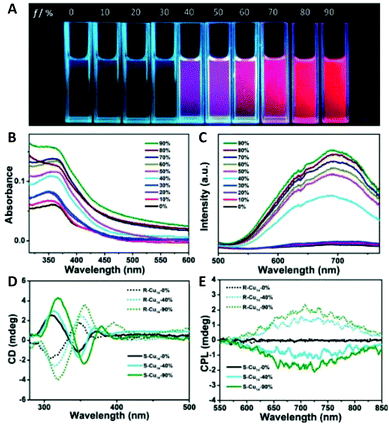 | ||
| Fig. 24 (A) Photographs of fluorescent R-Cu14 in solutions with different fractions of n-hexane. (B) UV/Vis absorption spectra, (C) emission spectra, (D) CD spectra, and (E) CPL spectra of R/S-Cu14 in DCM/n-hexane mixtures with different (fv) values. Adapted with permission from ref. 173. Copyright 2019 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. | ||
The results were interpreted within the frame of AIE. The restriction of intramolecular rotations and vibrations due to the electrostatic interactions, the C–H⋯π interactions and hydrogen-bonding interactions, activate the radiative transitions in the aggregated state and in the crystal. Thus, the AIE enhancement originates from the restricted intramolecular rotation (RIR) mechanism, which triggers strong PL and CPL properties.
4. Applications of CuNCs
4.1. Applications in sensing
The advantages of ultrasmall and atomically precise clusters with respect to their larger counterparts include the following: (i) the ability to tune the size-dependent properties by carefully choosing the protective ligand and/or the (bio) template; (ii) clusters are more biocompatible and environmentally friendly, some are non-toxic, however, some are quite toxic; (iii) some clusters show size-dependent and relatively strong photoluminescence. These characteristics, particularly the photoluminescence, make CuNCs attractive for applications in sensing. The number of yearly published CuNCs being applied as fluorescent probes is increasing exponentially due to the large demand for analytical tools in modern society, particularly in medicine and environmental control. Table 2 summarizes the recently published papers on the application of CuNCs as a new nanomaterial-based detection tool or bioassay. The easily scalable synthetic procedures facilitate real-life applications.| Template/ligand | Cluster size (d nm) | Ex/Em Wavelength (nm) | QY % | Read out | Target | LOD | Linear response | Ref. |
|---|---|---|---|---|---|---|---|---|
| Note, that in the third row of Table 2 the notations <′> and <′′> refer to the first and second emission peaks. In some cases, the excitation wavelength is not mentioned in the article, thus, a notation was used. | ||||||||
| Metal cation detection | ||||||||
| BSA | 1.5 ± 0.4 nm | 320/420 | — | Turn off | Hg2+ | 4.7 pM | 10 pM–10 μM | 56 |
| L-Cysteine | 2–3 nm | 375/480 | 14.3 | Turn off | Hg2+ | 24 nM | 0.1 μM–1 mM | 121 |
| GSH | 2–3 nm | 375/440 | 3.08 | Turn off | Hg2+ | 22 nM | 40 nM–60 μM | 128 |
| GSH | 2.4 ± 0.6 nm | 360/445 | 10.6 | Turn off | Hg2+ | 3.3 nM | 0.01–10 μM | 239 |
| DNA | <5 nm | 340/650 | — | Turn on | Hg2+ | 16 pM | 50 pM–500 μM | 240 |
| Curcuma roots | 2.3 ± 1.2 nm | 365/440 | 7.2 | Turn off | Hg2+ | 0.12 nM | 0.0005–25 μM | 241 |
| MT | <5 nm | — | — | Turn on | Hg2+ | 43.8 nM | 97 nM–2.3 μM | 242 |
| 3.1–15.6 μM | ||||||||
| dsDNA | 2–3 nm | 570/600 | — | Turn off | Hg2+ | 4 pM | 0.04–8 nM | 243 |
| PATP | 2.16 ± 0.36 nm | 365/602 | 24.6 | Turn off | Hg2+ | 10 ppt | 25 ppt–25 ppb | 244 |
| PEI | 2.5 nm | 360/430 | 2.1 | Turn off | Fe3+ | 340 nM | 0.5–1000 μM | 30 |
| TA | 2.2 ± 0.5 nm | 360/430 | 14 | Turn off | Fe3+ | 10 nM | 10 nM–10 μM | 145 |
| Dopamine | 8 nm | 320/390 | 9.6 | Turn off | Fe3+ | 1.2 μM | 5–300 μM | 148 |
| GSH | 2.2 nm | 340/430 | 6 | Turn off | Fe3+ | 25 nM | 0.01–1 μM | 135 |
| Cysteamine | 3 nm | 385/467 | 16 | Turn off | Fe3+ | 423 nM | 0–1000 μM | 138 |
| GSH | 1.7 ± 0.3 nm | 365/422 | 8.6 | Turn off | Fe3+ | 0.3 μM | 1–100 μM | 245 |
| CCCDL | 2 ± 0.53 nm | 365/425 | — | Turn off | Fe3+ | 20 nM | 0.05–30 μM | 246 |
| — | 3.5 nm | 350/430 | — | Turn off | Fe3+ | 2 μM | 0–30 μM | 247 |
| ACE | 30 nm | 280/340 | — | Turn off | Fe3+ | 23.4 nM | 0.2–100 μM | 248 |
| L-Histidine | 2 nm | 385/495 | — | Turn off | Fe3+ | 82 nM | 0.1–20 μM | 249 |
| BSA | 1 nm | 340/420 | — | Turn off | Cu2+ | 1 nM | 0.02–34 μM | 52 |
| D-Penicillamine | 1.2 ± 0.4 nm | 391/673 | — | Turn on | Cu2+ | 0.3 ppm | 0.95–6.35 ppm | 115 |
| PEI | 1.7 nm | 380/495 | 7.9 | Turn off | Cu2+ | 8.9 nM | 22 nM–8.8 μM | 250 |
| Poly(thymine) | — | 340/615 | 11.2 | Turn on | Cu2+ | 5.6 μM | 15–35 μM | 251 |
| Cytidine | 1.5 ± 0.5 nm | 300/380 | — | Turn on | Cu2+ | 0.032 μM | 0.05–2 μM | 252 |
| GSH | 4 nm | 330/615 | 1.2 | Turn on | Cu2+ | 0.17 μM | 0.25–10 μM | 253 |
| PEI | 2–3 nm | 365/495 | 2.63 | Turn off | Cu2+ | 0.12 μM | 0.2–8 μM | 254 |
| BSA | 2.8 ± 0.5 nm | 325/410 | 15 | Turn off | Pb2+ | — | — | 57 |
| Random dsDNA | — | 340/585 | — | Turn off | Pb2+ | 5 nM | 5–100 nM | 72 |
| TBAN | — | 312/408 | — | Turn off | Pb2+ | 4.9 μM | 48 nM–77 μM | 95 |
| GSH | 2.2 ± 0.2 nm | 365/606 | 5.3 | Turn off | Pb2+ | 1 nM | 1–160 nM | 137 |
| Poly(thymine) | — | 340/615 | — | Turn off | Pb2+ | 0.4 nM | 1–500 nM | 255 |
| MT | <5 nm | — | — | Turn on | Pb2+ | 142 nM | 0.7–96 μM | 242 |
| GSH | 2.3 nm | 340/585 | — | Turn on | Pb2+ | 30 μM | 40–105 μM | 127 |
| Turn on | Zr4+ | 40 μM | 60–250 μM | |||||
| BSA | 1.5 nm | 370/435 | — | Turn off | Co2+ | — | — | 59 |
| PEI | 1.1 ± 0.4 nm | 488/590 | 20 | Turn on | Ca2+ | 220 nM | 2–350 μM | 256 |
| DNA | 2.8 ± 0.5 nm | 354/561 | — | Turn on | Mn2+ | 10 μM | 100–250 μM | 257 |
| DHLA | 1.6 nm | 362/627 | 2.8 | Turn on | Zn2+ | — | 50–400 μM | 110 |
| GSH | 2.5 nm | 340/600 | 6.2 | Turn on | Zn2+ | 1.17 μM | 4.68–2240 μM | 225 |
| BSA | <3 nm | 440/650 | — | Turn off | Zn2+ | 15 nM | 0.66–5.3 μM | 258 |
| Cysteine | 2.8 ± 0.6 nm | 387/615 | — | Turn on | Zn2+ | 1 μM | 1.5–750 μM | 259 |
| L-Cysteine | 3.5 nm | 360/490 | 5.6 | Turn off | Cr6+ | 43 nM | 0.05–60 μM | 116 |
| TSA/BSA | — | 350/430 | 1.2 | Turn off | Cr6+ | 3.54 nM | 50–400 nM | 260 |
| Organic molecules | 3 nm | 468/620 | — | Turn off | Cr3+ | 3 μM | 0–50 μM | 261 |
| Poly(thymine) | 2–3 nm | 340/620 | — | Turn off | Cr3+ | 0.03 μM | 0–70 μM | 262 |
| DTT | 2.3 nm | 360/590 | — | Turn on | Al3+ | 0.01 μM | 0.01–7 μM | 263 |
| Cysteamine | 2.3 ± 0.5 nm | 330/430 | — | Turn on | Al3+ | 26.7 nM | 1–7 μM | 264 |
| DNA | — | 340/640 | — | Turn off | Al3+ | 62 nM | 0.1–30 μM | 265 |
| Anion detection | ||||||||
| PSS | 1.7 nm | 325/665 | 8 | Turn off | S2− | 650 nM | 1–20 μM | 35 |
| DNA | — | 340/585 | — | Turn off | S2− | 80 nM | 0.2–2 μM | 82 |
| Penicillamine | 2.7 ± 0.2 nm | 326/580 | 2 | Turn off | S2− | 500 nM | 1–100 μM | 113 |
| L-Cysteine | 2.5 nm | 382/460 | — | Turn on | S2− | 42 nM | 0.2–50 μM | 117 |
| Yeast extract | 2–4 nm | 370/450 | 9.3 | Turn on | S2− | 10 nM | 0.02–0.8 μM | 266 |
| CEW | 2.6 nm | 465/661 | 6.2 | Turn off | S2− | — | — | 226 |
| GSH | — | 365/622′′ | — | Turn off | S2− | 4.3 nM | 26–128 nM | 267 |
| GSH | <5 nm | 360/607 | — | Turn off | S2− | 0.5 μM | 0.5–20 μM | 268 |
| Silk fibroin | 2.8 ± 0.5 nm | 326/422′′ | 4.9 | Turn on | S2− | 0.286 μM | 5–110 μM | 269 |
| PEI | 2 nm | 270/430 | — | Turn off | I− | 100 nM | 0–10 μM | 29 |
| Cysteamine | 3 nm | 385/467 | 16 | Turn off | I− | 2.02 μM | 0–10 mM | 138 |
| TA | 2.3 nm | 380/460 | — | Turn off | I− | 18 nM | 20–100 μM | 270 |
| ssDNA | 2 nm | 345/637 | — | Turn off | I− | 15 nM | 0.05–40 μM | 271 |
| 40–80 μM | ||||||||
| PVP | 3–5 nm | 365/421 | — | Turn off | I− | 1 ng mL−1 | 10–70 ng mL−1 | 272 |
| PVP | 3.3 ± 0.4 nm | 350/430 | 13 | Turn off | ClO− | 0.1 μM | 1–30 μM | 33 |
| PVP | 0.96–5.11 nm | 340/416 | 9.4 | Turn off | ClO− | 19 nM | 0.3–2.5 μM | 34 |
| TSA | 3.7 ± 0.5 nm | 338/420 | 13.2 | Turn off | CN− | 5 nM | 0.01–1 μM | 273 |
| Turn off | NO2− | 5 μM | 15–50 μM | |||||
| L-Cys | 4 nm | 360/490 | — | Turn off | NO2− | 95.4 nM | 1–80 μM | 274 |
| AA | 2 nm | 370/454 | — | Turn off | NO2− | 3.6 nM | 0.0125–125 μM | 275 |
| TA | 2 nm | 360/430 | — | Turn on | PO43− | 9.6 nM | 0.07–80 μM | 144 |
| PDA | 53.7 ± 4 nm | 340/580 | 2.54 | Turn on | PO43− | 1.5 nM | 0.003–70 μM | 276 |
| Poly(thymine) | 2–3 nm | 340/620 | — | Turn on | P2O74− | 5 nM | 0–70 μM | 262 |
| DNA | — | 340/640 | — | Turn on | F− | 1 μM | 2–150 μM | 265 |
| Hydrogen peroxide and Glucose detection | ||||||||
| Papain | 2.3 ± 0.7 nm | 370/620 | 14.3 | Turn off | H2O2 | 0.2 μM | 1–50 μM | 54 |
| D-Penicillamine | 1 ± 0.4 nm | 345/640′′ | 14.1 | Turn off | H2O2 | 0.01 mM | 0.05–2 mM | 114 |
| DHLA | 1.8 nm | 342/590 | — | Turn off | H2O2 | 0.3 μM | 1–10 μM | 111 |
| MSA | 4.5 nm | — | — | — | H2O2 | 0.001 mM | 0.01–1 mM | 142 |
| BSA | — | 335/410 | — | — | H2O2 | 10 μM | 10 μM–1mM | 58 |
| Glucose | 100 μM | 100 μM–2mM | ||||||
| PEI | 1.8 nm | 355/480 | 3.8 | Turn off | H2O2 | 0.4 μM | 0.5–10 μM | 26 |
| Glucose | 8 μM | 10–100 μM | ||||||
| Poly(thymine) | 1.7–5.1 nm | 340/615 | — | Turn off | H2O2 | 0.55 μM | 0.55–110 μM | 70 |
| Glucose | 0.05 mM | 0.05–1.6 mM | 73 | |||||
| dsDNA | 3–5 nm | 340/570 | — | Turn off | H2O2 | 3 nM | 10 nM–2 μM | 277 |
| Glucose | 12 nM | 50 nM–100 μM | ||||||
| GSH | 2.3 nm | 340/585 | — | Turn off | H2O2 | 0.6 μM | 1–60 μM | 127 |
| Turn off | Glucose | 0.37 μM | 0.8–50 μM | |||||
| β-CD | 2 nm | 360/450 | — | — | H2O2 | 0.2 μM | 0.02–10 mM | 278 |
| Glucose | 0.4 μM | 0.04–20 mM | ||||||
| DNA | — | 340/640 | — | Turn on | H2O2 (color.) | 2.92 μM | 5–2000 μM | 279 |
| Turn off | H2O2 (fluor.) | 0.266 μM | 0.5–2000 μM | |||||
| Turn on | Glucose (color.) | 15 μM | 50–3000 μM | |||||
| Turn off | Glucose (fluor.) | 0.5 μM | 1–500 μM | |||||
| L-Cysteine/GOx | — | 330/628 | 5.4 | Turn off | Glucose | 1.5 μM | 5–100 μM | 219 |
| GSH | 1.5 nm | 358/625 | 2.1 | Turn on | Glucose | 32 μM | 0.1–2 mM | 280 |
| Small molecule detection | ||||||||
| TA | 3.75 ± 0.71 nm | 355/430 | — | Turn on | Ascorbic acid | 0.11 μM | 0.5–10 μM | 143 |
| BSA | 2.7 ± 0.8 nm | 330/407 | — | Turn off | Kojic acid | 0.07 μM | 0.2–50 μM | 47 |
| Poly(thymine) | — | 340/610 | — | Turn off | Humic acid | 0.4 mg L−1 | 0–8 mg L−1 | 281 |
| BSA | — | 325/400 | 15 | Turn off | Picric acid | 120 nM | 0.8–100 μM | 51 |
| GSH | 2.36 ± 0.67 nm | 360/625 | — | Turn off | Picric acid | 2.74 μM | 9.9–43 μM | 282 |
| DNA | — | 340/627 | — | Turn off | Picric acid | 0.03 μM | 0.1–100 μM | 283 |
| Cysteamine | 3–5 nm | 365/480 | 2.3 | Turn off | Picric acid | 139 nM | 1–80 μM | 284 |
| AA | 4 nm | 350/430 | — | Turn off | Picric acid | 0.98 μM | 2–40 μM | 285 |
| L-Cys | 2.2 nm | 370/494 | — | Turn off | Picric acid | 0.19 μM | 2.5–25 μM | 286 |
| L-Cys | 4 nm | 360/490 | — | Turn on | Folic acid | 69.8 nM | 0.1–10 μM | 274 |
| Ovalbumin | 1.8 nm | 348/625 | 3.95 | Turn off | Folic acid | 0.18 μM | 0.5–200 μM | 287 |
| PEI | 2.3 nm | 355/480 | — | Turn off | TNT | 14 pM | 0–8 nM | 288 |
| L-Cys | — | 375/525 | — | Turn on | m-DNB | 0.13 μM | 99 nM–1.3 μM | 289 |
| BSA | <3 nm | 440/650 | — | Turn on | RDX | 1.62 nM | 0–0.238 μM | 258 |
| PEI | 1.8 nm | 355/480 | 3.8 | Turn off | Sudan dyes I | 65 nM | 0.1–30 μM | 27 |
| Sudan dyes II | 70 nM | 0.1–30 μM | ||||||
| Sudan dyes III | 45 nM | 0.1–25 μM | ||||||
| Sudan dyes IV | 50 nM | 0.1–25 μM | ||||||
| L-Cysteine | 2.3 nm | 400/495 | Turn off | Quinoline yellow | 0.11 μM | 0.2–5.5 μM | 122 | |
| AA | 1.59 ± 0.23 nm | 393/505 | 33.6 | Turn off | Aniline yellow | 1.44 μM | 0–16 μM | 290 |
| dsDNA | 3–5 nm | 340/570 | 3.5 | Turn off | Ochratoxin A | 5 ng mL−1 | 0–0.1 μg mL−1 | 291 |
| Poly(thymine) | <5 nm | 340/625 | — | Turn on | Ochratoxin A | 2 nM | 2.5–250 nM | 292 |
| His-pAT | 12–25 nM | 350/600 | — | Turn on | Aflatoxin B1 | 0.15 pg mL−1 | 0.46–400 pg mL−1 | 293 |
| hpDNA | 2 nm | 340/575 | — | Turn off | MC-LR | 3 pg L−1 | 0.005–1200 μg L−1 | 294 |
| dsDNA | 2.5 nm | 340/575 | 42 | Turn off | MC-LR | 4.8 ng L−1 | 0.01–1000 μg L−1 | 295 |
| BSA | — | — | — | — | Bisphenol A | 0.12 nM | 1 nM–10 μM | 46 |
| Poly(thymine) | 1–2 nm | 345/598 | — | Turn on | Melamine | 95 nM | 0.1–6 μM | 88 |
| MBA | — | — | — | — | Hydrazine | 1.04 μM | 1.04–425 μM | 125 |
| GSH | 6 nm | 334/432 | — | Turn on | OPD | 93 ng L−1 | 0.15–110 μg L−1 | 296 |
| L-Histidine | 2 nm | 393/492 | 2.4 | Turn off | Fluazinam | 0.25 μM | 1–40 μM | 297 |
| dsDNA | <5 nm | 340/560 | — | Turn on | Acetamiprid | 2.37 nM | 5–500 nM | 298 |
| GSH | 2.61 nm | 330/430′ | — | Turn on | Dinotefuran | 7.04 μM | 10–500 μM | 299 |
| CTAB | 6.28 nm | 254/620 | — | Turn off | Metam sodium | 0.63 mg kg−1 | 1–100 mg kg−1 | 300 |
| L-Cysteine | 5.6 nm | 360/460 | 76 | Turn off | Biliverdin | 233 nM | 0.5–40 μM | 120 |
| Bilirubin | 229 nM | 1–10 μM | ||||||
| BSA | <4 nm | 330/404 | — | Turn off | Bilirubin | 257 fM | 10–150 pM | 60 |
| BSA | 3.3 ± 0.8 nm | 320/405 | 6.4 | Turn on | Bilirubin | 6.62 nM | 10–60 nM | 61 |
| HSA | 3 ± 0.3 nm | 390/646 | 3.6 | Turn off | Bilirubin | 35 nM | 1.25–7.5 μM | 301 |
| 145 nM | 5–28.75 μM | |||||||
| BSA | 3.15 ± 0.38 nm | 355/410 | — | Turn off | Dopamine | 0.1637 pM | 0.1–0.6 nM | 62 |
| dsDNA | — | 340/565 | — | Turn off | Dopamine | 20 pM | 0.1–10 nM | 302 |
| BSA | 2.5 nm | 325/406 | — | Turn on | Dopamine | 0.28 μM | 0.5–50 μM | 303 |
| BSA | 2 nm | 365/640′′ | — | Turn off | Dopamine | 32 nM | 0.1–100 μM | 304 |
| D-Penicillamine | — | 280/650 | 7.16 | Turn off | Histamine | 30 nM | 0.05–5 μM | 305 |
| TFTP | <5 nm | 325/590 | 43 | Turn off | Histamine | 60 nM | 0.1–10 μM | 229 |
| GSH | 2–2.5 nm | 360/595 | — | Turn on | Heparin | 26 nM | 0.1–10 μM | 126 |
| dBSA | 1.7 nm | 350/642 | 2.32 | Turn off | Heparin | 0.26 ng mL−1 | 1.25–250 ng mL−1 | 306 |
| BSA | <4 nm | 330/410 | — | Turn off | Heparin | 0.0406 ng mL−1 | 6–9 ng mL−1 | 307 |
| CTAB | <8 nm | 290/480 | — | Turn on | Carbamazepine | 0.08 μg mL−1 | 0.2–20 μg mL−1 | 308 |
| BSA | <5 nm | 325/420 | — | — | Paraoxon | 12.8 nM | 50 nM–0.5 μM | 309 |
| 0.5–35 μM | ||||||||
| L-Cysteine | 2 nm | 368/493 | 5.8 | Turn off | Tetracycline | 5.6 μM | 0–20 μM | 310 |
| GSH | 373/595 | 3.6 | Turn off | 8.4 μM | 0–20 μM | |||
| GSH | 1–3 nm | 380/600 | 3.6 | Turn off | Vitamin B1 | 4.6 nM | 20 nM–0.1 mM | 311 |
| Ovalbumin | 3.6 ± 1.1 nm | 350/560 | 5.8 | Turn off | Vitamin B1 | 380 pM | 1–1000 nM | 312 |
| Turn on | Doxycycline | 270 nM | 1–1000 μM | |||||
| Adenosine | 2.4 ± 0.3 nm | 285/417 | — | Turn off | Nitrofurantoin | 30 nM | 0.05–4 μM | 313 |
| BSA | 5 nm | 325/400 | — | Turn on | D-Penicillamine | 0.54 μg mL−1 | 0.6–30 μg mL−1 | 314 |
| Lysozyme | 3.4 nm | 335/550 | — | Turn on | Choline | 25 nM | 0.1–80 μM | 315 |
| BSA | 2.1 nm | 320/640 | — | Turn off | Rutin | 0.02 μM | 0.1–100 μM | 41 |
| BSA | 2 nm | 320/400 | — | Turn off | Gossypol | 25 nM | 1–100 μM | 316 |
| Cytidine | 1.5 ± 0.5 nm | 300/380 | — | Turn off | Hemin | 0.045 μM | 0.05–4 μM | 252 |
| — | 3.5 nm | 350/430 | — | Turn off | Hemin | 68 nM | 0–1 μM | 247 |
| AA | 2 nm | 390/455 | — | Turn off | Quercetin | 0.19 μM | 0.7–50 μM | 317 |
| L-Proline | 2.1 nm | 373/453 | — | Turn off | Quercetin | 12 nM | 0.5–50 μM | 318 |
| DNA | 2.8 ± 0.2 nm | 350/670′′ | — | Turn on | Acetaminophen | 0.26 μM | 1–80 μM | 319 |
| BSA | 1–2 nm | x/547 | Turn on | Tramadol | 0.8 nM | 0.003–2.5 μM | 320 | |
| BSA | 1.4 ± 0.2 nm | 545/640 | — | Turn off | Mangiferin | 210 nM | 3–180 μM | 321 |
| GSH | 2.4 ± 0.4 nm | 360/585 | 3 | Turn off | Creatinine | 0.63 μg L−1 | 2.5–34 μg L−1 | 222 |
| BSA | 2.5 nm | 525/643 | — | Turn off | Creatinine | 50 nM | 5–60 μM | 322 |
| GSH | 2.3 nm | 340/585 | — | Turn off | Cholesterol | 2.7 μM | 6–80 μM | 127 |
| PVP | 2.3 nm | 380/425 | — | Turn on | GSH | 17 μM | 0–140 μM | 323 |
| PVP | 3 nm | 380/425 | — | Turn on | GSH | 3 μM | 5–100 μM | 324 |
| dsDNA | — | 340/580 | — | Turn off | GSH | 2 μM | 2–80 μM | 80 |
| Cys | 2 μM | 2 μM–0.1 mM | ||||||
| Hcy | 5 μM | 5 μM–0.2 mM | ||||||
| Poly(thymine) | 5 nm | 340/615 | — | Turn on | GSH | 15 nM | 0–1000 nM | 325 |
| Cys | 12.5 nM | 0–1000 nM | ||||||
| Hcy | 20 nM | 0–1000 nM | ||||||
| DNA nanoribbon | 0.63 nm | 351/594 | 12.3 | Turn off | GSH | 0.43 μM | 24–100 μM | 326 |
| Cys | 0.31 μM | 24–100 μM | ||||||
| PEI | 2–3 nm | 365/495 | 2.63 | Turn on | GSH | 0.26 μM | 0.5–25 μM | 254 |
| Turn on | Cys | 0.34 μM | 1–25 μM | |||||
| Ovalbumin | 5.6 ± 1.1 nm | 370/440 | — | Turn on | L-Lysine | 5.5 μM | 10 μM –1 mM | 327 |
| DNA | 2.8 ± 0.2 nm | 350/670′′ | — | Turn off | Arginine | 0.35 μM | 1–100 μM | 319 |
| Cysteine | 2 nm | 360/490 | — | Turn on | Tryptophan | 75 nM | 0.1–30 μM | 328 |
| Phenylalanine | 840 nM | 1–27 μM | ||||||
| Adenosine | 3.7 ± 0.7 nm | 285/380 | 1.34 | — | Nucleosides | 20 μM | 20 μM–0.5 mM | 329 |
| Cytidine | 1.5 ± 0.5 nm | 300/380 | 0.44 | |||||
| Guanosine | 1.5 ± 0.4 nm | 300/380 | 0.27 | |||||
| DNA | 5 nm | 340/615 | — | Turn on | ATP | 93 nM | 0.2–50 μM | 330 |
| dsDNA | 3–4 nm | 340/598 | — | Turn on | ATP | 5 pM | 0.01–100 nM | 331 |
| Poly(thymine) | <5 nm | 350/620 | — | Turn on | ATP | 10.29 nM | 100 nM–100 μM | 332 |
| GSH | 5.5 nm | 365/605 | — | Turn off | ATP | 0.67 μM | 1–2000 μM | 333 |
| Histidine | — | 350//456 | 1.6 | Turn off | GTP | — | — | 147 |
| dsDNA | 5 nm | 340/600 | — | Turn off | NAD+ | 0.2 nM | 0.2–20 nM | 334 |
| hpDNA | — | 345/635 | — | Turn on | NAD+ | 5 nM | 5 nM–0.5 μM | 335 |
| Silyl | 2–6 | 410/695 | — | Turn on | H2O | 0.0018% v/v | 0.0062–0.19% v/v | 112 |
| AA | 1.2 ± 0.3 nm | 385/498 | 6.63 | Turn on | Urea | 0.01 mM | 0.25–5 mM | 19 |
| GSH | 2.17 nm | 340/585 | 13.91 | Turn off | Urea | 5.86 μM | 20–150 μM | 336 |
| dsDNA | — | 340/596 | — | Turn off | DNA aptamer | 28 nM | 0.05–500 μM | 70 |
| Cocaine | 0.1 μM | 0.5–100 μM | ||||||
| PAA-g-MEA | 0.8 ± 0.2 nm | 360/600 | 5.7 | Turn off | APDC | — | — | 36 |
| DNA | — | 349/617 | — | Turn on | Biotin | 3.1 nM | 10–1000 nM | 337 |
| RNA detection | ||||||||
| dsDNA | — | 340/608 | — | Turn on | microRNA | 1pM | 1pM–10nM | 75 |
| dsDNA | 3–4 nm | 340/608 | — | Turn on | microRNA | 10 pM | 10–400 pM | 76 |
| DNA | — | — | — | — | microRNA | 8.2 fM | 25–300 fM | 81 |
| dsDNA | 2.5 nm | — | — | Turn on | microRNA-21 | 19.05 aM | 100 aM–100 pM | 338 |
| DNA | — | 340/605 | — | Turn on | microRNA-21 | 18.7 pM | 50–1000 pM | 339 |
| DNA | 2.5 nm | 340/580 | — | Turn on | microRNA-21 | 10 aM | 0.1 fM–10 pM | 340 |
| Poly(thymine) | — | 350/630 | — | Turn on | microRNA-122 | 44 fM | 100 fM–1 nM | 341 |
| DNA | 2 nm | 400/490 | — | Turn off | microRNA-155 | 0.6 pM | 1 pM–10 nM | 342 |
| DNA | 2 nm | 400/580 | — | Turn on | microRNA-155 | 11 pM | 50 pm–10 nM | 343 |
| dsDNA | 1.5 nm | x/390 | — | Turn off | microRNA-155 | 36 aM | 100 aM–100 pM | 344 |
| DNA | 5 nm | x/616 | — | Turn on | microRNA-222 | 0.03 fM | 0.5 fM–70 nM | 345 |
| Enzymatic activity detection | ||||||||
| 4-MTP | 2–3 nm | 345/625 | 9 | Turn off | β-Gal | 0.9 U L−1 | 2.5–212 U L−1 | 141 |
| GSH | 2–5 nm | 410/640 | 8.1 | Turn off | β-Gal | 0.7 U L−1 | 2.3–96 U L−1 | 227 |
| dsDNA | — | 340/574 | — | Turn off | Exo III | 0.02 U mL−1 | 0.05–2 U mL−1 | 65 |
| DNA | 5 nm | 340/610 | — | Turn off | Exo III | 0.02 U mL−1 | 0.02–10 U mL−1 | 346 |
| DNA | — | 340/575 | — | Turn off | EcoRI | 870 μU μL−1 | 0.002–0.1 U μL−1 | 347 |
| DNA | 3.58 nm | — | — | — | MMP7 | 5.3 pg mL−1 | 0.01–100 ng mL−1 | 348 |
| DNA | 2.5 nm | 350/620 | — | Turn on | MTase | 0.17 mU μL−1 | 0.1–0.2 U μL−1 | 349 |
| dsDNA | — | 340/590 | — | Turn off | Dam MTase | 0.5 U mL−1 | 0.5–10 U mL−1 | 350 |
| DNA | 2.5 nm | 340/570 | — | Turn off | MNase | 1 mU mL−1 | 1–50 mU mL−1 | 351 |
| TA | 2.04 nm | 360/438 | — | Turn off | PPase | 0.19 U L−1 | 0.5–18 U L−1 | 352 |
| GSH | 2.1 nm | 360/615 | — | Turn on | PPase | 1.3 mU mL−1 | 3–40 mU mL−1 | 223 |
| Poly(thymine) | — | 340/615 | 6.8 | Turn off | S1 nuclease | 0.5 μU μL−1 | 5 μU μL−1–2 mU μL−1 | 353 |
| dsDNA | 5 nm | x/610 | — | Turn off | S1 nuclease | 3 mU mL−1 | 5–80 mU mL−1 | 354 |
| D-Penicillamine | 2–5 nm | 302/646 | 11.2 | Turn off | ACP | 1.3 U L−1 | 3.8–22.8 U L−1 | 220 |
| PDA | 53.7 ± 4 nm | 340/580 | 2.54 | Turn on | ACP | 1 mU L−1 | 1.2 mU L−1–25 U L−1 | 276 |
| dsDNA | — | 335/565 | — | Turn on | ALP | 0.1 nM | 0.1–2.5 nM | 74 |
| GSH | — | 360/610 | — | Turn on | ALP | 0.15 mU mL−1 | 0.5–25 mU mL−1 | 224 |
| BSA | <5 nm | 335/410 | 15.2 | Turn on | ALP | 0.1 mU mL−1 | 0.5–150 mU mL−1 | 355 |
| L-Histidine | 3–4 nm | 390/485 | — | Turn off | ALP | 45 μU mL−1 | 0.5–40 mU mL−1 | 356 |
| dsDNA | 3.2 nm | 340/575 | — | Turn on | ALP | 6 mU L−1 | 0.04–8 U L−1 | 357 |
| Poly(thymine) | 2–3 nm | 340/620 | — | Turn off | ALP | 125 μU mL−1 | 0–62.5 mU mL−1 | 262 |
| dsDNA | 4–5 nm | 340/570 | 3.9 | Turn off | UDG | 0.5 mU mL−1 | 1 mU mL−1–0.1 U mL−1 | 358 |
| Poly(thymine) | 2.87 nm | 400/602 | — | Turn on | UDG | 50 μU mL−1 | 100 μU mL−1–0.01 U mL−1 | 359 |
| Poly(thymine) | 2.5–5.3 nm | 345/650 | — | Turn on | UDG | 50 μU mL−1 | 50 μU mL−1–2 mU mL−1 | 360 |
| Poly(thymine) | 2 nm | 340/520 | — | Turn off | T4 PNKP | 0.25 U mL−1 | 0.25–25 U mL−1 | 361 |
| Poly(thymine) | 2 nm | 340/620 | — | Turn on | T4 PNKP | 0.02 U mL−1 | 0.02–20 U mL−1 | 362 |
| DNA | 3.3 nm | 340/570 | 3.4 | Turn on | T4 PNKP | 0.06 U mL−1 | 0.07–15 U mL−1 | 363 |
| dsDNA | — | 340/565 | — | Turn off | PNK | 0.49 U mL−1 | 0–3 U mL−1 | 364 |
| PEI | 2–3 nm | 365/495 | 2.63 | Turn on | AChE | 1.38 mU mL−1 | 3–200 mU mL−1 | 254 |
| PVP | 2.72 nm | 370/438′ | — | Turn on | AChE | 0.56 U L−1 | 2–70 U L−1 | 365 |
| Poly(thymine) | — | 340/600 | — | Turn off | AChE | 0.05 mU mL−1 | 0.11–2.78 mU mL−1 | 366 |
| DNA | 2.5 nm | 340/570 | 11.2 | Turn on | TdT | 60 mU L−1 | 0.7–14 U L−1 | 367 |
| DNA/RNA | 3 nm | 340/570 | — | Turn off | RNase H | 0.55 mU mL−1 | 0.6 mU mL−1–30 U mL−1 | 368 |
| GSH | 2.4 nm | 300/500 | 17.55 | Turn off | GLU | 0.22 U L−1 | 0.5–6 U L−1 | 369 |
| BSA/Gly–Gly | 3 nm | 330/415′ | — | Turn off | TYR | 44 U L−1 | 0.1–7 U L−1 | 370 |
| DNA | 2.28 ± 0.56 nm | 340/590 | — | Turn on | Thrombin | 0.9 nM | 1–50 nM | 371 |
| GSH | 3 nm | 350/560 | 14.27 | Turn on | HAase | 14 mU mL−1 | 0–0.4 U mL−1 | 372 |
| CS/GSH | <10 nm | 350/574 | 5.2 | Turn off | Lysozyme | 1.6 nM | 5–110 nM | 373 |
| PEI | 2 nm | 374/515 | — | Turn on | PKA | 38 mU mL−1 | 0.1–6 U mL−1 | 374 |
| Protein detection | ||||||||
| DNA | — | 349/617 | — | Turn on | Streptavidin | 0.47 nM | 1–200 nM | 337 |
| Poly(thymine) | 5 nm | 340/600 | — | Turn on | Streptavidin | 0.1 nM | 0.5–1000 nM | 375 |
| Poly(thymine) | — | 340/630 | — | Turn on | Streptavidin | 20 pM | 69.2 pM–17.3 nM | 376 |
| BSA | 1.9 nm | 330/630 | 3.7 | Turn off | Glycoprotein | 2.6 nM | 5–220 nM | 377 |
| DNA | 5 nm | 340/585 | — | Turn on | Actin | 0.12 μg mL−1 | 0–12 μg mL−1 | 378 |
| Poly(thymine) | — | 340/615 | — | Turn off | Trypsin | 42 ng mL−1 | 0.25–1000 μg mL−1 | 379 |
| GSH | 4.5 nm | 340/610 | — | Turn on | Trypsin | 2 ng mL−1 | 2–20 ng mL−1 | 380 |
| dsDNA | 2–3 nm | 350/591 | — | Turn off | Trypsin | 48 pg mL−1 | 1–1000 ng mL−1 | 381 |
| PSS | 3–4 nm | 390/645 | — | Turn on | Trypsin | 20 ng mL−1 | 0.1–6 μg mL−1 | 382 |
| Turn off | Cytochrome C | 0.83 nM | 8–680 nM | |||||
| — | 3.5 nm | 350/430 | — | Turn off | Cytochrome C | 0.8 μM | 1–8 μM | 247 |
| Ferritin | 16.5 nM | 0–0.25 μM | ||||||
| Aptamer | 17.8 nm | 332/463 | 8.2 | Turn on | VEGF165 | 12 pM | 10–800 pM | 383 |
| BSA | <4 nm | 330/410 | — | Turn on | Protamine | 0.12 ng mL−1 | 3–12 ng mL−1 | 307 |
| dsDNA | — | 340/565 | — | Turn off | CEA | 6.5 pg mL−1 | 0.01–2 ng mL−1 | 384 |
| BSA | 9 nm | — | — | Turn on | PSA | 145.7 fg mL−1 | 0.5 pg mL−1–100 ng mL−1 | 385 |
| dsDNA | 3.2 nm | 340/575 | — | Turn on | IgG | 7 pg mL−1 | 0.05–12 ng mL−1 | 357 |
| Temperature sensing | ||||||||
| HSA | 3 nm | 330/414 | 4 | Turn off | Temp | — | — | 50 |
| GSH | 2.3 nm | 410/610 | 5 | Turn off | Temp | — | — | 130 |
| CLEDNN | 1.7 ± 0.4 nm | 373/454 | 7.3 | Turn off | Temp | — | 10–55 °C | 134 |
| GSH | 2.4 ± 0.4 nm | 400/610 | 4.5 | Turn off | Temp | — | 20–45 °C | 386 |
| pH sensing | ||||||||
| Trypsin | 2.03 ± 0.46 nm | 363/455 | 1.1 | Turn off | pH | — | — | 39 |
| BSA | 1.6 ± 0.5 nm | 320/420 | — | Turn on | pH | — | — | 49 |
| BSA | 2.5 nm | 325/406 | — | Turn on | pH | — | — | 303 |
| Cysteine | 2.3 nm | 365/490 | 8.8 | Turn off | pH | — | — | 133 |
| GSH | 1–3 nm | 380/600 | 3.6 | Turn off | pH | — | 4–12 | 311 |
| PEI | 2 nm | 380/498 | 7.5 | Turn on | pH | — | — | 387 |
| CEW | 2.4 ± 0.2 nm | 337/417 | 0.98 | Turn on | pH | — | — | 388 |
| L-Tyrosine | 1–2 nm | 430/477 | — | Turn off | pH | — | 5.02–11.92 | 389 |
| Infections detection | ||||||||
| DNA | 6 nm | — | — | Turn on | HBV | 12 × 109 molecules | 12 × 109–12 × 1013 molecules | 390 |
| dsDNA | <4 nm | 340/600 | — | — | TB | 5 fg μL−1 | 10–100 fg μL−1 | 79 |
| CP25 | 2.3 ± 0.3 nm | 460/630 | — | Turn off | CTV | 220 pg mL−1 | 400 pg mL−1–25 ng mL−1 | 391 |
As mentioned earlier, highly fluorescent, and biocompatible CuNCs have found applications in the sensing of metal cations such as mercury, iron and lead, not only for environmental control, but also in clinical practice. The advances in synthetic procedures have led to sensors with high sensitivity and low detection limits, even down to fM concentrations of a target material in some cases with negligible amounts of the sensor. The possible bio-applications of CuNCs are countless. In most applications, the detection of a certain molecule is based on fluorescence quenching. In the following, we describe some sensing applications, and we do this according to the target species to be detected. More detailed descriptions and applications of CuNCs can be found in the reviews by Yi Lv,85 Chuanxi Wang,392 Bo Feng,393 Baisong Chang394 and Yuming Huang.395 Ronghua Yang396 and Erkang Wang397 summarized the recent progress in biosensing applications of CuNCs utilizing DNA scaffolds.
Iron is an essential element for red blood cells for the transport of oxygen. Ferrous/ferric iron is a redox system, and it is involved in electron transport in bioprocesses. Under normal conditions, the iron content for a healthy person is 4–5 g and both lower and higher amounts are problematic. Iron is toxic upon overload and can cause severe diseases such as hemochromatosis. At the same time, insufficient iron levels in the blood can lead to the development of iron deficiency anaemia. In addition to clinical blood tests and attempts to prepare nanomaterial-based sensors, CuNCs have also been extensively applied in ferric ion sensing in water, urine and serum samples based on the fluorescence quenching of a sensor signal.30,135,138,145,148,245–249 For example, a dopamine-protected CuNCs-based biosensor could properly operate in the range of 5–300 μM with a LOD of 1.2 μM.148 However, the extensive research in the field resulted in even lower LOD values, down to 10 nM, when tannic acid (TA)-protected CuNCs were implemented as a sensor.145 Note that the CuNCs-based ferric ion sensors mentioned above showed impressive sensitivities, however, none of them have made it to clinical practice so far.
Copper is required for the proper functioning of several Cu-containing enzymes such as ceruloplasmin, cytochrome-c oxidase, etc. According to the “WHO Guidelines for Drinking-water Quality”, the allowed concentration of copper in drinking water is 2 mg L−1 and the tolerable uptake is 10 mg per day per kg of body weight.398 During short- and long-term exposure from dietary supplements or drinking water, the excessive intake of copper builds up in the kidneys, brain and eyes and can cause severe damage. Apart from standard analytical techniques, CuNCs-based sensors have found important applications for detecting Cu ions in drinking water samples.115,250,252–254 Zhihe Qing et al.251 prepared poly(thymine)-templated and highly fluorescent (QY 11.2%) CuNCs as a turn-on sensor for Cu2+ detection and its toxicide screening. The sensor showed a good performance and could detect 5.6 μM of copper in contaminated water samples. However, the ongoing research and the race for preparing template-selective and ion-sensitive/selective sensors with excellent detecting capabilities led to a sensor with a LOD as low as 1 nM with a linear concentration range of 0.02–34 μM.52
Lead contamination became a global threat to human health and upon exposure, it can accumulate in the brain, liver, kidney, and bones. The WHO states that blood lead concentrations below 5 μg dL−1 can reduce the IQ of children, cause behavioural and learning difficulties, and attack the brain and central nervous system to cause coma, convulsions and even death. The guideline value of lead in drinking water is 10 μg L−1.399 On top of clinically accepted blood testing, researchers have developed CuNCs-based fluorescent sensors. For example, Chuanxi Wang et al. reported the synthesis and application of GSH-protected CuNCs as a label-free fluorescent probe to detect Pb2+ in CAL-27 cells.137 The proposed fluorescent probe showed excellent turn-off detection ability down to 1 nM with a linear response in the range of 1–160 nM. Xia Chu and co-workers prepared another sensor that was highly selective and sensitive and could operate at even lower concentrations (LOD 0.4 nM)255 than the lead ion sensors previously reported by Junhua Chen et al. (LOD 5 nM),72 N. Vilar-Vidal et al. (LOD 4.9 μM),95 Ran Liu et al. (LOD 142 nM)242 and Fei Qu et al. (LOD 30 μM).127
Chromium salts have many applications in industry and vary in their toxicological profiles from one oxidation state to another. Cr(VI) is extremely toxic and is a carcinogen, whereas Cr(III) is an essential nutrient for organisms to maintain normal physiological functioning.400 Thus, for the clinical assessment and diagnosis of chromium poisoning via inhalation or ingestion, fast and sensitive analytical methods are required. CuNCs applications were extended towards that goal too. For instance, Malin Cui et al. prepared cys-CuNCs as a fluorescent probe for Cr+6 with a LOD of 43 nM.116 Mixed ligand shell BSA/thiosalicylic acid (TSA)-CuNCs showed much better sensitivity towards Cr+6 with a LOD of 3.54 nM.260 For Cr+3 detection, Ajnesh Singh et al.261 and Can Chen et al.262 developed a hybrid CuNCs@organic nanoparticle complex and poly(thymine)-CuNCs sensor with minimum detection ability of 3 μM and 30 nM, respectively.
Aluminum is found in consumer products like antacids, food additives, cosmetics, etc. However, the FDA set a limit for bottled water of 0.2 mg L−1. Dithiothreitol (DTT)-protected CuNCs were suggested as an accessible sensor for the detection of aluminum in food samples.263 Under optimized conditions, the fluorescent probe exhibited a good linear relationship in the range of 0.01–7 μM with a detection limit of 0.01 μM. Later, Chanida Boonmee et al.264 and Jiawei Pang et al.265 developed sensors with largely improved LODs of 26.7 nM and 62 nM, respectively.
Zinc is an essential mineral, and it is involved in DNA synthesis and normal taste perception, supports wound healing, immune function, and reproductive health. The WHO set a provisional maximum tolerable daily intake (PMTDI) of 1.0 mg per kg of body weight.401 High doses of zinc can cause acute or chronic issues upon short or long-term exposure. Thus, the estimation and detection of zinc levels in water are of great importance.110 Liyun Lin et al.225 and Xueqing Gao et al.259 demonstrated the applicability of GSH- and Cys-CuNCs as fluorometric probes for zinc ion detection with LODs of 1.17 μM and 1 μM, respectively. However, BSA-CuNCs proved to be a better Zn2+ sensor with a LOD of 15 nM.258
CuNCs-based sensors were also applied for the detection of Co(II),59 Ca(II)256 and Mn(II).257 Sensing applications of metal cations using CuNCs are also summarized in the review by Jintong Song.212
Iodine is an important micronutrient for human growth and metabolism. Both iodine deficiency and surplus are harmful for the human body and can cause severe diseases (increased infant mortality and hypothyroidism in the case of deficiency, and hyperthyroidism in the case of excess of iodine). Reported fatal doses of iodine during ingestion are 200 mg to 20 g. CuNCs-based sensors were applied for iodine detection in water and urine samples.272 Very recently Yu-Ting Tai et al.138 reported the detection of iodine in urine samples with LOD of 2.02 μM using fluorescent CuNCs. However, much earlier, Yaping Zhong et al.29 reported PEI-protected CuNCs with much higher sensitivity, and a LOD value of 100 nM. TA-CuNCs as an iodine biosensor further lowered the LOD to 18 nM.270 However, ssDNA-CuNCs showed excellent analytical performance for iodine with LOD as low as 15 nM and linear ranges of 0.05–40 μM and 40–80 μM with two different slopes.271
Hypochlorous acid (HOCl) belongs to the family of ROSs, has good microbicidal activity in the cells and is involved in the fight against pathogens and infections. Apart from being an essential chemical for biological processes, an excess of the acid can cause different diseases like atherosclerosis, arthritis and even cancer. Thus, the precise determination and sensing of hypochlorous acid concentrations in water samples or biota are crucial and of great importance. CuNCs protected with PVP polymer are found to be excellent fluorescent probes for hypochlorite detection. In an earlier report by Qin Tang et al., the PVP-protected CuNCs showed high sensitivity towards ClO− and a LOD of 0.1 μM.33 However, very recently, Weiru Dong et al. applied an iodide-enhanced approach to increase the detection sensibility and surpassed the previously reported LOD value (LOD 19 nM).34
Nitrite (NO2−) ions are a part of the nitrogen cycle and are involved in the oxidation of haemoglobin (Hb) to methaemoglobin (metHb). The impairment of oxygen transport due to the elevated concentrations of nitrite ions in the bloodstream leads to the development of a condition called methaemoglobinaemia. It is also believed to cause gastric cancer and other severe diseases. The WHO “Guidelines for Drinking-water Quality” set 1 mg L−1 as the maximum contaminant level for nitrite ions in drinking water.403 Considering the impact of exposure to nitrite ions (short/long term), several research groups have attempted to outperform the detection limit normally obtained by routine spectroscopic analysis and monitoring of water quality (standard LOD of 0.01 to 1 mg L−1). Three independent research groups reported the applicability of CuNCs as a sensitive probe for nitrite ions. For instance, Jinshun Cang et al. developed a fluorometric sensor for the detection of nitrite and cyanide anions in water samples with LODs of 5 μM and 5 nM, respectively.273 Improved sensitivity was achieved by Suqin Han et al. with a LOD of 95.4 nM.274 Even better sensitivity was reported for stabilizer-free CuNCs as fluorometric probes for nitrite ions.275 A remarkable sensitivity of 3.6 nM was achieved with a probe operating in a wide analytical range (12.5 nM–125 μM).
Phosphates are essential electrolytes/minerals for the body and are involved in many processes such as bone and teeth formation. The normal serum phosphate concentration ranges from 2.5 to 4.5 mg dL−1 (0.81 to 1.45 mmol L−1). High phosphate levels (hyperphosphatemia) or very low phosphate levels (hypophosphatemia) in the blood indicate certain conditions, including chronic kidney disease, which require special medical attention.404 Taking advantage of their fluorescence properties, Haiyan Cao et al. demonstrated the applicability of TA-CuNCs as a turn-on sensor for phosphate ions in water samples with LOD of 9.6 nM.144 Six-fold enhancement in sensitivity for phosphate ions was achieved when polydopamine-CuNCs with AIE properties were applied as a fluorometric sensor.276 The minimum detectable phosphate ion concentration in tap water and human serum was 1.5 nM. Can Chen et al. developed poly(thymine)-CuNCs for the detection of pyrophosphate (PPi, P2O74−) ions in human serum samples with LOD of 5 nM.262
The applications of CuNCs were also extended towards the detection of fluoride ions.265
On the other hand, H2O2, a reactive oxygen species (ROS), is crucial for oxidative cellular signalling, it induces intracellular stress and accelerates aging. Hence, the early diagnosis of H2O2 can prevent the development of certain diseases. Therefore, the detection of hydrogen peroxide is of great importance and CuNCs-based sensors have already been reported.54,87,90,114,127,142,279,280 For example, Lianzhe Hu et al. reported the synthesis of BSA-capped CuNCs to detect hydrogen peroxide and glucose with a detectable minimum concentration or limit of detection (LOD) of 10 and 100 μM, respectively.58 Hong Qun Luo and co-workers succeeded in the preparation of PEI-CuNCs,26 which could operate in a linear range of 0.5–10 μM and 10–100 μM for H2O2 and glucose, respectively. The sensor could detect the target at concentrations as low as 0.4 and 8 μM for H2O2 and glucose, respectively. Tingyao Zhou et al.111 and Xuxian Su et al.219 reported the synthesis of DHLA-CuNCs and L-cys/GOx-CuNCs, which showed better sensitivity towards hydrogen peroxide (LOD 0.3 μM)111 and glucose (LOD 1.5 μM),219 respectively. However, Hai-Bo Wang et al. reported the synthesis of dsDNA-templated CuNCs, which is so far the most sensitive fluorescent probe for the detection of hydrogen peroxide and glucose.277 The sensor can detect H2O2 and glucose with LODs of 3 nM and 12 nM, respectively with a wide dynamic range (10 nM–2 μM for hydrogen peroxide and 50 nM–100 μM for glucose). In general, the detection and PL quenching of a sensor is believed to be caused by the oxidation of CuNCs by hydrogen peroxide.
Yaping Zhong et al. made use of 6-thio-β-cyclodextrin (β-CD)-templated CuNCs as a peroxidase mimetic to substitute for GOx in biocatalytic glucose oxidation reaction.278 β-CD on the cluster surface acts as a pocket for host-guest interactions and provides a catalytic site for the non-enzymatic oxidation of glucose. Within the proposed method, the glucose and H2O2 detection limits were 0.4 μM and 0.2 μM, respectively.
4.1.4.1. Acid detection. Picric acid (2,4,6-trinitrophenol, TNP) is a pale yellow and odourless solid with poor solubility in water. Normally, it is safe to handle when hydrated but it becomes extremely dangerous and explosive when dry. Besides, it readily forms even more sensitive explosives when in contact with many metals (copper, lead, mercury, zinc, etc.) when subjected to heat, friction, or impact. Upon inhalation or absorption through the skin, it can cause severe health problems. The determination of picric acid in drinking water and its impact on the environment became an issue for communities. In addition to the common detection methods, CuNCs have attracted much interest due to the fast, effective, and sensitive detection of TNP with low LOD values. So far, most of the reported (published in 2018) fluorometric CuNCs sensors are based on fluorescence quenching due to TNP. GSH-,282 DNA-,283 cysteamine-,284 and ascorbic acid-templated285 CuNCs were reported with LODs of 2.74 μM, 0.03 μM, 139 nM, 0.98 μM, respectively. The PL quenching of the clusters due to the electrostatic interaction between the electron acceptor (TNP) and electron donor clusters was ascribed to the FRET phenomenon in BSA-CuNCs.51 The sensor was able to detect the lowest concentration of TNP (LOD 120 nM) as compared to the other sensors, with a larger analytical range (0.8–100 μM). In another report by Krishnamoorthy Shanmugaraj et al., the reduction of fluorescence for L-cys-protected CuNCs286 was ascribed to the inner filter effect.
Folic acid (FA), a synthetic form of Vitamin B, is very crucial for early development during pregnancy; it is involved in many physiological processes promoting the formation and regeneration of blood cells and preventing neurovascular defects. The importance of FA will be described in more detail in section 4.2.1. Two reports in the literature are devoted to the detection of folic acid using L-cys (chemiluminescence (CL) method)274 and ovalbumin-protected CuNCs (fluorometric method).287 Based on the changes in the CL intensity time profiles upon the addition of FA, the deducted LOD value was 69.8 nM and the method had an application range of 0.1–10 μM.274 A much wider linear response towards FA was obtained by fluorometric detection using ovalbumin-CuNCs (0.5–200 μM); however, the method was slightly less sensitive (LOD 0.18 μM).287
Ascorbic acid (AA), also known as vitamin C, is a vital nutrient for the body and is involved in the production of collagen, in the metabolism of proteins, and in the formation of blood vessels to heal wounds and form scar tissue, etc. Ascorbic acid is widely used as a common reducing agent in cluster chemistry, and also became a detection target in biosensing. Among the commonly used diagnostic methods, a novel fluorometric method based on the application of CuNCs became very popular. Hanbing Rao et al. reported the applicability of TA-CuNCs as a turn-on fluorometric sensor for AA.143 The sensor could operate in the linear range of 0.5–10 μM with LOD of 0.11 μM. The sensing of humic acid (found in soil and water, LOD 0.4 mg L−1)281 and kojic acid (used in cosmetics and food colouring, LOD 0.07 μM)47 were also successfully conducted using poly(thymine)- and BSA-CuNCs, respectively.
The applicability of CuNCs was extended towards the sensing of the explosives trinitrotoluene (TNT)288 and m-dinitrobenzene (m-DNB)289 as analogue molecules to picric acid. The LODs were 14 pM and 0.13 μM, respectively.
4.1.4.2. Organic dyes and toxins detection. Sudan dyes are synthetic red azo dyes that were widely used in foodstuff and commercial manufacturing for colouring purposes. However, they are now classified as potential carcinogens and are banned from use in many countries. Nevertheless, the contamination of food with Sudan dyes continues to be a problem. Even though several tests have already been developed for these kinds of dyes (formally known as industrial solvent dyes), fast and extremely efficient and sensitive fluorometric sensors are still in demand. Yu Ling et al. prepared PEI-CuNCs as a sensitive and selective fluorescence probe for Sudan I–IV dyes based on the PL quenching of blue-emitting clusters (as a consequence of FRET).27 With the developed sensing principle, the LODs for Sudan I–IV dyes were 65 nM, 70 nM, 45 nM and 50 nM, respectively.
Quinoline yellow is another artificial colouring dye used in the food and beverage market to impart an intense greenish-yellow colour. However, some countries including Canada and the USA have banned its use and other countries have agreed to reduce the limit of daily intake to several micrograms. Thus, the regular monitoring of the quinoline yellow concentrations in food and drinks is of great importance for food safety administration. Unni Sivasankaran et al. applied L-cys-CuNCs as a fluorometric probe to detect the amount of the dye present in soft drinks and sweets.122 The developed sensor could operate in a range of 0.2–5.5 μM with LOD of 0.11 μM.
4-Aminoazobenzene also known as aniline yellow, is a textile dye mainly used to colour lacquer, varnish, wax products, oil stains and styrene resins. It is among the 24 prohibited textile dyes and is considered as a carcinogen. Thus, the detection of aniline yellow in water samples is of crucial importance. For monitoring the water contamination with the dye, Qiang Li et al. have prepared a highly luminescent (QY of 33.6%) CuNCs-based turn-off fluorometric sensor with LOD of 1.44 μM.290
Ochratoxin A (OTA) is a natural mycotoxin produced by several fungi. The poor storage of agricultural commodities such as cereal grains, dried fruits and drinks can lead to the growth of fungi and consequently, release OTA into foodstuff. Thus, the detection of OTA toxin is of major concern for food safety regulations. Chunxia Song et al. have developed dsDNA-CuNCs to detect OTA in corn samples with LOD of 5 ng mL−1.291 Yue He et al. prepared polyT-CuNCs as a fluorometric sensor for OTA in spiked diluted red wine with a LOD of 2 nM.292
Another foodborne toxin called aflatoxin B1 (AFB1) is produced by fungi and is found in food crops. The ingestion of products contaminated with AFB1 is potentially life-threatening. The WHO classifies AFB1 as a potent carcinogen that can affect all organ systems.405 A plethora of methods is now available to detect AFB1 and related toxins, however, more sensitive and selective tests are yet to be developed. For this reason, Ying Xiong et al. developed a novel fluorometric sensor for AFB1, utilizing histone-ds-poly(AT)-CuNCs.293 With the designed sensing strategy, the minimum detectable AFB1 concentration was 0.15 pg mL−1 with a rather large analytical range of 0.46–400 pg mL−1.
Microcystin-leucine-arginine (MC-LR) is the most toxic among the microcystins produced by cyanobacteria in water systems. The contamination of water ecosystems with MC-LR represents a global problem and can cause a series of related health problems. Apart from traditional methods, Yanli Zhang et al. have prepared novel fluorometric probes based on hairpin-CuNCs294 and dsDNA-CuNCs.295 The developed sensors were able to detect the toxin in real water samples with LODs of 3 pg L−1 and 4.8 ng L−1, respectively.
4.1.4.3. Industrial and agricultural products detection. Industrial safety is one of the top challenges to ensure a smooth-running operation in the workplace and limit the exposure to toxic chemicals. Although high safety standards are fundamental for every aspect of production in many countries, many industrial chemicals, even at low concentrations, remain a risk and present a danger to workers and customers. Among the potentially risky chemicals is bisphenol A, which is widely used in the production of polycarbonate, epoxy resins and plastic bottles. However, at high temperatures and pressures, the released bisphenol A from plastic bottles contaminates the water, which presents a potential risk related to cancers and cardiovascular diseases. Shuangjiao Xu et al. reported the applicability of BSA-CuNCs as a chemiluminescence probe to detect bisphenol A in water samples with LOD of 0.12 nM.46
Melamine has many industrial applications including the production of melamine resins, laminates, glues, fertilizers and in food, for example in milk to increase the apparent protein content (melamine is a nitrogen-rich compound). However, high concentrations or exposure to melamine causes inflammatory reactions and hyperplasia in the urinary bladder in animals. Although no apparent data are available about the toxicity in humans, the WHO guidelines strongly limit the tolerable daily intake of melamine in animal and infant food.406 Therefore, it is of crucial importance to regularly monitor the amount of melamine and its analogues in food. Hong-Wei Zhu et al. applied poly(thymine)-CuNCs as a fluorometric probe for the determination of melamine in milk products.88 The designed probe showed excellent analytical performance with the highest reported sensitivity towards melamine, so far (LOD of 95 nM).
Hydrazine is highly used in materials synthesis as a reducing agent. Its industrial applications range from the preparation of polymer foams to pesticides, it is also a precursor for many pharmaceuticals and has recently been used as a rocket fuel (as a safer alternative to hydrogen). As a chemical, it is highly toxic and can cause irritation to the eyes and skin, and damage the liver and kidneys. Thus, the detection of hydrazine in the environment is of great importance. Xiaohui Gao et al. studied the electrocatalytic performance of mercaptobenzoic acid (MBA)-CuNCs loaded on activated carbon to oxidize and detect hydrazine.125 From the linear curve fitting between the current density and the hydrazine concentration, the LOD value of 1.04 μM was successfully derived.
Among the industrial amines, o-phenylenediamine (OPD) was widely used in cosmetics; however, it has been banned due to its toxicity, carcinogenicity, and possible disruption of genetics. Motivated by these issues, Yujun Ma et al. have developed GSH-CuNCs as a fluorometric probe to detect OPD in rivers and textile dye wastewater samples.296 The developed sensor was very sensitive to OPD, with a detection limit of 93 ng L−1.
Recently, Zhifeng Cai et al. reported the applicability of L-histidine-CuNCs as a potent fluorometric probe for the detection of fluazinam (agricultural fungicide) in lake and tap waters.297 The sensor could operate in a wide analytical range of 1–40 μM with LOD of 0.25 μM. Kaimei Fan et al.298 and Yue Yang et al.299 applied CuNCs for the detection of neonicotinoid insecticides such as acetamiprid (brand name: Assail) and dinotefuran in water and food samples. The lowest detectable amounts of acetamiprid and dinotefuran were 2.37 nM (ref. 298) and 7.04 μM (ref. 299) using dsDNA- and GSH-CuNCs-based fluorometric probes, respectively. CTAB-encapsulated CuNCs were successfully applied in the detection of metam sodium (fungicide) in real samples with a LOD of 0.63 mg kg−1.300
4.1.4.4. Bilirubin/biliverdin detection. Bilirubin (yellowish compound, strong antioxidant) is a metabolite produced in the catabolic pathway of the heme breakdown to clear the body from waste products due to the death of aging red blood cells. The catabolism of heme proteins happens under the action of heme oxygenase to produce biliverdin (green tetrapyrrolic bile pigment), which is later reduced to bilirubin by biliverdin reductase. It is important for the body to excrete bilirubin as it is potentially toxic. High bilirubin levels indicate an unusual rate of red blood cell breakdown or that the liver is not clearing the bilirubin from the bloodstream properly. With this in mind, Sanu K. Anand et al. prepared L-cys-CuNCs as a point of care analysis probe for the detection of both biliverdin and bilirubin.120 The fluorescence quenching of the clusters’ emission upon the gradual addition of bile pigments was ascribed to the static quenching process. The limits of detection for biliverdin and bilirubin with the designed sensor were 233 nM and 229 nM, respectively. When the human serum albumin (HSA) was used as a templating matrix for the preparation of clusters, the resulting red emissive HSA-CuNCs could detect the bilirubin in two linear concentration ranges with LODs of 35 nM and 145 nM.301 Although this sensor significantly improved the detection sensitivity (with respect to laboratory tests), it was further greatly increased when BSA was implemented as a template. With the assistance of Fe3+, the fluorescence quenching of the system can be restored by the addition of bilirubin by taking advantage of its antioxidant properties.61 The minimum detectable bilirubin concentration was 6.62 nM. Without the addition of Fe3+, the BSA-CuNCs showed excellent sensing properties for bilirubin with LOD as low as 257 fM.60
4.1.4.5. Neurotransmitter detection. Neurotransmitters are molecules used by the nervous system to transmit messages between neurons, or from neurons to muscles. Most neurotransmitters are either small amines, amino acids, or neuropeptides. Many neuromodulators, such as dopamine (catecholamine family), are monoamines. Dopamine is involved in many functions, including motor control, reward and reinforcement, and motivation. Histamine, one of the major monoamines, plays a role in metabolism, temperature control, regulating various hormones, and controlling the sleep-wake cycle, among other functions. Dopamine deficiency is related to certain medical conditions, including depression and Parkinson's disease. Thus, the detection of dopamine levels for the diagnosis of neurological diseases is of great importance. High levels of histamine are associated with allergic reactions and the disruption of normal bodily functions. Normal dopamine and histamine levels in the blood are 0 to 30 pg mL−1 (195.8 pmol L−1) and 25–65 ng mL−1, respectively. For the detection of dopamine levels in both serum and urine samples, CuNCs prepared in various templating matrices have been applied as a sensing probe. Among them, BSA-CuNCs were the most applied. R. S. Aparna et al.,62 Zhuang Miao et al.,303 and Weijie He et al.304 reported the applicability of BSA-CuNCs as a sensor to detect dopamine with LODs of 0.1637 pM, 0.28 pM and 32 nM, respectively. dsDNA-CuNCs302 were also applied as a dopamine sensor, however, the sensitivity was lower as compared to BSA-CuNCs. Ailing Han et al. prepared 2,3,5,6-tetrafluorothiophenol (TFTP)-protected CuNCs with self-assembled induced emission (SAIE) properties (QY 43%) and tested them for histamine sensing.229 The developed sensor could detect histamine at its lowest concentration of 60 nM in the range of 0.1–10 μM. Furthermore, the histamine detection sensitivity was increased twice (LOD 30 nM) when D-penicillamine-CuNCs were applied as a fluorometric sensor with AIE enhancement properties.305
4.1.4.6. Drugs, antibiotics, and other pharmaceuticals detection. Heparin (Hep) is a naturally occurring, highly sulphated polysaccharide (highly negatively charged macromolecule) that is widely used as a potent anticoagulant therapeutic drug for the treatment and prevention of thrombotic diseases and for maintaining blood fluidity in extracorporeal devices. Bleeding and heparin-induced thrombocytopenia are the main adverse reactions associated with heparin therapy. Laboratory monitoring of the antithrombotic efficacy of heparin can reduce the risk of haemorrhage. Thus, a fast and efficient clinical test for heparin is highly desired. Taking advantage of the fluorescence properties of CuNCs, they were applied as a point-of-care diagnostic sensor to monitor the heparin concentrations in serum samples. Bo Yin et al. made use of GSH-CuNCs attached to the surface of functionalized reduced graphene oxide (RGO) in the form of a CuNCs/RGO nanocomposite as a turn-on sensor for heparin.126 The weakly emissive nanocomposite (due to the photoinduced electron transfer from CuNCs to RGO) became highly emissive upon the addition of Hep. The competitive interaction of heparin with RGO released the CuNCs from the composite and restored its original fluorescence. Thus, by following the PL intensity enhancement of the sensor, the concentrations of heparin can be quantitively determined. As such, with the above-mentioned sensing principle, the minimum detectable amount of Hep was found to be 26 nM.126 In contrast, BSA-CuNCs were applied as a turn-off sensor for Hep.306,307 The quenching of the PL exhibited a linear correlation upon the gradual increase in heparin concentration. Moreover, the sensing of Hep was very much dependent on whether denatured or native BSA was used to template CuNCs. With denatured dBSA-CuNCs as a sensor, the lowest detectable Hep concentration was 0.26 ng mL−1.306 However, the native BSA-CuNCs showed much better analytical performance with LOD of 0.0406 ng mL−1.307
The applicability of CuNCs was extended towards the sensing of other drugs such as carbamazepine (anticonvulsant drug, used to reduce nerve impulses during seizures and nerve pain) with LOD of 0.08 μg mL−1,308 and paraoxon (ophthalmological drug against glaucoma) with LOD of 12.8 nM.309
Antibiotics and similar drugs are used to treat bacterial infections by killing or inhibiting the growth of disease-causing microorganisms. However, the inappropriate use or overuse of antibiotic drugs can cause the microorganisms to develop resistance to the antibiotics, which is a serious problem. Several techniques have been developed and are widely applied in clinical trials to detect the residues of different antibiotics in the environment. Tetracyclines are a class of antibiotics used to treat respiratory tract infections and certain infections of the skin, eye, lymphatic, intestinal and urinary systems. Zhuosen Wang et al. have developed L-cys- and GSH-CuNCs as tetracycline sensors with LODs of 5.6 μM and 8.4 μM, respectively.310 Ovalbumin- and GSH-CuNCs, on the other hand, were applied as sensors for doxycycline with LOD of 270 nM (ref. 312) as well as for the detection of vitamin B1 with LODs of 380 pM (ref. 312) and 4.6 nM.311 CuNCs have also been also applied to detect nitrofurantoin (also used as a fungicide, LOD 30 nM),313D-penicillamine (LOD 0.54 μg mL−1),314 choline (LOD 25 nM),315 rutin (LOD 0.02 μM),41 gossypol (LOD 25 nM),316 hemin (brand name: Panhematin, LODs 0.045 μM (ref. 244) and 68 nM (ref. 248)), quercetin (LODs 0.19 μM (ref. 317) and 12 nM (ref. 318)), acetaminophen (brand name: Paracetamol, LOD 0.26 μM),319 tramadol (LOD 0.8 nM)320 and mangiferin (LOD 210 nM).321
4.1.4.7. Disease marker detection. Creatinine is a waste product of muscle tissue breakdown; it is filtered through the kidneys and excreted in the urine. The creatinine levels in the blood, as well as the rate of creatinine clearance, indicate the kidney function and test for kidney diseases. Clinical tests for creatinine are time-consuming, and novel, fast methods with high precision are still in demand. Roghayeh Jalili et al. have reported the use of GSH-CuNCs as a creatinine sensor taking advantage of the Al3+-triggered AIE of the original clusters.222 The sensing is based on the fluorescence quenching of the signal upon the addition of creatinine due to the coordination with Al3+. A linear dependence of PL versus concentration has been observed with LOD of 0.63 μg L−1. A LOD of 50 nM was reached when BSA-CuNCs were implemented as a turn-off sensing platform for creatinine, which is directly applicable for clinical samples.322
Cholesterol is an essential substance for the body, and it is involved in the production of hormones and vitamin D, as well as in the digestion of food. Nevertheless, the high levels of cholesterol in the blood (mostly due to the diet) represent a silent danger and can cause heart attacks (due to the build-up of plaques that can block or narrow arteries). Thus, the analysis of cholesterol levels in the blood is a prerequisite for the assessment of heart disease. Although clinically available cholesterol tests called a lipid panel or a lipid profile are quite precise for the determination of the cholesterol levels in the blood, with the advancement of technology, novel and fast tests could be an attractive alternative. Considering the necessity to develop novel fluorometric tests, Fei Qu et al. developed a turn-off sensor based on the AIE of GSH-CuNCs.127 The synergistic effect of Pb2+ and Zr4+ on GSH-CuNCs greatly enhanced the PL properties and the latter was applied in the detection of cholesterol with the detection limit down to 2.7 μM.
4.1.4.8. Biothiol and biogenic amino acid detection. Cysteine and lysine are among 20 naturally occurring biogenic amino acids that serve as building blocks to form polypeptides and proteins. Like the other amino acids, cysteine and lysine are abundant in the L-form. Homocysteine (Hcy) is a sulphur-containing amino acid that is formed during the metabolism of methionine (Met, essential amino acid) to cysteine (Cys). These biothiols are necessary for many bodily functions. A high homocysteine level, also called hyperhomocysteinemia, can contribute to arterial damage and blood clots in blood vessels and can indicate a deficiency in vitamin B-12 or folate. The concentration of total cysteine (tCys) in serum/plasma is normally 250 μmol L−1, which is 20-fold higher than the plasma tHcy level. Hyperlysinemia is a genetic condition caused by the deficiency of the aminoadipic semialdehyde synthase enzyme that breaks down the lysine. Glutathione is a naturally occurring tripeptide composed of glutamine, cysteine, and glycine. The thiol group from the Cys is a potent reducing agent. As an antioxidant, GSH plays a role in detoxification, cellular reactions, the regulation of protein and gene expressions, etc. A deficiency of GSH puts the cell at risk for oxidative damage. Several clinically proved methods are used for the detection of biothiols in the blood, including chromatographic and mass spectrometric methods. However, a facile and rapid detection method is still required for faster clinical diagnosis. On this note, Tianzi Li et al.323 and Yu-e Shi et al.324 implemented the PVP-CuNCs for the detection of GSH in human serum samples with LODs of 17 μM and 3 μM, respectively. Yihui Hu et al. have developed a dsDNA-templated CuNCs as a fluorescence probe for the detection of GSH, Cys and Hcy in biological fluids.80 The LODs for GSH and Cys were 2 μM, and for Hcy it was 5 μM. Jinlan Yang et al. developed the UV-light-induced synthesis of PEI-CuNCs and applied them in the detection of GSH and Cys with LODs of 0.26 μM and 0.34 μM, respectively.254 However, much better sensitivity and analytical performance for biothiols were achieved when poly(thymine)-CuNCs were applied as a probe.325 The minimum detectable concentrations of GSH, Cys and Hcy were 15 nM, 12.5 nM and 20 nM, respectively. Another biothiol sensor based on DNA nanoribbon-templated CuNCs with QY as high as 12.3% has been applied for the detection of GSH and Cys.326 Within the developed sensing strategy, the lowest detectable concentrations of analytes were 0.43 and 0.31 μM for GSH and Cys, respectively. A detection platform for biogenic amino acids was also established using templated CuNCs. For example, Xiangning Bu et al. developed a hybrid DNA-CuNCs/carbon dots sensor for the detection of arginine with LOD of 0.35 μM.319 Mingming Zhang et al. have developed ovalbumin-CuNCs as a turn-on fluorescence probe to detect L-lysine with LOD of 5.5 μM for a wide analytical range (10 μM–1 mM).327 Yasaman-Sadat Borghei et al. applied Cys-CuNCs as a chemiluminescence probe for the detection of tryptophan and phenylalanine with LODs of 75 nM and 840 nM, respectively.328
4.1.4.9. Nucleosides/nucleotides detection. Nucleosides are a structural subunit of nucleic acids, consisting of a molecule of sugar (ribose or deoxyribose) linked to a nitrogen-containing organic ring compound (pyrimidine-based: cytosine, thymine, uracil, or purine-based: adenine or guanine). Nucleotides are composed of nucleosides and phosphate groups. Apart from their intracellular functions as building blocks for the genetic code, nucleosides and nucleotides play an important role in extracellular compartments as signalling molecules to regulate various disease outcomes including inflammation, ischemia and reperfusion injury, infectious diseases, etc. Yong Wang et al. prepared adenosine, cytidine, and guanosine-protected CuNCs with blue emissions, where the QY of the clusters decreased in the above series of nucleosides.329 More importantly, by implementing the multivariate chemometrics analysis (principal component analysis (PCA) and hierarchical cluster analysis (HCA)), the authors were able to discriminate between different nucleosides. The detection limits for all three mentioned nucleosides were 20 μM for the range of 20 μM–0.5 mM.329 For the detection of nucleotides, specifically ATP and GTP, DNA,330,331 poly(thymine),332 GSH333 and histidine147 were applied as templating ligands. The LODs for ATP using DNA as a template were 93 nM (ref. 330) and 5 pM;331 with poly(thymine)-CuNCs,332 and GSH-CuNCs333 the LOD values were 10.29 pM and 0.67 μM, respectively. Jia Ge et al.334 and Yinan Wang et al.335 applied dsDNA-CuNCs and hpDNA-CuNCs for the detection of nicotinamide adenine dinucleotide (NAD+) with LODs of 0.2 nM and 5 nM, respectively.
4.1.6.1. β-Galactosidase activity detection. β-Galactosidase or lactase (shortly called β-gal, EC 3.2.1.23) is a glycoside hydrolase enzyme that catalyses the hydrolysis of β-galactosides. It is involved in the glycolysis pathway for breaking down the glycosidic bond in lactose into galactose and glucose. β-Gal is important for energy production and is used as a marker for gene expression and target for cancers. Thus, the detection of its enzymatic activity in cells is crucial. Among other commercially available assays for the detection of β-gal activity, copper clusters were also applied as a fluorometric sensor to probe and monitor the β-gal activity, taking advantage of the AIE properties of clusters. For instance, the hydrophobicity-controlled assembly of 4-methylthiophenol-protected CuNCs with AIE characteristics was applied for the β-gal activity detection under physiological conditions.141 The highly emissive cluster assemblies (QY of 9%) as a sensor could operate in a wide analytical range of 2.5–212 U L−1 with LOD of 0.9 U L−1 (1 U is the amount of enzyme that catalyses the reaction of 1 μmol substrate per minute). However, when the Al3+-driven self-assembly of GSH-protected clusters (QY of 8.1%) was applied as a sensor, the analytical performance range was slightly narrower (2.3–96 U L−1), yet an improved LOD value (0.7 U L−1) was achieved.227
4.1.6.2. Exonuclease III activity detection. The exonuclease III (Exo III, EC 3.1.11.2) enzyme from the exonuclease family catalyses DNA degradation and the removal of mononucleotides from the 3′-hydroxyl termini of double-stranded DNA. It is involved in the damage-control and repair of DNA strand breaks, contributing to DNA fidelity, etc. As a matter of fact, the detection of Exo III activity is crucial for disease diagnosis, especially in the early stages. Alternative methods for Exo III activity detection include the use of fluorescent metal nanoclusters, in particular CuNCs. A label-free fluorescence probe based on dsDNA-templated CuNCs has been achieved and applied as a turn-off fluorometric sensor for Exo III. Two groups have independently prepared dsDNA-CuNCs sensors for Exo III.65,346 In both cases, the detection limit was 0.02 U mL−1, however, the analytical range varied significantly, e.g., 0.05–2 U mL−1 in ref. 65 and 0.02–10 U mL−1 in ref. 346.
4.1.6.3. Methyltransferase activity detection. Methyltransferase enzymes (MTases, family EC 2.1.1) regulate the methylation of substrates, which is as an epigenetic modification that is very important for gene transcription as well as gene silencing/activation. Abnormal methylation events of DNA provide a basis for the development of diseases. Apart from the well-established colorimetric, immune-based techniques for the MTase activity detection, nanomaterial-based fluorometric probes recently became very popular. Jingxian Yin et al. prepared a biosensor for MTase activity detection based on the utilization of CuNCs.349 Dumbbell DNA with a locked circle consisting of two poly T loops and a poly (AT-TA) stem has been designed as an effective template. In the presence of MTase, the digestion of the DNA strands by exo/endonucleases has been prevented and the PL signal was observed. However, the enzymatic digestion of the DNA was not avoidable in the absence of MTase; thus, with the degradation of the template, the PL of the cluster cannot be seen. Based on the proposed detection method, the CuNCs-based sensor could operate in the linear range of 0.1–0.2 U μL−1 with LOD of 0.17 mU μL−1.349 The reverse strategy has been implemented by Dengpeng Gao et al. who designed another label-free and sensitive fluorescence method for the detection of DNA MTase activity.350 Herein, (AT/TA)-rich dsDNA containing the methylation-responsive sequence was used as a template for cluster formation. In the absence of DNA adenine methylation methyltransferase (Dam MTase), the relatively long (AT/TA)-rich dsDNA is an effective template and resulted in the formation of CuNCs with bright fluorescence. In the presence of Dam MTase, the specific methylation-sensitive restriction endonuclease Dpn I cleaves the methylated dsDNA and produces shorter dsDNA, which fails to template fluorescent CuNCs. In this case, the sensor could operate in a wider analytical range of 0.5–10 U mL−1 with a detection limit of 0.5 U mL−1.350
4.1.6.4. Inorganic pyrophosphatase activity detection. Inorganic pyrophosphatase (PPase) enzyme (EC 3.6.1.1) is capable of hydrolysing and catalysing the conversion of one pyrophosphate (PPi) to two phosphates (Pi). It is involved in lipid metabolism, calcium absorption and other biochemical transformations. Considering its huge impact on biotransformation, it has recently been shown that the expression of PPase is closely related to certain diseases. Alongside traditional detection methods, CuNCs-based probes were also examined in this field by Mingshu Ye et al.223 and Qing Liu et al.352 Al3+-triggered AIE enhancement of GSH-protected CuNCs has been considered as a turn-on detection scheme for PPi activity screening.223 In the presence of PPi, the fluorescence of the CuNCs/Al3+ system was quenched due to the stronger coordination between Al3+ and PPi. Upon the addition of PPase, PPi was consumed by PPase, thus the native PL of the clusters was preserved. Based on this sensing principle, the PPase activity can be detected in the linear range of 3–40 mU mL−1 with LOD of 1.3 mU mL−1.223 Similarly, in the presence of Fe3+, the PL of TA-CuNCs was quenched.352 The addition of PPi and coordination with iron restored the PL of the cluster. When the PPi was hydrolysed to Pi in the presence of PPase, the release of Fe3+ again triggered the quenching of the cluster's PL signal. Thus, under optimized sensing conditions, the linear range for PPase activity sensing was 0.5–18 U L−1 with LOD of 0.19 U L−1.352
4.1.6.5. S1 nuclease activity detection. S1 nuclease (EC 3.1.30.1) belongs to the family of endonuclease enzymes that catalyse the endonucleolytic cleavage of ssDNA and RNA into oligo- or mononucleotides and hydrolysis loops in dsDNA. Besides their importance in various cellular processes, they are also of paramount interest for biotechnology applications. The S1 activity assay is normally based on PAGE, HPLC and ELISA techniques, which are time-consuming. Fluorometric probes based on CuNCs could offer much faster detection schemes. In this context, poly(thymine)353 and hairpin354 loop-templated CuNCs with bright PL properties were developed for the S1 nuclease activity detection. In the absence of S1 nuclease, polyT of 30-mer (T30) was able to effectively template and produce emissive clusters.353 However, in the presence of an enzyme, the digestion of the single strand led to smaller fragments and therefore cluster formation was not effective. Thus, just by monitoring the PL changes, it is possible to screen and assay the S1 nuclease activity.353 With this method, the lowest detectable amount of S1 nuclease was 0.5 μU μL−1. It was also found that the hairpin DNA template with a specific AT sequence stem and a hairpin loop can serve as a template to result in the formation of highly intense red-emitting CuNCs.354 However, in the presence of S1 nuclease, the hairpin loop was digested, leading to a significant decrease in the fluorescence intensity.354 The sensor operating range was 5–80 mU mL−1 with LOD of 3 mU mL−1. These results demonstrated the applicability of CuNCs as a new, simple, low-cost, and effective probe for S1 nuclease detection.
4.1.6.6. Acid phosphatase activity detection. Acid phosphatase (ACP, EC 3.1.3.2) is an enzyme that liberates phosphate groups during digestion under acidic conditions. The enzyme is synthesized in the liver, spleen, bone marrow, and prostate gland. Abnormally high serum levels of ACP indicate infection, injury, or cancer of the prostate. Thus, apart from clinically accepted ACP tests, CuNCs as novel fluorometric probes were also investigated and applied in the detection and evaluation of prostate activity and function. In this regard, Yuanyuan Huang et al. prepared a CuNCs-based fluorometric probe to detect the concentration of ACP in human serum and seminal plasma with LOD of 1.3 U L−1.220 Much better analytical performance for ACP detection was achieved when polydopamine (PDA)-CuNCs were applied as a sensor with a LOD value of 1 mU L−1.276
4.1.6.7. Alkaline phosphatase activity detection. Alkaline phosphatase (ALP, EC 3.1.3.1) or basic phosphatase is a membrane-bound glycoprotein that catalyses the breakdown of proteins through the hydrolysis of phosphate monoesters. It plays a particularly important role in liver function and bone development. Severely abnormal levels of ALP can signify a severe underlying medical condition, typically one relating to the liver, bones, or gallbladder. In clinical practice, the diagnosis of those diseases is via blood-based analysis through the determination of ALP levels. CuNCs were also applied as an assay for ALP activity detection.355,357 The sensing principle is based on the use of PPi as a natural substrate for the ALP, which in the cells/tissues hydrolyses the PPi to Pi to maintain normal levels in the bone. Considering the strong interaction of metal cations with PPi, the formation of fluorescent CuNCs is hampered even in the presence of the DNA template.74 However, in the presence of ALP, the PPi hydrolyses to Pi, which cannot strongly interact with Cu2+; thus, during the reduction process, the metal cations tend to cluster inside the DNA strands and form fluorescent CuNCs. Based on this strategy, the ALP levels can be quantitatively determined, i.e., the lowest detectable concentration was 0.1 nM.74 GSH-CuNCs with Al3+-triggered AIE enhancement were also applied in the ALP activity detection.224 The addition of Al3+ to the cluster solution caused aggregation and thus, PL enhancement. In the presence of PPi, due to the Al3+-PPi complexation, the Pl was quenched. However, the hydrolysis of PPi with ALP, liberated the Al3+ and restored the PL intensity. The developed biosensor could operate in a wide linear range (0.5–25 mU mL−1) with LOD of 0.15 mU mL−1.224 The opposite effect was observed when poly(thymine)-CuNCs were applied in the detection of ALP.262 In brief, the quenching of PL (in the presence of Cr3+ ions) was restored by the addition of PPi, due to the strong binding affinity of pyrophosphate towards the chromium ions. Finally, in the presence of ALP, PPi is hydrolysed, the Cr3+-PPi complex degrades and the release of Cr3+ induces fluorescence quenching again.262 Yanling Hu et al. studied another ALP substrate, p-nitrophenylphosphate (PNPP), which was hydrolysed to p-nitrophenol (PNP).356 The hydrolyzation product caused significant quenching of the original fluorescence signal of the L-His-CuNCs. Based on the sensing method, the ALP activity can be straightforwardly determined with the lowest reported value of 45 μU mL−1 in the analytical performance range of 0.5–40 mU mL−1.356
4.1.6.8. Uracil-DNA glycosylase activity detection. DNA glycosylases are enzymes that remove the damaged nitrogenous base while keeping the sugar–phosphate backbone in DNA intact. Among different kinds of DNA glycosylases that are responsible for the excision of different bases in the DNA, uracil-DNA glycosylase (UDG, EC 3.2.2.27) specifically recognizes and removes uracil lesions in both double- and single-stranded DNAs and initiates the downstream repairing process. The abnormal expression of DNA glycosylases is closely related to various human diseases, such as human immunodeficiency, bloom syndrome, cancers, etc. Therefore, the detection of UDG levels and activity is very important for disease diagnosis and the proper functioning of base excision repair (BER) processes. Besides the highly applicable radiographic method, CuNCs were also exploited in this field as a low-risk and labour-free UDG assay. dsDNA strands have been shown to efficiently template the metal to form fluorescent CuNCs. However, the strand with uracil deoxyribonucleotides represents a substrate for UDG and recognition by the enzyme results in the spontaneous detachment of the double helix into ssDNAs. Consequently, the formed ssDNAs cannot effectively template the cluster, which leads to the weakening of the emission intensity. By monitoring the PL changes, the UDG activity can be quantitatively determined. As such, Manman Cao et al. prepared dsDNA-CuNCs for UDG activity detection.358 The turn-off fluorometric sensor could operate in a range of 1 mU mL−1–0.1 U mL−1 with LOD of 0.5 mU mL−1. Poly(thymine)-CuNCs, on the other hand, manifest much better performances for UDG analysis.359,360 Here, the developed turn-on sensors covered a vast detection ranges from 100 μU mL−1 to 0.01 U mL−1 (ref. 359) and 50 μU mL−1 to 2 mU mL−1 (ref. 360) with LOD of 50 μU mL−1 (in both cases).
4.1.6.9. Polynucleotide kinase phosphatase activity detection. The DNA damage caused by exposure to genotoxic agents results in the formation of DNA with a 3′-phosphate terminal. However, for the proper biochemistry of DNA, the removal of the 3′-phosphate and attachment of a hydroxyl group to the 3′-tail end of the DNA backbone is essential. T4 polynucleotide kinase phosphatase (T4 PNKP, EC 2.7.1.78) is a popular enzyme that catalyses the hydrolytic removal of the 3′-phosphoryl of polynucleotides and at the same time phosphorylates DNA at the 5′-hydroxyl end. For the T4 PNKP activity detection, poly(thymine)-templated CuNCs were exploited as a turn-off361 and a turn-on362 biosensor by using T-rich and A-rich DNA probes as a substrate for T4 PNKP. The 3′-end of the ssDNA probe (T-rich) modified with a phosphate group is hydrolysed into a 3′-hydroxyl, which can further be extended into dsDNA by the DNA polymerase in the presence of dNTPs.361 The polymerization product is unable to template and form fluorescent clusters. In the case of an A-rich probe, an additional step is required. Briefly, the formed dsDNA in the process described above is digested via a restriction enzyme followed by the amplification cycle, polymerization and nicking to produce T-rich ssDNA, which can later template the clusters.362 Thus, by following the changes in PL upon cluster formation, the T4 PNKP activity can be detected. The detectable minimum amounts of T4 PNKP enzyme with those sensors were 0.25 U mL−1 (ref. 361) and 0.02 U mL−1. (ref. 362) Similarly, Xingxing Zhang et al. developed a dsDNA-CuNCs sensor based on the hybridization of two sequence-dependent ssDNA strands to form a partial dsDNA duplex.363 This DNA serves as a substrate for T4 PNKP to be further used for the synthesis of full dsDNA. With this sensor, the LOD of T4 PNKP activity detection was 0.06 U mL−1. Another report by Ruqin Yu and co-workers established a detection platform for polynucleotide kinase (PNK) activity sensing with the LOD of 0.49 U mL−1.364
4.1.6.10. Acetylcholinesterase activity detection. Acetylcholinesterase (AChE, EC 3.1.1.7) catalyses the breakdown of the acetylcholine neurotransmitter into acetate and choline upon the termination of impulse transmission at cholinergic synapses. The abnormal concentrations of acetylcholine have been linked to the development of several neurodegenerative diseases such as Parkinson's disease, myasthenia gravis and Alzheimer's disease (AD). Thus, the detection of AChE activity is of great importance in clinical diagnostics. Apart from colorimetric and electrochemical detection methods, fluorometric methods have recently attracted much interest due to their simplicity and faster analysis. Taking advantage of their fluorescence properties, Jinlan Yang et al.,254 Mengke Wang et al.365 and Pengjuan Ni et al.366 developed CuNCs-based fluorometric probes to detect AChE activity in human serum samples. Within the proposed sensing strategies, the LODs of the sensors were 1.38 mU mL−1,254 0.56 U L−1,365 and 0.05 mU mL−1.366
4.1.6.11. Other enzyme activity detection. The potential applications of CuNCs towards the enzyme activity assay have also been explored for restriction enzyme EcoRI (LOD 870 μU μL−1),347 matrix metalloproteinase-7 (MMP-7, LOD 5.3 pg mL−1),348 micrococcal nuclease (MNase, LOD 1 mU mL−1),351 terminal deoxynucleotidyl transferase (TdT, LOD 60 mU L−1),367 reverse transcription-associated ribonuclease H (RNase H, LOD 0.55 mU mL−1),368 β glucuronidase (GLU, LOD 0.22 U L−1),369 tyrosinase (TYR, LOD 44 U L−1),370 thrombin (LOD 0.9 nM),371 hyaluronidase (HAase, LOD 14 mU mL−1),372 lysozyme (LOD 1.6 nM)373 and protein kinase A (PKA, LOD 38 mU mL−1).374
Streptavidin (SA)-biotin is the strongest noncovalent biological interaction known in biology. The tetrameric unit of the SA protein can bind four biotin molecules. The bioconjugation of SA to biomolecules or a labelled target (fluorophore, enzyme etc.), has been applied in modern detection techniques and bioassays like Western blotting, ELISA, and Flow Cytometry. Thus, taking advantage of a strong and specific SA–biotin interaction, a novel detection method has been employed based on the interaction between the CuNCs–SA–biotin system. For example, Hai-Bo Wang et al.375 have shown that the interaction of SA with poly(thymine)-templated CuNCs attached to biotin can prevent the degradation and enzymolysis of DNA by Exo I. On the other hand, fluorescent CuNCs, can be used to determine the SA concentration from calibration curves. Jiepei Cao et al. reported the use of fluorescent CuNCs for the study of SA–biotin interactions under magnetic separations.337 Within the developed strategy, the LODs of SA and biotin were 0.47 nM and 3.1 nM, respectively. A better SA sensing platform was developed by Yue He et al.376 based on the utilization of poly(thymine)-CuNCs as a turn-on sensor (LOD 20 pM). Thus, as a classic model, SA-biotin bioconjugated CuNCs can be exploited as a sensitive, highly selective, and low-cost protein-binding assay.
Li-Juan Ou et al.379 and Yanling Hu et al.382 have reported the applications of poly(thymine)-CuNCs and PSS-CuNCs as a fluorometric assay for the detection of trypsin with LOD of 42 ng mL−1 and 20 ng mL−1, respectively. Cytochrome c (Cyt c) is a well-known electron transfer mediator and thus a quencher, and it can quench the PL of CuNCs.247 However, in the presence of trypsin, Cyt c hydrolyses into peptide fragments and thus inhibits the quenching of PL from CuNCs.382 Better sensitivity towards trypsin was achieved when GSH-CuNCs together with Cyt c (LOD 2 ng mL−1)380 and dsDNA-CuNCs (LOD 48 pg mL−1)381 were implemented as fluorometric sensors.
Xin-Ge Li et al.377 and Quanwei Song et al.378 prepared CuNCs-based bio probes for the detection of glycoproteins and actin, respectively. The LODs of glycoproteins and actin were 2.6 nM and 0.12 μg mL−1, respectively. CuNCs-based detection of VEGF165,383 protamine,307 ferritin,247 carcinoembryonic antigen (CEA),384 prostate-specific antigen (PSA)385 and human immunoglobulin G (IgG)357 were also successfully demonstrated with LODs of 12 pM, 0.12 ng mL−1, 16.5 nM, 6.5 pg mL−1, 145.7 fg mL−1 and 7 pg mL−1, respectively.
Jiu-Ju Feng and co-workers established a CuNCs-based nanothermometer that displayed a temperature dependence of PL intensity in the range of 10–55 °C (Fig. 25A).134 The PL quenching was reversible and could be switched on and off for at least 6 cycles with negligible fatigue. Moreover, it was shown that the decrease in PL intensity was caused by the activation of nonradiative pathways.
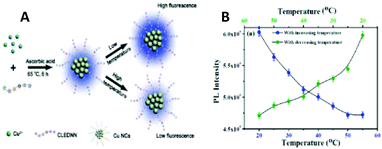 | ||
| Fig. 25 (A) Schematic illustration of the formation mechanism of CuNCs with temperature-dependent fluorescence. Republished with permission from ref. 134. Copyright 2014 Royal Society of Chemistry. (B) PL reversibility exhibited by the HSA-CuNCs as a function of temperature. Reprinted with permission from ref. 50. Copyright 2015 American Chemical Society. | ||
In another case, Saptarshi Mukherjee and co-workers studied the temperature dependence of the PL properties of HSA-CuNCs.50 With increasing temperature, the PL intensity of the clusters was quenched, mostly due to thermal instability/unfolding caused in the stabilizing scaffolds of HSA protein (Fig. 25B). A similar effect was been observed when GSH-CuNCs were heated to 80 °C.130 The increase in temperature led to the decrease in PL intensity because of the increase in collision rate and activation of nonradiative pathways. The decrease in temperature resulted in the complete restoration of PL properties in all mentioned cases. Furthermore, GSH-CuNCs were applied as a nanothermometer to detect intracellular temperature differences in MC3T3-E1130 and MDA-MB-231386 cancer cells. As expected, with increasing the temperature, the red fluorescence of internalized clusters faded away.
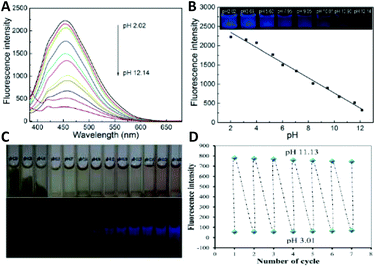 | ||
| Fig. 26 (A) Fluorescence emission spectra of CuNCs in Britton–Robinson (BR) buffers of different pH values. (B) The calibration curve of the pH values in the range of 2.02–12.14 versus the fluorescence intensity of the CuNCs in BR buffers. Republished with permission from ref. 39. Copyright 2014 Royal Society of Chemistry. (C) Photographs showing the colour changes of the CuNCs in BR buffer at different pH values under visible light (upper) and UV light (lower). (D) The fluorescence intensity of the CuNCs reversibly went up and down when alternating the pH value. Republished with permission from ref. 49. Copyright 2015 Royal Society of Chemistry. | ||
In contrast, BSA-templated CuNCs were highly fluorescent in alkaline medium and less emissive under acidic conditions.49,303 Again, the sensor could operate effectively for 7 cycles with no fatigue (Fig. 26C and D).49 A similar phenomenon has been observed with green-emitting cys-CuNCs.133 Compared to the emission peak intensity at pH 4, about 66-fold higher intensities were observed at pH 9. As for PEI-capped CuNCs, the fluorescence emission reached its maximum at pH 13 and it was about 20-fold higher than that at pH 5.2.387
CuNCs prepared directly from chicken egg white (CEW) demonstrated pH-reversible features mostly ascribed to conformational changes in the ovalbumin protein in CEW with pH variations from acidic to basic.388 The intense green emission was observed in strongly basic conditions, whereas at lower and mostly in acidic buffers, the PL decreased, consistent with the precipitation of the CuNCs.
A colorimetric detection platform for hepatitis B virus (HBV) was established by using DNA-CuNCs.390 The designed sensor can specifically target and sense the HBV DNA with LOD of 12 × 109 molecules. Acceptable recoveries were detected from serum samples containing HBV DNA.
Ehsan Shokri et al. reported the virus-detected synthesis of fluorescent CuNCs and tested its applicability in the detection of Citrus Tristeza Virus (CTV) in real samples with LOD of 220 pg mL−1.391
Thus, dsDNA-templated CuNCs can be applied as a portable and digital diagnostic tool in combination with sensor technologies.
A large number of studies have been conducted for preparing water-soluble and highly fluorescent CuNCs as a novel fluorometric/colorimetric sensing platform. Note that CuNCs-based fluorometric sensors (mostly turn-off) sometimes show impressive sensitivity towards the target (LOD in the fM or aM range) and can operate in a wide analytical range. Nevertheless, the main drawback is that most of them are not atomically precise and less attention has been paid to the selectivity. The developed sensors have not been commercialized and applied in real clinical diagnosis.
4.2. Applications in biomedicine
Therefore, highly fluorescent, and FA-conjugated CuNCs have found applications in the early diagnosis of cancers via targeted bioimaging of FR overexpressed cancerous cells. Chuanxi Wang et al.132 demonstrated the potential of FA-conjugated and GSH-protected CuNCs as a diagnostic tool to detect gastric HeLa cells (FR-positive cell line) in physiological conditions with no adverse effect on the cell line. Moreover, the clusters were highly biocompatible and showed very small cytotoxicity.
Jun-Mei Xia et al. prepared FA-protected CuNCs (direct synthesis) and tested them against HeLa and A549 cells (human lung carcinoma cells, FR-negative cell line).407 No cluster-induced cytotoxicity was observed when the cells were incubated in the presence of different CuNCs concentrations. However, significant fluorescence was observed in the case of HeLa cells and almost no fluorescence for A549 cells. Since HeLa cells express higher levels of FR, it initiates faster internalization of FA-CuNCs in the cells through receptor-mediated endocytosis. Yaling Wang et al. demonstrated the one/two-photon fluorescence imaging of HeLa and A549 cells using bifunctional peptide (Sv)-functionalized CuNCs.408 Interestingly, the clusters were found to be mainly localized in the nucleus and less in the cytoplasm. Moreover, the two-photon (TP) images were less affected by the auto-fluorescence of the cell, thus making TP superior to one-photon (OP) imaging (Fig. 27A).
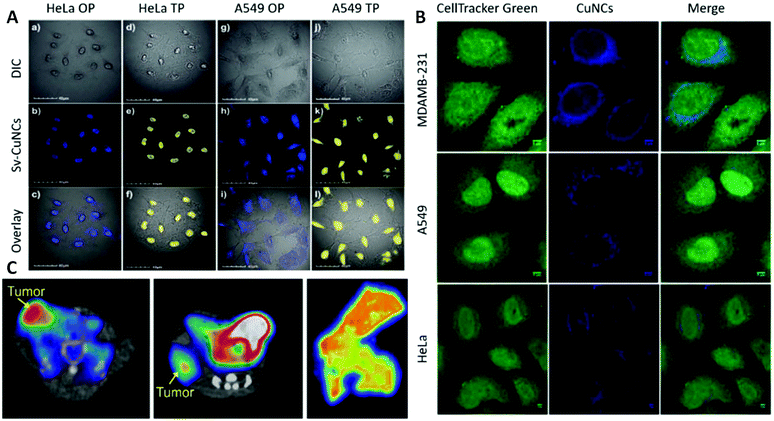 | ||
| Fig. 27 (A) One- and two-photon confocal images of Sv–CuNCs for HeLa and A549 cells: (a)–(c) and (d)–(f) are OP and TP images of HeLa cells, respectively; (g)–(i) and (j)–(l) are OP and TP images of A549 cells, respectively. Differential interference contrast (top panel), fluorescence image of Sv–Cu cluster (middle panel, blue and yellow represent OP and TP emission, respectively) and merged view (bottom panel). Republished with permission from ref. 408. Copyright 2013 Royal Society of Chemistry. (B) Subcellular localization of CuNCs: HeLa, MDAMB-231, and A549 cells were incubated with CuNCs for 12 h at 37 °C. Reprinted with permission from ref. 135. Copyright 2015 American Chemical Society. (C) PET imaging of tumours in the mouse cell line xenograft model (left), mouse PDX model (middle) and human tumour tissue (right). Reprinted with permission from ref. 410. Copyright 2019 American Chemical Society. | ||
The preferential localization of CuNCs in the nucleus was also reported by Chan Wang et al.45 and Markus J. Barthel et al.409 It was found that the cluster internalization is due to the crossing of the membrane since the endocytosis is inhibited at 4 °C. However, at 37 °C the accumulation of CuNCs in the perinuclear area additionally pinpointed the endocytotic uptake.409 Chan Wang et al. showed that CAL-27 cells incubated with BSA-CuNCs kept their morphology over time and at different doses of the cluster.45 As mentioned before, the fluorescence signal was located mostly in the cellular nucleus. Therefore, BSA-CuNCs displayed real potential for labelling the oligonucleotide and the detection of nucleic acids.
Similar results were observed when 293T cells were treated and incubated with PEI-CuNCs.387 According to the MTT assay, 75% of cells stayed viable after 24 h of incubation. Confocal microscopy imaging revealed that the cells kept their normal morphology after the treatment and showed the internalization of the cluster into the cytoplasm and the nucleus. The energy-dependent, clathrin-mediated endocytosis has been proposed as an internalization mechanism/pathway. On the contrary, Hong Huang et al.134 have shown that on fluorescence imaging of HeLa cells with CuNCs protected with a peptide (amino acid sequence: CLEDNN), most clusters were localized in the cell membrane and cytoplasmic area. A very weak blue fluorescence was detected in the nucleus. Therefore, the clusters are bio-safe and do not cause genetic disruption.
Jigna R. Bhamore et al. have demonstrated that CuNCs prepared from the curcuma root extract showed pronounced cytotoxicity in RIN5F and MDAMB231 cancerous cell lines.241 The cell cycle analyses revealed that the G2/M phase was more affected in MDAMB231 cells, which suggests that CuNCs might contribute to both G1/S arrest and cellular apoptosis in cancer cells. Upon fungal cell imaging (Penicillium citrinum cells), the clusters were found to be localized near the nuclear region. Saptarshi Mukherjee and co-workers demonstrated that the cellular internalization and localization of GSH-CuNCs strongly depend on the cell line.135 For instance, in HeLa (malignant immortal cell line derived from cervical cancer) and MDAMB-231 (human breast adenocarcinoma) cells the clusters were preferentially localized near the nuclear region, whereas in the case of A549 (human lung carcinoma), the clusters were more distributed in the cytoplasm (Fig. 27B).
Transferrin-templated CuNCs have been applied for the imaging of a human cancer cell line and a normal mouse cell line (3T3) through the detection of transferrin receptors.53 Because HeLa cells have a higher level of transferrin receptor expression, the cells treated with CuNCs showed an intense red fluorescence, whereas week fluorescence was observed in the case of 3T3 cells.
Gyu Seong Heo et al.410 demonstrated the applicability of radiolabelled and peptide bioconjugated CuNCs (64Cu-CuNCs-FC131) for the sensitive and accurate detection of tumours in triple-negative breast cancer (TNBC) patient-derived xenograft mouse models and human TNBC tissues (Fig. 27C). 64Cu-CuNCs showed weak tumour uptakes, whereas CXCR4 targeted 64Cu-CuNCs-FC131 demonstrated significant tumour uptake. Considering the biocompatibility and low toxicity of clusters, the proposed strategy can be applied in clinical settings for the detection of overexpressed CXCR4 biomarkers in both mouse models and human TNBC tissues, for positron emission tomography (PET) imaging as well as for therapy. The obtained results demonstrated the advantages of radiolabelled CuNCs as a potential translational imaging and drug delivery machinery for cancer theranostics. A recent review by Rogach and co-workers summarizes the theranostic applications of CuNCs in both in vitro and in vivo platforms.411
Moving forward, Fuping Gao et al.40 demonstrated the applicability of radiolabelled 64Cu-BSA-CuNCs as an efficient and sensitive radiotracer for PET imaging. The bioconjugation of LHRH receptor peptide to the clusters ensured the effective deposition and clearance from the tumour in vivo. Receptor-targeted clusters showed higher tumour uptake properties than that of nontargeted clusters in A549 human lung tumour. Tomographic images revealed that after 0.5 h post-injection, the clusters were distributed all over the body (Fig. 28A). However, at higher post-injection times, a strong radioactivity signal was coming from the kidneys and bladder, thus the clusters were mainly accumulating in those organs (Fig. 28B). This is indicative of the cluster clearance through the renal route. Additionally, fluorescence imaging of cryosections of tissues has shown that targeted clusters were mostly localized in the tumour and kidneys, whereas nontargeted clusters were in the kidneys (Fig. 28C). GSH-CuNCs were applied in the imaging of MC3T3-E1130 and MGC-803 cells;225 the latter report is based on the aggregation-induced emission (AIE) effect. In the presence of Zn2+, the aggregation of the clusters resulted in bright red emission and thus, Zn2+-triggered AIE of CuNCs has been applied as a “light-up” nanoprobe for the detection/imaging of zinc ions in living cells.
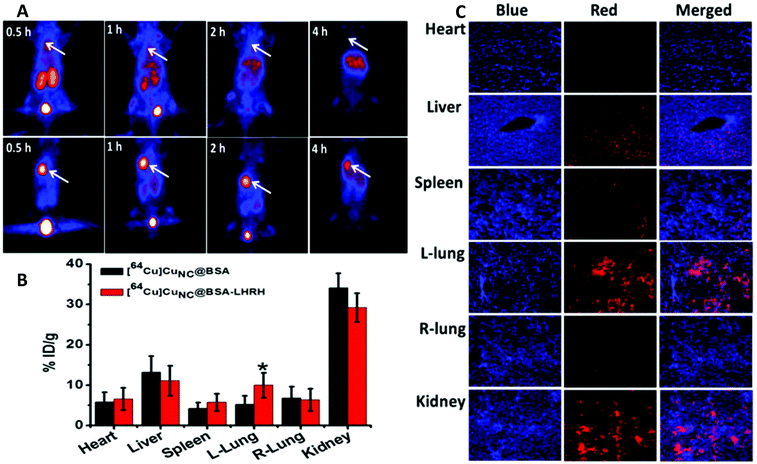 | ||
| Fig. 28 (A) In vivo PET images of coronal single slices on orthotopic A549 lung tumour-bearing mice after intravenous injection of 6.7 MBq of [64Cu]CuNC@BSA (top) and [64Cu]CuNC@BSA-LHRH (bottom). Images were acquired at 0.5, 1, 2, and 4 h. White arrows indicate the lung tumour. (B) The corresponding organ biodistribution of [64Cu]CuNC@BSA and [64Cu]CuNC@BSA-LHRH at 4 h after iv injection in mice bearing orthotopic A549 lung tumour. (C) Fluorescence microscopy images of tissue sections. The tissues were taken at 4 h after the injection of CF680R-conjugated [64Cu]CuNC@BSA-LHRH via the tail vein. Blue fluorescence shows nuclei stained with DAPI, and red fluorescence shows the location of [64Cu]CuNC@BSA-LHRH. Reprinted with permission from ref. 40. Copyright 2015 American Chemical Society. | ||
Arun Chattopadhyay and co-workers implemented the AIE-based enhancement of PL to monitor in vitro pH variations between cancerous (human breast adenocarcinoma cells: MCF-7) and non-cancerous (human embryonic kidney cells: HEK-293) cell lines.123 The as-synthesized clusters showed bright orange-red emission in acidic conditions (pH 4.5) and cyan-green emission in basic medium (pH 7.4, Fig. 29). The study demonstrated that higher expression levels of various proteins and biomolecules in MCF-7 cells triggered faster aggregation kinetics in the cells at higher pH (pH 7.4). On the contrary, HEK-293 cells lack the overexpression of such biomolecules in the intracellular environment, thus prohibiting the AIE-induced PL of the clusters.
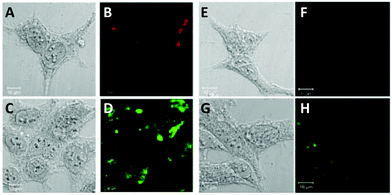 | ||
| Fig. 29 (A) Bright-field image and (B) fluorescence image in the red channel of MCF-7 cells treated with the NC probe for 1 h in culture medium at pH 4.5. (C) Bright-field image and (D) fluorescence image acquired in the green channel of MCF-7 cells treated with the NC probe for 1 h in culture media at pH 7.8. (E) Bright-field image and (F) fluorescence image in the red channel of HEK-293 cells treated with the NC probe for 1 h in culture media at pH 4.5. (G) Bright field image and (H) fluorescence image acquired in the green channel of HEK-293 cells treated with the NC probe for 1 h in culture media at pH 7.4. Reprinted with permission from ref. 123. Copyright 2018 American Chemical Society. | ||
Zhichao Liu et al. prepared a CuNCs-based fluorescence probe for the real-time sensing and multicolour imaging of calcium ions in neurons.256
The intracellular calcium ion detection revealed the inhomogeneous localization and accumulation of calcium in the neuronal cytoplasm (Fig. 30). Besides, the inhomogeneity caused the formation of bright spots in the overlay image resulting from Ca2+-rich domains. More importantly, the study demonstrated that ROS-induced neuronal death is directly proportional to calcium ion overload. Thus, these findings provide a solid basis for the mechanistic studies of oxidative stress-related diseases using CuNCs.
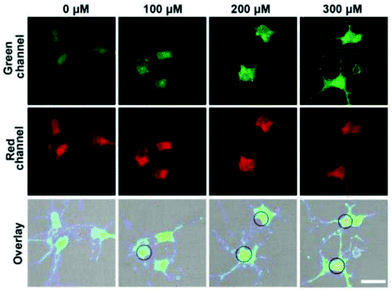 | ||
| Fig. 30 Confocal fluorescence microscopy images of neurons collected from different channels after the neurons were co-incubated with a CuNC@AF660 probe (90 μg mL−1) in the presence of different concentrations of Ca2+ (0, 100, 200, and 300 μM), respectively. Scale = 25 μm. Reprinted with permission from ref. 256. Copyright 2019 American Chemical Society. | ||
Blue-emitting lysozyme-CuNCs (QY 18%) prepared by Rama Ghosh et al. have been considered as fluorescent agents for cell labelling.42 In this regard, HeLa cells (cervical cancer) incubated with CuNCs were detected via fluorescence microscopy imaging, where a bright blue colour was attributed to the clusters since control cells and cells treated only with the ligand lacked intense PL properties. The clusters showed no cytotoxicity towards the cells.
Jinbin Liu and co-workers demonstrated that CuNCs assemblies with enhanced AIE properties can be used to image HeLa cells with green fluorescence protein (GFP)-labelled lysosomes.218 Both red emission (from the CuNCs assemblies) and green emission (from GFP) were observed at the incubation time of 24 h, which means that the cluster assemblies are highly stable and keep their PL properties under a lysosomal acidic microenvironment (pH ≈ 4.5–6).
The latest review by Baisong Chang discusses the recent advances in the bioimaging applications of CuNCs.394
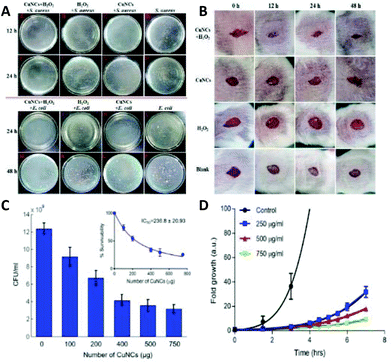 | ||
| Fig. 31 (A) The influence of CuNCs on the growth of Gram-positive (S. aureus) and Gram-negative (E. coli) bacteria. Gram-positive bacteria were recorded at 12 h and 24 h: S. aureus (105 cfu mL−1) co-incubated with CuNCs plus H2O2 (A and E), H2O2 (B and F), CuNCs (C and G), null (D and H), respectively; Gram-negative bacteria were observed at 24 h and 48 h: E. coli (105 cfu mL−1) co-incubated with CuNCs plus H2O2 (I and M), H2O2 (J and N), CuNCs (K and O), null (L and P), respectively. The concentrations of CuNCs and H2O2 were 10 mg mL−1 and 1 mM, respectively in all cases. (B) Images of the wounded mice treated with CuNCs plus H2O2, CuNCs, H2O2 and null, respectively. Republished with permission from ref. 54. Copyright 2015 Royal Society of Chemistry. (C) Antimicrobial activity of GSH-CuNCs (inset: IC50 value). (D) Fold growth of E. coli, DH5α at different concentrations of GSH-CuNCs. Reprinted with permission from ref. 136. Copyright 2018 American Chemical Society. | ||
Baghdasaryan et al. reported the synthesis and antibacterial properties of GSH-CuNCs toward E. coli, DH5α cells.136 The antibacterial properties were studied at different cluster concentrations. It was found that with increasing cluster concentration, much lower bacterial growth was observed as compared to control bacteria (Fig. 31C). Moreover, the studies of the growth kinetics of DH5α cells treated with CuNCs revealed that at low concentrations, the clusters showed a bacteriostatic effect, whereas, at higher concentrations, there was a transition from bacteriostatic to bactericidal toward E. coli, DH5α cells (Fig. 31D).
Hong Yan Zou et al. have also studied the antimicrobial properties of dopamine-capped CuNCs using Gram-positive S. aureus bacterial strains.148 The clusters showed dose-dependent antimicrobial properties with a minimum inhibitory concentration of 158 μg mL−1.
The bacterial death associated with the bacterial cell membrane damage was considered as an antimicrobial mechanism for Gram-positive bacteria S. aureus and B. subtilis (Bacillus subtilis) and Gram-negative bacteria E. coli and P. aeruginosa (Pseudomonas aeruginosa).412 Moreover, TA-CuNCs possessed selective antibacterial activity against Gram-positive bacteria by inhibiting the bacterial growth.
A combined bioimaging/drug delivery/antibacterial multimodal CuNCs-based system was recently developed by Anitha T. Simon et al.413 BSA-CuNCs together with the drug kanamycin loaded inside the luminescent hydroxyapatite nanoparticles (HAP) (Fig. 32) were applied first to image the bacteria, then to transport the drug and finally to explore their antibacterial activity against Gram-positive and Gram-negative bacteria.
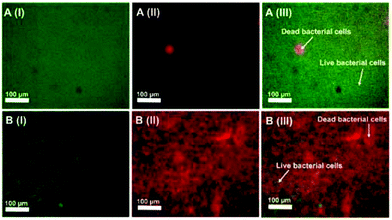 | ||
| Fig. 32 [A(I–III)] Epifluorescence images of AO/PI stained control biofilm: [A(I)] AO-stained, [A(II)] PI-stained, and [A(III)] merged image. The arrows in the merged image of the control biofilm denote the regions with dead and live bacterial cells. [B(I–III)] Biofilm treated with kanamycin-loaded doped HAP NPs: [B(I)] AO-stained, [B(II)] PI-stained, and [B(III)] merged image. The arrows in the merged image of the treated biofilm denote the regions with dead and live bacterial cells. Reprinted with permission from ref. 413. Copyright 2019 American Chemical Society. | ||
The drug-free nanocarrier showed no bactericidal activity, whereas kanamycin-loaded cargo exposed antibacterial properties. The system was also effective in eradicating P. aeruginosa biofilm.
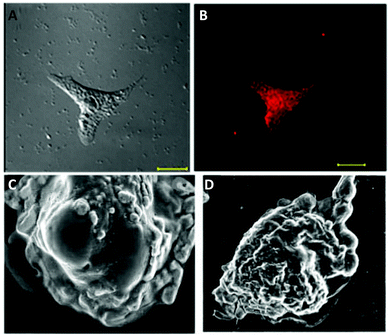 | ||
| Fig. 33 Confocal microscopic images of HeLa cells treated with the CP-loaded CuNC–hydrogel composite, recorded after 4 h of incubation. (A) Image under bright-field and (B) fluorescence image of HeLa cells under green light. Scale bar is 20 μm. (C and D) FESEM images of HeLa cells, treated with the CuNC–hydrogel composite and CP-loaded CuNC–hydrogel composite for 24 h, revealing apoptotic cell deaths. Reprinted with permission from ref. 32. Copyright 2015 American Chemical Society. | ||
As already discussed in section 4.4.3, hydroxyapatite nanoparticles doped with fluorescent BSA-CuNCs have been designed as an effective kanamycin drug delivery carrier.413 The disruption of the bacterial cell wall after treating with the nanocarrier suggested the release of the drug from the cavity of the carrier and interaction with the bacteria through the depletion of NADH, thus causing unprevented cell damage.
The real-time monitoring of transferrin-templated CuNCs in conjugation with the anticancer drug doxorubicin (Tf-CuNCs-DOX) was performed in cancerous and normal cells in vitro for drug localization and release purposes (Fig. 34).414 It was observed that TfR overexpressed cells showed higher drug release profiles with pronounced therapeutic activity. In vivo studies on Swiss albino mice bearing a tumour of Dalton's lymphoma ascites (DLA cells) demonstrated tumour volume reduction after the administration of the Tf-CuNCs-DOX nanodrug, mainly due to the targeted internalization and longer circulation time.
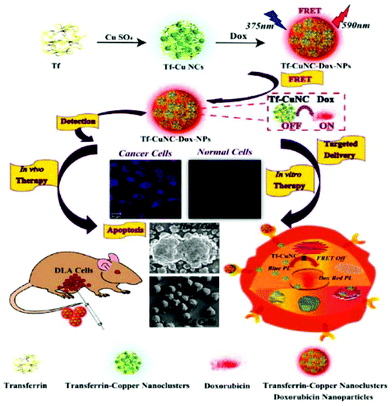 | ||
| Fig. 34 Scheme showing the formation of a targeted nanodrug for FRET-assisted bioimaging, targeted delivery and synergistic therapeutic activity (in vitro and in vivo). Reprinted with permission from ref. 414. Copyright 2018 American Chemical Society. | ||
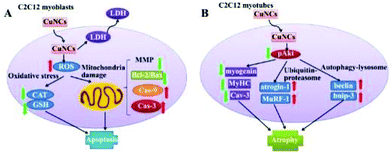 | ||
| Fig. 35 Schematic illustration of CuNC-induced cytotoxicity, ROS production, apoptosis, and atrophy in muscle cells. CAT, catalase; CuNCs, copper nanoclusters; GSH, glutathione; LDH, lactic dehydrogenase; MMP, mitochondrial membrane potential; ROS, reactive oxygen species. Adapted with permission from ref. 415. Copyright 2015 John Wiley & Sons, Ltd. | ||
4.3. Applications in catalysis
Catalytic processes based on the use of nanomaterials as catalysts are currently a dynamic research field. The high interest in clusters is governed by their size-dependent and molecular-like properties. On decreasing the size of the clusters to the subnanometer range, the surface energy increases drastically. This, on the other hand, makes the clusters highly catalytically active for chemical transformations. The literature is replete with examples of very small clusters (Cun, where n < 5), that are very active and selective catalysts in many homogeneous and heterogeneous catalytic conversions.416 For instance, Cu3 and Cu4 clusters prepared using size-selection in combination with a cluster source were applied in methanol synthesis from CO2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 417,418 and in CO oxidation.419 Together with extensive DFT calculations, the catalytic activity of these clusters in various reactions was found to be strongly size-dependent. Cu5 cluster species were reported to be highly active in propene oxidation420 and CO2 decomposition.421 More thorough examples of catalytically active tiny copper clusters can be found in the review by Corma.422
417,418 and in CO oxidation.419 Together with extensive DFT calculations, the catalytic activity of these clusters in various reactions was found to be strongly size-dependent. Cu5 cluster species were reported to be highly active in propene oxidation420 and CO2 decomposition.421 More thorough examples of catalytically active tiny copper clusters can be found in the review by Corma.422
In the following (sub)sections, we restrict our discussion to the catalytic conversions utilizing copper nanoclusters (containing more than 6 metal atoms) as heterogeneous/homogeneous catalysts prepared by “wet chemical” methods.
Noelia Vilar-Vidal et al. studied the size-dependent catalytic properties of CuNCs, namely Cu5, Cu13 and Cu20.423 The catalytic reduction of methylene blue (MB) to leucomethylene blue (LMB) by hydrazine was performed in the presence of CuNCs.114,423 It was found that small clusters were catalytically very active, whereas with increasing the cluster size, the catalytic activity of the cluster was reduced. Cu20 clusters did not catalyse the reaction at all. The reaction mechanism is explained as a cluster-mediated electron transfer from the donor (N2H4) to the acceptor (MB) through the conduction band of the CuNCs. Consequently, with increasing the size of the cluster, the band gap became smaller and thus, the cluster became catalytically less active, as is schematically depicted in Fig. 36.
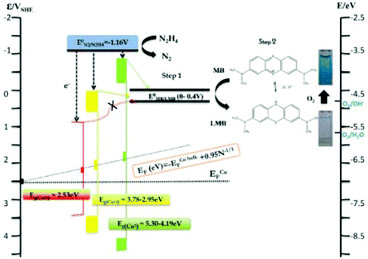 | ||
| Fig. 36 Schematic energy diagram showing the catalytic activity of different CuNCs used for MB reduction by N2H4. Reprinted with permission from ref. 423. Copyright 2012 American Chemical Society. | ||
De-en Jiang and co-workers studied the catalytic CO2 reduction to (i) HCOOH or (ii) CO when using a polyhydrido [Cu32H20{S2P(OiPr)2}12] cluster as an active catalyst.188 According to DFT calculations, in the first reaction, after CO2 adsorption, the addition of a negative hydrogen to the positive carbon of CO2 leads to the formation of an intermediate HCOO* species through the lattice hydride mechanism. The addition of a second hydrogen results in the formation of the final product leaving the cluster with two hydrogen vacancies. The loss of lattice hydrides is rapidly compensated via subsequent proton-reduction steps (Scheme 9A). In the second reaction, again via lattice hydride mechanism (Scheme 9B), the addition of a first hydride to an oxygen atom of CO2 resulted in the formation of an intermediate COOH* species bonded to the cluster. The cleavage of the CO–OH bond in the intermediate species leads to the release of CO and the formation of a OH* intermediate. The release of water and an electrochemical proton-reduction process recovers the lost lattice hydrides in the catalytic cycle and brings the cluster back to its native composition. In accordance with theoretical calculations, the experimental electrocatalytic CO2 reduction to HCOOH was favoured at low overpotentials.
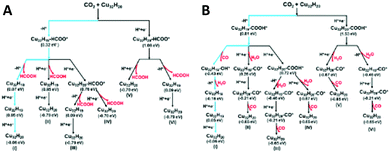 | ||
| Scheme 9 Theoretical reaction schemes for CO2 electroreduction on [Cu32H20{S2P(OiPr)2}12] to form (A) HCOOH and (B) CO via six different pathways as described in ref. 188 (Reprinted with permission, Copyright 2017 American Chemical Society). The calculated free energy difference (ΔG) for each elementary step is shown under each intermediate state. The most favourable reaction pathway is highlighted in blue. | ||
Tomoaki Tanase and co-workers studied the hydrosilylation of CO2 with tertiary hydrosilanes catalysed by [Cu8(μ-H)6(μ-dppm)5](PF6)2.157 The corresponding silyl formats were obtained after 48 h in acetonitrile, at 45 °C and 1 atm pressure of CO2.
Recently Kingshuk Basu et al.118 prepared red-emitting cys-CuNCs and demonstrated its catalytic activity for the reduction of 4-nitrophenol to 4-aminophenol at room temperature. The catalyst can be recycled without loss in activity.
Jiangfei Cao and co-workers studied the catalytic degradation reaction of amido black 10B (AB-10B) dye using L-cys-CuNCs supported on red globe flower carbon.424 Unlike control catalysts (carbon and clusters alone), up to 98% degradation efficiency was reached when the Fenton-like CuNCs/C-H2O2 system was used as a catalyst. Several intermediates were confirmed by LC-MS analysis for the degradation reaction of AB-10B dye.
4.3.2.1. Oxidation reactions. Chan Wang et al. studied the catalytic oxidation of styrene in the presence and absence of BSA-templated CuNCs.45 In the presence of CuNCs, the styrene oxidation resulted in several reaction products with benzaldehyde as the major product and with 70% conversion of styrene. In the absence of catalyst, the oxidation reaction occurred very slowly with 7% of styrene conversion.
Finally, CuNCs were found to catalyse the oxidation of peroxidase substrate 3,3′,5,5′-tetramethylbenzidine (TMB) (ox-TMB)148 and ethanol.172
4.3.2.2. “Click” reactions. Cu-catalysed [3 + 2] Huisgen cycloadditions of terminal alkynes and organic azides (CuAAC) are classic examples of “click” chemistry, where copper compounds are applied as heterogeneous or homogeneous catalysts in 5-membered heteroatom ring formation reactions. The review by Rajender S. Varma425 discusses the catalytic applications of Cu-based compounds (including click reactions), whereas the review by Francisco Alonso426 is focused on the “click” chemistry of nanostructured copper compounds. In this section, we will mainly consider the applications of atomically precise copper clusters as both supported and unsupported catalysts in “click” reactions. In this context, Hayton and co-workers studied the catalytic activity of supported (on silica) and unsupported [Cu20(CCPh)12(OAc)6] clusters for the [3 + 2] cycloaddition of alkynes (HCCR: R = Ph, CO2Et, and tBu) with benzyl azide.177 Although comparable triazole yields were obtained with both homogeneous and heterogeneous catalysts, the supported cluster remained stable under applied reaction conditions, whereas marked precipitation and colour change were detected for the unsupported cluster.
Didier Astruc and co-workers studied another set of “click” reactions when applying PEG-CuNCs as homogeneous and heterogeneous catalysts.427 According to the preliminary results, the supported cluster on a Santa Barbara Amorphous-15 (SBA-15) mesoporous silica sieve showed better catalytic performance than the unsupported one (Scheme 10). Besides, the exposure of the catalyst to air led to the formation of Cu2O species on the cluster surface, which, on the other hand, boosted the catalytic activity of the cluster.
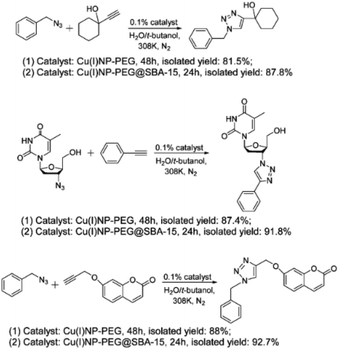 | ||
| Scheme 10 CuAAC reactions catalysed by supported and unsupported CuNP-PEGs as described in ref. 427. Republished with permission, Copyright 2017 Royal Society of Chemistry. | ||
4.3.2.3. Hydrogenation of ketones. Nanfeng Zheng and co-workers studied the catalytic hydrogenation reactions of 2-hexanone and 3-hexanone to the corresponding alcohols by using a [Cu25H10(SPhCl2)18]3− cluster as an active catalyst at room temperature.182 Combined with DFT studies, a single-site hydrogenation mechanism was proposed, where HCHO (a model ketone chosen for the calculations) was interacting with the cluster through the formation of an intermediate alkoxy group with a μ3-H atom from the cluster. The neighbouring μ6-H from the cluster diffused close to the O atom of the alkoxy group to form the reaction product. Two hydrogen vacancies formed during this process were filled by the dissociation of molecular hydrogen to complete the catalytic cycle.
4.3.2.4. Carbonylation reactions. The catalytic activity of the [Cu32(PET)24H8Cl2](PPh4)2 cluster was studied for the carbonylation reaction of anilines.189 The reactions of substituted anilines with diisopropyl azodicarboxylate in the presence of a catalyst showed 100% conversions to carbamate products (Scheme 11). Although complete aniline conversions were also reached with a copper salt and a CuPET complex as catalysts, and the yields of carbamate products were significantly reduced as compared to the cluster.
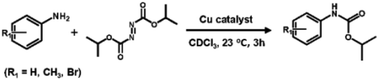 | ||
| Scheme 11 Carbonylation of anilines as described in ref. 189. Reprinted with permission, Copyright 2020 American Chemical Society. | ||
4.4. Applications in optoelectronic devices
Metal nanoclusters, particularly CuNCs, have been considered as a new generation of light-emitting diodes (LEDs) with high colour rendering index (CRI) and wide-gamut display lighting. Although commercially available LED emitters mostly use the combination of red, green, and blue (RGB) LED phosphors, orange and yellow are also occasionally applied. The main goal is to mix or convert blue or near-violet LED-sources into white LED devices (WLEDs).Benefiting from the strong and tunable PL properties of CuNCs, the Rogach group has developed a method for preparing down-conversion white light-emitting devices (LEDs).428 The synthetic method involves the formation of PVP-stabilized CuNCs and further treatment with various electron-rich ligands, such as GSH, cysteine, cysteamine, etc. They showed that the initial QY of CuNCs reported to be 8%, can be greatly enhanced (for several folds) with the treatment of the above-mentioned ligands.
The highest QY and stability were detected after GSH treatment (QY of 27%). This was attributed to the presence of electron-rich groups (–SH, –NH2, –COOH) of the ligand. Furthermore, the pre-treatment of the GSH-CuNCs powder with thermally-curable silicone resin and hardener, followed by mixing with green and red phosphors and finally coating the final mixture onto a commercial UV GaN chip, resulted in the fabrication of a white LED device (Fig. 37A). When excited at 380 nm, the PL peaks of the cluster, green and red phosphors were positioned at 434, 538 and 620 nm, respectively (Fig. 37Aa). Pictures of blue and white LEDs, and their emission spectra, are presented in Fig. 37A(b, c and d). The typical CIE (Commission Internationale de l'Eclairage) chromaticity coordinates of the device were 0.350, 0.334 (Fig. 37Ae), with a high colour rendering index (CRI) value of 92 (Fig. 37Af), correlated colour temperature (CCT) of 4742 K and a luminous efficiency of 9.8 lm W−1.
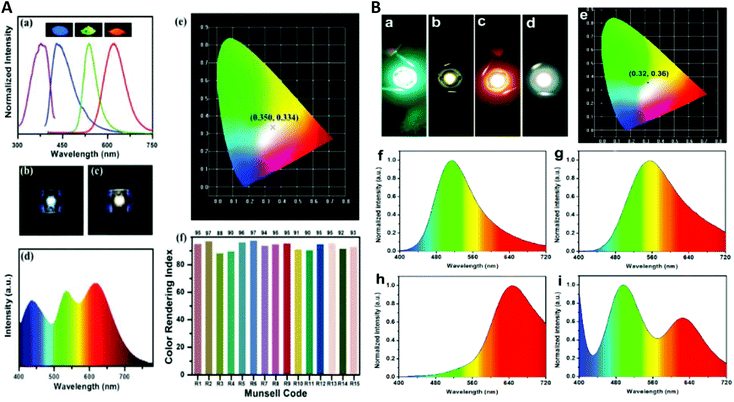 | ||
| Fig. 37 (A) (a) PLE spectrum of powdered GSH-treated CuNCs (purple line); the PL spectra of powdered GSH-treated Cu NCs (blue line), VG61E (green line) and MPR635 (red line) phosphors, with the photographs of the respective powders. Photographs of (b) blue LED and (c) white LED. (d) Emission spectrum, (e) CIE chromaticity coordinates and (f) CRI at various Munsell codes of white LEDs. Republished with permission from ref. 428. Copyright 2016 Royal Society of Chemistry. (B) Fluorescence images of the LEDs from CuNCs self-assembled ribbons (a), sheets (b), and AuNCs sheets (c). (d) Fluorescence images of the WLED from CuNCs ribbons, CuNCs sheets, and AuNCs sheets. (e) The CIE chromaticity coordinates of the WLED in (d). The corresponding emission spectra are shown in (f), (g), (h), and (i). Reprinted with permission from ref. 139. Copyright 2015 American Chemical Society. | ||
Going beyond, the same group fabricated another white LED by utilizing only CuNCs and no RGB phosphors.429 In brief, the orange-emitting CuNCs were prepared in water from the reduction with GSH. The latter was used as both reducing and capping ligand. The solvent (ethanol)-driven AIE effect was applied to improve the QY of the as-prepared CuNCs (in solution 24%, solid state 43%). Blue emitting CuNCs were prepared as mentioned before.428 The integration of orange and blue CuNCs onto commercially available GaN LED chips, resulted in the fabrication of all-CuNCs-based WLEDs. The device emitted white light with CIE, CRI, and CCT of (0.36, 0.31), 92, and 4163 K, respectively. In another case, Rogach and co-workers prepared a colour down-conversion device based on composite polymer/CuNC films.430 The copolymerization of chitosan with hypromellose resulted in the formation of a biocompatible graft copolymer, called hypromellose grafted chitosan (HGC). The latter was further applied as a depositing template to load GSH-CuNCs. The electrostatic interaction between the positively charged polymer network and negatively charged CuNCs resulted in the aggregation of the clusters showing enhanced AIE with QY as high as 42%. The obtained nanocomposite HGC/CuNCs film was used as a remote phosphor in a UV-LED chip with CIE, CRI and CCT of (0.493, 0.441), 70.1, and 2505 K, respectively. Another WLED with a colour coordinate at (0.32, 0.36) was fabricated by Yang and co-workers by combining blue-green-emitting CuNCs ribbons, yellow-emitting CuNCs sheets and AuNCs sheets with red emission (Fig. 37B).139 Note that the synthesis and AIE enhancement of these clusters were described in the section 3.3.5.
In 2020, Qiong Jia and co-workers reported a novel orange LED by covering a commercial UV LED chip with GSH-CuNCs/Zn-HDS powder.369 The developed LED with CIE coordinate of (0.51, 0.45), CCT of 2461 and CRI of 73.5 proved to be an excellent nanocomposite for light-emitting devices.
Nanfeng Zheng and co-workers reported the synthesis, crystal structure and applications of Cu53 clusters as a precursor to CuI films for perovskite solar cells.194 In brief, the iodination of Cu53 clusters at room temperature resulted in the formation of CuI films that were used as hole transport layers (HTO) for solar cells. More importantly, the developed perovskite solar cell featured a high power conversion efficiency (PCE) of 14.3%, had negligible hysteresis, and high stability (Fig. 38).
 | ||
| Fig. 38 Device structure and cross-sectional SEM image of (A) Cu53 and (B) CuI on perovskite films. (C) Energy-level diagram of CuI-based perovskite solar cell (PSC). (D) Best J–V curves of devices with CuI (w-CuI) and without CuI (w/o-CuI). FS = forward scan, RS = reverse scan. Adapted with permission from ref. 194. Copyright 2019 Wiley-VCH Verlag GmbH &Co. KGaA, Weinheim. | ||
5. Summary, challenges, and outlook
In summary, this review is a testimony to the ever-growing interest and importance of atomically precise copper nanoclusters in the era of modern nanotechnology. As a growing and fascinating era of nanosciences, the field of atomically precise metal nanoclusters, in particular CuNCs, is of paramount importance due to its unique chemical and physical properties.431 As we have already discussed in previous sections of this review, CuNCs show excellent biocompatibility and are highly photoluminescent, which are important advantages with respect to noble metal clusters. Unlike gold clusters, CuNCs have major stability issues that limit their prospective applications. However, the list of stable, compositionally precise, and highly luminescent clusters is growing continuously. In the following, we address the challenges that remain and the knowledge gaps that need to be filled.5.1. Synthesis
As we have shown in the previous sections, for the preparation of water-soluble CuNCs, both the bottom-up and top-down methods are straightforward and can be easily scaled up for mass production. The critical point here is the selection of an appropriate matrix (polymers, proteins, DNA, etc.) that will provide effective protection against the aggregation of the clusters and against the oxidation of the metallic core upon exposure to air. Although the water solubility imparts promising features for biomedical applications and biocompatibility, the main drawback is the control of the cluster size. The selective preparation of clusters with certain sizes and shapes, avoiding polydisperse mixtures, is uncontrollable to say at least. Normally, the synthesis of hydrophilic clusters in matrices results in the formation of polydisperse clusters from which certain sizes can be separated and collected via electrophoretic methods or column chromatography techniques. Note that these separation techniques require longer separation times and are not very effective, especially when neutral ligands are used as a protective shell (in the case of electrophoretic separations). This problem can somehow be overcome when top-down methods are applied. Herein, the initial mixture is etched in the presence of an excessive ligand with a greater binding affinity toward the metallic core to yield the selective formation of certain sizes. Thanks to the crystallization of some clusters, not only can their size and composition be determined, but the extraction of crystallized clusters from the mother liquor makes the separation and purification from other by-products simple. Besides, taking advantage of their solubility differences in polar and non-polar organic solvents, a mixture of two or more clusters can be separated and extracted into the corresponding solvent medium. This property is a huge advantage with respect to aqueous phase soluble clusters. Also, the knowledge of size and composition at the molecular levels is a blueprint for understanding the evolution of the structure and properties with size. This, in turn, is the basis for tuning their size-dependent properties for suitable applications. Thus, future work should be directed towards the development of synthetic procedures and protocols for selectively obtaining certain cluster sizes with high selectivities and yields. Besides, the field of CuNCs could profit more from atomically precise clusters in order to gain a better understanding of the structure-property relationship.5.2. Characterization
A variety of analytical and spectroscopic methods have been applied for the characterization of CuNCs. The importance of UV–vis spectroscopy as a “first glance” spectroscopic technique for the analysis and characterization of the clusters has been introduced in section 3.1. We will shortly discuss other techniques, their advantages, and disadvantages for the characterization of small CuNCs.A relatively simple way to obtain the size distribution and the morphology of the nanoscale objects is to “see” the nanostructure using transmission electron microscopy (TEM). The basic concept relies on the use of an electron beam passing through the thin layer of the deposited sample on a coated grid. However, the imaging of the small nanoclusters of a few nanometres in size is very challenging. Intense electron beams easily damage the clusters, leading to decomposition or aggregation. That is why sometimes the size distribution observed from TEM is not in agreement with the actual size of the cluster. However, this does not throw shade on the importance of TEM as a microscopic technique. Among other characterization techniques, TEM analysis comes in handy for having a clue about their size distribution.56,71,110,145 Although it is impossible to “see” the organic layer around the cluster core in TEM, it provides a hint about the overall core size. For copper nanoclusters, it was even more difficult and challenging to image and obtain the exact size-distribution histograms due to the low stability of the clusters in the solution or under the electron beam.132,136 In many reported cases, the discrepancy between the observed size from the TEM microscopic imaging and alternative methods (dynamic light scattering for example)136 was of major concern. Nonetheless, it is still a powerful technique for estimating the approximate size of a monodisperse sample and the size distribution of polydisperse mixtures of clusters. Thus, future efforts need to be directed toward solving the problem of the stability of the clusters under electron beams for imaging purposes. Aberration-Corrected Scanning Transmission Electron Microscopy (AC-STEM)432 or similar techniques are more appropriate for the imaging of ultra-small clusters since they are (i) non-destructive (the applied voltages are considerably lower as compared to the conventional TEM technique), and (ii) they provide ultra-high resolution images as well as allow the determination of the number of metal atoms inside the cluster. On this note, STEM techniques have been applied in the size determination (as well as the number of metal atoms) of copper31,218,321,326,409,410 and gold433,434 clusters, and copper/gold bimetallic clusters435 in a more accurate and controllable way.
X-ray photoelectron spectroscopy is a surface-sensitive analytical technique for determining the composition, oxidation state and empirical formula of the material being analysed. The concept is based on the interaction of the core electrons of the sample with an incident X-ray beam. The ejected electrons have characteristic energies for each energy level for every element present in the sample. Thus, by obtaining a survey XPS spectrum of the analyte, a complete picture of the composition and oxidation state can be extracted. XPS is extensively used for the characterization of CuNCs. Due to the difficulties in preparing and crystallizing stable copper nanoclusters, XPS has become a paramount analytical tool for determining the oxidation state of the metal inside the cluster composition. The binding energies of characteristic Cu 2p1/2 and Cu 2p3/2 appear at 952.3 and 932.3 eV, respectively, which are due to Cu0.32,33,121 However, the binding energy of Cu 2p3/2 for Cu+1 is only 0.1 eV apart; thus, it is impossible to distinguish Cu+1 from Cu0 in the cluster. In contrast, Cu2+ shows a satellite peak at around 942 eV and hence, the absence of this signal in the survey spectrum indicates the successful reduction of the copper ion. Depending on the ligand protecting the metallic core, specific elements can also be analysed. In the S 2p spectrum, the presence of a strong peak at 165.7 eV indicates the chemisorption of sulphur on the cluster surface. The peaks at the 284.6, 399.4, 530.9 eV can be assigned to C 1s, N 1s and O 1s, respectively.18,109 Thus, herein, the main drawback of the technique is the inability to distinguish metallic copper from copper(I).
Infrared (IR) spectroscopy is one of the most common and widely employed spectroscopic techniques for determining structures and identifying functional groups present in organic and hybrid organic/inorganic materials. It is especially handy in characterizing the MNCs. An indication of the successful anchoring of the thiol group on the cluster surface can be inferred by the absence of the characteristic S–H stretching vibrational mode at 2559 cm−1. Other functional groups such as carboxylates and amines can be directly tested and identified. However, with this technique, we can neither obtain information about the size of the cluster nor judge whether the sample is monodisperse or polydisperse.
Mass spectrometry (MS) is a crucial analytical technique for the determination of the chemical composition and to check the molecular purity of the cluster sample. It furthermore allows the monitoring of ligand exchange reactions in situ and the analysis of the intermediate products of such processes at any time.436 Thus, it can provide insight into reaction mechanisms. Several mass spectrometry techniques based on soft and hard ionization modes have been used for the determination of cluster composition.172,201 However, most of such analyses were conducted using matrix-assisted laser desorption/ionization (MALDI) and electrospray ionization (ESI). Both techniques are well suited for the needs in the cluster field.437 By choosing a proper MALDI matrix and ionization conditions in ESI, a mass detection of atomically precise CuNCs becomes a blueprint for assigning their compositions and following the reaction kinetics. The mass determination of organic phase-soluble clusters with precisely determined molecular compositions was more successful as compared to aqueous phase-soluble clusters (for the majority of reported cases). For instance, the chemical compositions and molecular formulas of Cu14(C2B10H10S2)6(CH3CN)8,172 [Cu25H22(PPh3)12]Cl,183 and [Cu81(PhS)46(tBuNH2)10(H)32]3+ (ref. 196) clusters determined by ESI-MS fully agree with the composition established by single-crystal X-ray studies. However, for hydrophilic clusters produced within biomolecular templates, the determination of the composition and the chemical formula is far more challenging due to fragmentation and the consequent detection of small species rather than the original molecular ion species.35,136 However, some examples have shown the successful determination of the composition when glutathione17 or other biomolecules50,55 were used as templating scaffolds.
X-ray crystallography is arguably the most important and informative technique in the field of ligand-protected metal clusters. The X-ray structure of the cluster is of paramount importance because it provides information on the “construction” of the cluster core, the arrangement of the ligands, chirality, etc. However, the main challenge remains the growth of suitable crystals. Technically, the issue is often the small quantities and purity of the sample. Nevertheless, during the last decade, a large number of crystal structures of copper clusters have been solved, as evidenced by section 2.2. In this context, there is still the need to (i) improve the crystallization methods by finding the best solvent and the best conditions, and (ii) apply more sophisticated and automated “robots” for the understanding of the fundamental crystal nucleation processes.
5.3. Size-separation
Size-exclusion chromatography (SEC) was among the first methods applied for obtaining the narrow size (shape) distribution of nanomaterials when passing through a compact gel medium with various pore sizes. The SEC separation mechanism of nanomaterials is directly related to their sizes. The bigger particles/clusters elute faster in early fractions since they cannot penetrate the small pores and thus, their progress through the gel is not hindered. In contrast, smaller clusters have longer eluting times, because they can penetrate all the pores of the gel that are larger than their sizes. Thus, depending on the nature and properties of the nanomaterial, various gels, and beads compatible with organic and aqueous solvents can be applied in the SEC separation.438 Although it is easily scalable for the separation of large quantities of clusters, the method is time-consuming, and requires the use of special gels suitable for the properties of a cluster of interest. On the other hand, SEC gels can be applied in the separation of clusters, regardless of their charge state. Electrophoretic separations, on the other hand, are limited to water-soluble and charged clusters;136 agarose and polyacrylamide (PAGE) gels are widely used as matrices for horizontal/vertical electrophoresis in slab gels. These matrices act as molecular sieves for the separation of molecules based on their size and charge. The gel porosity and hardness as well as temperature affect the separation. The main disadvantages of PAGE separations are the long separation times and the comparably large efforts for the preparation and casting of the gels prior to the separations. In capillary electrophoresis (CE), the ionized species move across the anti-convective gel-free narrow capillaries (normally made of fused silica, FS).439 However, unlike PAGE, the driving force for CE is electroosmotic flow (EOF), which arises on the walls of the capillary tube due to the applied electric field. The main advantages here are the separation of charged (negatively and positively) and neutral species in a single run and very short running/separation times. Electrophoresis and CE, are quite straightforward for the separation of water-soluble clusters; however, the stability of the clusters in the buffers can be an issue in both techniques.5.4. Unexplored compositions
Over the years, there have been impressive achievements in the field of copper nanoclusters. Nevertheless, the synthetic procedures for the preparation of CuNCs are not at the level where one can design a synthesis to obtain a certain size and composition. It is mainly based on the “trial-error” concept but it would be better to have insight into how to obtain some specific clusters. Copper clusters from low nuclearity (Cu6) to high nuclearity (Cu136) have already been discovered during the last few decades (Table 1). The majority of the discovered structures do not possess metallic cores; they are rather oxidized and contain chalcogenides as part of their core. Only in the last few years have CuNCs with partial Cu(0) character been discovered, showing unique structural properties.168,169,172,177,183,192,194 These are the only known clusters that fulfill the superatom theory with magic numbers for closed electron shells (only 2-electron systems are known for copper). In contrast, for gold nanoclusters, examples with magic electron counts from 2-electron to 58-electron systems are known and further examples are continuously being added.11,12 We are still awaiting the discovery of CuNCs with higher magic numbers. Possibly, this issue is related to the lower stability of CuNCs as compared to their gold counterparts.5.5. Stability issues
Biocompatibility, nontoxicity, catalytic activity, and strong photoluminescence are desired properties for any nanomaterials in view of the potential applications. However, a nanomaterial with these properties will not be suitable for applications unless it is sufficiently stable. The stability of the nanomaterials is of foremost importance and without solving stability issues, it will be difficult to proceed towards applications. Hereafter, the discussion is mostly made for small clusters but it also applies to larger particles and assemblies. Several factors need to be considered and carefully analysed to overcome the stability issue. Firstly, the choice of the matrix or protective ligands is of paramount importance. The type of the ligand (thiolate, phosphane, etc.) affects the stability of the cluster due to the affinity for the metal, and the overall structure of the ligand is also important. Normally, larger ligands, such as proteins, polymers, and DNA oligomers, have a greater potential to protect the cluster from aggregation due to the steric hindrance. Smaller ligands normally cannot effectively protect the core and the clusters are more prone to aggregation. On the other hand, under certain conditions (temperature, pH, solvent, etc.) the aggregation of these clusters imparts strong PL properties (AIE enhancement).18,141Secondly, the stability of the clusters depends on the external conditions like the temperature of the medium/environment, the pH of the medium/buffer when dispersed in the aqueous phase and finally the presence of ions. It is important to establish a better understanding of the cluster's stability against such factors.
5.6. Size-composition-property relationships at the nanoscale level
The quantized nano regime in ultra-small MNCs results in rich molecular-like features in their optical absorption spectra that are strongly dependent on the cluster size. This means that by tuning the size of the cluster, the physicochemical properties can be changed in a controlled manner. This concept has been vastly utilized in the case of gold nanoclusters; however, it remains a complicated task for CuNCs. This is partially because of the difficulty in preparing a series of atomically precise CuNCs with varying sizes. On the other hand, this limits our understanding of the size-dependent properties of clusters of a similar kind. Perhaps, in the future, after having gained enough knowledge on the topic, we will be able to precisely control and tune the properties of CuNCs on demand for specific applications.5.7. Challenges in applications
Nanomaterials with various structures, shapes, and morphologies, represent possibilities for applications due to their unique properties. CuNCs have found remarkable applications in catalysis, bioimaging and sensing as we have tried to elaborate in this review. The improvement of CuNCs needs to be the focus in order to find better, inexpensive and stable catalysts, to synthesize highly fluorescent, stable and biocompatible nanodyes that can be commercialized and applied in daily clinical diagnosis. There is certainly a long way to go, however, with the combined efforts of both experimental and theoretical studies, we will be able to synthesize, design and fabricate CuNCs-based universal catalysts for catalytic reactions. There is room for improvement in sensing probes, to ensure high sensitivity and selectivity towards toxic analytes, to ensure a safe and protected environment. Although CuNCs-based fluorescent probes sometimes show impressive sensitivity, they have never been commercialized and applied in real clinical practice. Finally, for biomedical applications, it is necessary to have biocompatible and nontoxic fluorescent dyes to detect any pathogens and disease markers to facilitate the clinical diagnosis, especially in the earlier stages of the disease. Although a vast amount of work has been done in this field, the research for the best is still on.We are hopeful that the field of atomically precise CuNCs will expand beyond recognition and will aid enormously in the understanding of the size-dependent properties of materials at the nanoscale.
Author contributions
All authors contributed equally.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial support from the Swiss National Science Foundation (Grant 200020_172511) and the University of Geneva is kindly acknowledged.References
- S. Dehnen, A. Eichhöfer and D. Fenske, Eur. J. Inorg. Chem., 2002, 2002, 279 CrossRef.
- O. Fuhr, S. Dehnen and D. Fenske, Chem. Soc. Rev., 2013, 42, 1871–1906 RSC.
- C. Coughlan, M. Ibáñez, O. Dobrozhan, A. Singh, A. Cabot and K. M. Ryan, Chem. Rev., 2017, 117, 5865–6109 CrossRef CAS PubMed.
- M. Walter, J. Akola, O. Lopez-Acevedo, P. D. Jadzinsky, G. Calero, C. J. Ackerson, R. L. Whetten, H. Gronbeck and H. Hakkinen, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 9157–9162 CrossRef CAS PubMed.
- H. Häkkinen, in Frontiers of Nanoscience, Elsevier Ltd, 1st edn, 2012, vol. 3, pp. 129–157 Search PubMed.
- Z. Lin, T. Slee and D. M. P. Mingos, Chem. Phys., 1990, 142, 321–334 CrossRef CAS.
- T. P. Martin, T. Bergmann, H. Goehlich and T. Lange, J. Phys. Chem., 1991, 95, 6421–6429 CrossRef CAS.
- H. Häkkinen, Adv. Phys.: X, 2016, 1, 467–491 Search PubMed.
- C. Lavenn, F. Albrieux, G. Bergeret, R. Chiriac, P. Delichère, A. Tuel and A. Demessence, Nanoscale, 2012, 4, 7334 RSC.
- P. D. Jadzinsky, G. Calero, C. J. Ackerson, D. A. Bushnell and R. D. Kornberg, Science, 2007, 318, 430–433 CrossRef CAS PubMed.
- I. Chakraborty and T. Pradeep, Chem. Rev., 2017, 117, 8208–8271 CrossRef CAS PubMed.
- R. Jin, C. Zeng, M. Zhou and Y. Chen, Chem. Rev., 2016, 116, 10346–10413 CrossRef CAS PubMed.
- S. Shahsavari, S. Hadian-Ghazvini, F. Hooriabad Saboor, I. Menbari Oskouie, M. Hasany, A. Simchi and A. L. Rogach, Mater. Chem. Front., 2019, 3, 2326–2356 RSC.
- Y. Lu, W. Wei and W. Chen, Chin. Sci. Bull., 2012, 57, 41–47 CrossRef CAS.
- N. K. Das and S. Mukherjee, Phys. Sci. Rev., 2018, 3, 1–22 CrossRef.
- S. Sharma, K. K. Chakrahari, J.-Y. Saillard and C. W. Liu, Acc. Chem. Res., 2018, 51, 2475–2483 CrossRef CAS PubMed.
- S. Maity, D. Bain and A. Patra, J. Phys. Chem. C, 2019, 123, 2506–2515 CrossRef CAS.
- X. Jia, J. Li and E. Wang, Small, 2013, 9, 3873–3879 CrossRef CAS PubMed.
- H.-H. Deng, K.-L. Li, Q.-Q. Zhuang, H.-P. Peng, Q.-Q. Zhuang, A.-L. Liu, X.-H. Xia and W. Chen, Nanoscale, 2018, 10, 6467–6473 RSC.
- A. Huczko, Appl. Phys. A: Mater. Sci. Process., 2000, 70, 365–376 CrossRef CAS.
- Y. Liu, J. Goebl and Y. Yin, Chem. Soc. Rev., 2013, 42, 2610–2653 RSC.
- Y. Xie, D. Kocaefe, C. Chen and Y. Kocaefe, J. Nanomater., 2016, 2016, 1–10 CrossRef.
- M. Cui, Y. Zhao and Q. Song, TrAC, Trends Anal. Chem., 2014, 57, 73–82 CrossRef CAS.
- Z. Wang, B. Chen and A. L. Rogach, Nanoscale Horiz., 2017, 2, 135–146 RSC.
- M. Zhao, L. Sun and R. M. Crooks, J. Am. Chem. Soc., 1998, 120, 4877–4878 CrossRef CAS.
- Y. Ling, N. Zhang, F. Qu, T. Wen, Z. F. Gao, N. B. Li and H. Q. Luo, Spectrochim. Acta, Part A, 2014, 118, 315–320 CrossRef CAS PubMed.
- Y. Ling, J. X. Li, F. Qu, N. B. Li and H. Q. Luo, Microchim. Acta, 2014, 181, 1069–1075 CrossRef CAS.
- Y. Ling, J. J. Wu, Z. F. Gao, N. B. Li and H. Q. Luo, J. Phys. Chem. C, 2015, 119, 27173–27177 CrossRef CAS.
- Y. Zhong, Q. Wang, Y. He, Y. Ge and G. Song, Sens. Actuators, B, 2015, 209, 147–153 CrossRef CAS.
- J. Feng, Y. Ju, J. Liu, H. Zhang and X. Chen, Anal. Chim. Acta, 2015, 854, 153–160 CrossRef CAS PubMed.
- M. Fernández-Ujados, L. Trapiella-Alfonso, J. M. Costa-Fernández, R. Pereiro and A. Sanz-Medel, Nanotechnology, 2013, 24, 495601 CrossRef PubMed.
- R. Ghosh, U. Goswami, S. S. Ghosh, A. Paul and A. Chattopadhyay, ACS Appl. Mater. Interfaces, 2015, 7, 209–222 CrossRef CAS PubMed.
- Q. Tang, T. Yang and Y. Huang, Microchim. Acta, 2015, 182, 2337–2343 CrossRef CAS.
- W. Dong, C. Sun, M. Sun, H. Ge, A. M. Asiri, H. M. Marwani, R. Ni and S. Wang, ACS Appl. Nano Mater., 2020, 3, 312–318 CrossRef CAS.
- P.-C. Chen, Y.-C. Li, J.-Y. Ma, J.-Y. Huang, C.-F. Chen and H.-T. Chang, Sci. Rep., 2016, 6, 24882 CrossRef CAS PubMed.
- R. Gui, J. Sun, X. Cao, Y. Wang and H. Jin, RSC Adv., 2014, 4, 29083 RSC.
- H. Zhang, X. Huang, L. Li, G. Zhang, I. Hussain, Z. Li and B. Tan, Chem. Commun., 2012, 48, 567–569 RSC.
- S. Xu, F. Chen, M. Deng and Y. Sui, RSC Adv., 2014, 4, 15664–15670 RSC.
- W. Wang, F. Leng, L. Zhan, Y. Chang, X. X. Yang, J. Lan and C. Z. Huang, Analyst, 2014, 139, 2990–2993 RSC.
- F. Gao, P. Cai, W. Yang, J. Xue, L. Gao, R. Liu, Y. Wang, Y. Zhao, X. He, L. Zhao, G. Huang, F. Wu, Y. Zhao, Z. Chai and X. Gao, ACS Nano, 2015, 9, 4976–4986 CrossRef CAS PubMed.
- B. Wang, R. Gui, H. Jin, W. He and Z. Wang, Talanta, 2018, 178, 1006–1010 CrossRef CAS PubMed.
- R. Ghosh, A. K. Sahoo, S. S. Ghosh, A. Paul and A. Chattopadhyay, ACS Appl. Mater. Interfaces, 2014, 6, 3822–3828 CrossRef CAS PubMed.
- M. Zhao, A.-Y. Chen, D. Huang, Y. Zhuo, Y.-Q. Chai and R. Yuan, Anal. Chem., 2016, 88, 11527–11532 CrossRef CAS PubMed.
- C. Wang, S. Shu, Y. Yao and Q. Song, RSC Adv., 2015, 5, 101599–101606 RSC.
- C. Wang, C. Wang, L. Xu, H. Cheng, Q. Lin and C. Zhang, Nanoscale, 2014, 6, 1775–1781 RSC.
- S. Xu, M. Deng, Y. Sui, Y. Zhang and F. Chen, RSC Adv., 2014, 4, 44644–44649 RSC.
- Z. Gao, R. Su, W. Qi, L. Wang and Z. He, Sens. Actuators, B, 2014, 195, 359–364 CrossRef CAS.
- H. Chen, L. Lin, H. Li, J. Li and J.-M. Lin, ACS Nano, 2015, 9, 2173–2183 CrossRef CAS PubMed.
- L. Xiaoqing, L. Ruiyi, L. Xiaohuan and L. Zaijun, RSC Adv., 2015, 5, 48835–48841 RSC.
- S. Ghosh, N. K. Das, U. Anand and S. Mukherjee, J. Phys. Chem. Lett., 2015, 6, 1293–1298 CrossRef CAS PubMed.
- X. Deng, X. Huang and D. Wu, Anal. Bioanal. Chem., 2015, 407, 4607–4613 CrossRef CAS PubMed.
- Y. Zhong, J. Zhu, Q. Wang, Y. He, Y. Ge and C. Song, Microchim. Acta, 2015, 182, 909–915 CrossRef CAS.
- T. Zhao, X.-W. He, W.-Y. Li and Y.-K. Zhang, J. Mater. Chem. B, 2015, 3, 2388–2394 RSC.
- H. Miao, D. Zhong, Z. Zhou and X. Yang, Nanoscale, 2015, 7, 19066–19072 RSC.
- A. Singh, T. Rai and D. Panda, RSC Adv., 2016, 6, 55539–55545 RSC.
- L. Xiaoqing, L. Ruiyi, L. Zaijun, S. Xiulan, W. Zhouping and L. Junkang, New J. Chem., 2015, 39, 5240–5248 RSC.
- N. Goswami, A. Giri, M. S. Bootharaju, P. L. Xavier, T. Pradeep and S. K. Pal, Anal. Chem., 2011, 83, 9676–9680 CrossRef CAS PubMed.
- L. Hu, Y. Yuan, L. Zhang, J. Zhao, S. Majeed and G. Xu, Anal. Chim. Acta, 2013, 762, 83–86 CrossRef CAS PubMed.
- A. S. Patel, H. Sahoo and T. Mohanty, Appl. Phys. Lett., 2014, 105, 063112 CrossRef.
- R. S. Aparna, A. D. Anjali, N. John, K. Abha, S. S. Syamchand and S. George, Spectrochim. Acta, Part A, 2018, 199, 123–129 CrossRef CAS PubMed.
- M. Jayasree, R. S. Aparna, R. R. Anjana, J. S. Anjali Devi, N. John, K. Abha, A. Manikandan and S. George, Anal. Chim. Acta, 2018, 1031, 152–160 CrossRef CAS PubMed.
- R. S. Aparna, J. S. A. Devi, J. Nebu, S. S. Syamchand and S. George, J. Photochem. Photobiol., A, 2019, 379, 63–71 CrossRef CAS.
- C. F. Monson and A. T. Woolley, Nano Lett., 2003, 3, 359–363 CrossRef CAS.
- A. Rotaru, S. Dutta, E. Jentzsch, K. Gothelf and A. Mokhir, Angew. Chem., Int. Ed., 2010, 49, 5665–5667 CrossRef CAS PubMed.
- H. Zhang, Z. Lin and X. Su, Talanta, 2015, 131, 59–63 CrossRef CAS.
- X. Zhu, H. Shi, Y. Shen, B. Zhang, J. Zhao and G. Li, Nano Res., 2015, 8, 2714–2720 CrossRef CAS.
- Y.-S. Borghei, M. Hosseini, M. R. Ganjali and S. Hosseinkhani, Sens. Actuators, B, 2017, 248, 133–139 CrossRef CAS.
- S. Kim, J. H. Kim, W. Y. Kwon, S. H. Hwang, B. S. Cha, J. M. Kim, S. S. Oh and K. S. Park, Microchim. Acta, 2019, 186, 479 CrossRef.
- S. Singh, M. K. Singh and P. Das, Sens. Actuators, B, 2018, 255, 763–774 CrossRef CAS.
- Z. Zhou, Y. Du and S. Dong, Anal. Chem., 2011, 83, 5122–5127 CrossRef CAS PubMed.
- X. Jia, J. Li, L. Han, J. Ren, X. Yang and E. Wang, ACS Nano, 2012, 6, 3311–3317 CrossRef CAS PubMed.
- J. Chen, J. Liu, Z. Fang and L. Zeng, Chem. Commun., 2012, 48, 1057–1059 RSC.
- G. Liu, Y. Shao, J. Peng, W. Dai, L. Liu, S. Xu, F. Wu and X. Wu, Nanotechnology, 2013, 24, 345502 CrossRef.
- L. Zhang, J. Zhao, M. Duan, H. Zhang, J. Jiang and R. Yu, Anal. Chem., 2013, 85, 3797–3801 CrossRef CAS.
- X.-P. Wang, B.-C. Yin and B.-C. Ye, RSC Adv., 2013, 3, 8633 RSC.
- F. Xu, H. Shi, X. He, K. Wang, D. He, Q. Guo, Z. Qing, L. Yan, X. Ye, D. Li and J. Tang, Anal. Chem., 2014, 86, 6976–6982 CrossRef CAS PubMed.
- C.-A. Chen, C.-C. Wang, Y.-J. Jong and S.-M. Wu, Anal. Chem., 2015, 87, 6228–6232 CrossRef CAS PubMed.
- H. Qian, C. Liu and R. Jin, Sci. China: Chem., 2012, 55, 2359–2365 CrossRef CAS.
- T. T. Tsai, C. A. Chen, N. Y. J. Ho, S. Yang and C. F. Chen, ACS Sens., 2019, 4, 2885–2892 CrossRef CAS PubMed.
- Y. Hu, Y. Wu, T. Chen, X. Chu and R. Yu, Anal. Methods, 2013, 5, 3577 RSC.
- Z. Wang, L. Si, J. Bao and Z. Dai, Chem. Commun., 2015, 51, 6305–6307 RSC.
- J. Liu, J. Chen, Z. Fang and L. Zeng, Analyst, 2012, 137, 5502–5505 RSC.
- T. Qing, Z. Qing, Z. Mao, X. He, F. Xu, L. Wen, D. He, H. Shi and K. Wang, RSC Adv., 2014, 4, 61092–61095 RSC.
- Q. Song, Y. Shi, D. He, S. Xu and J. Ouyang, Chem. – Eur. J., 2015, 21, 2417–2422 CrossRef CAS.
- R. Liu, C. Wang, J. Hu, Y. Su and Y. Lv, TrAC, Trends Anal. Chem., 2018, 105, 436–452 CrossRef CAS.
- C. A. Chen, S. C. Chen, M. J. A. Shiddiky, C. F. Chen and K. C. W. Wu, Chem. Rec., 2020, 20, 174–186 CrossRef CAS PubMed.
- Z. Mao, Z. Qing, T. Qing, F. Xu, L. Wen, X. He, D. He, H. Shi and K. Wang, Anal. Chem., 2015, 87, 7454–7460 CrossRef CAS PubMed.
- H.-W. Zhu, W.-X. Dai, X.-D. Yu, J.-J. Xu and H.-Y. Chen, Talanta, 2015, 144, 642–647 CrossRef CAS PubMed.
- D. Yang, Z. Guo, Y. Tang and P. Miao, ACS Appl. Nano Mater., 2018, 1, 168–174 CrossRef CAS.
- Z. Qing, X. He, D. He, K. Wang, F. Xu, T. Qing and X. Yang, Angew. Chem., Int. Ed., 2013, 52, 9719–9722 CrossRef CAS PubMed.
- M. Raja, J. Subha, F. B. Ali and S. H. Ryu, Mater. Manuf. Processes, 2008, 23, 782–785 CrossRef CAS.
- X. Yang, S. Chen, S. Zhao, D. Li and H. Ma, J. Serb. Chem. Soc., 2003, 68, 843–847 CrossRef CAS.
- Q. B. Zhang and Y. X. Hua, Phys. Chem. Chem. Phys., 2014, 16, 27088–27095 RSC.
- N. Vilar-Vidal, M. C. Blanco, M. A. López-Quintela, J. Rivas and C. Serra, J. Phys. Chem. C, 2010, 114, 15924–15930 CrossRef CAS.
- N. Vilar-Vidal, J. Rivas and M. A. López-Quintela, Phys. Chem. Chem. Phys., 2014, 16, 26427–26430 RSC.
- S. Huseyinova, J. Blanco, F. G. Requejo, J. M. Ramallo-López, M. C. Blanco, D. Buceta and M. A. López-Quintela, J. Phys. Chem. C, 2016, 120, 15902–15908 CrossRef CAS.
- P. Concepción, M. Boronat, S. García-García, E. Fernández and A. Corma, ACS Catal., 2017, 7, 3560–3568 CrossRef.
- I. Lisiecki and M. P. Pileni, J. Am. Chem. Soc., 1993, 115, 3887–3896 CrossRef CAS.
- J. N. Solanki, R. Sengupta and Z. V. P. Murthy, Solid State Sci., 2010, 12, 1560–1566 CrossRef CAS.
- I. Lisiecki and M. P. Pileni, J. Phys. Chem., 1995, 99, 5077–5082 CrossRef CAS.
- C. Vázquez-Vázquez, M. Bañobre-López, A. Mitra, M. A. López-Quintela and J. Rivas, Langmuir, 2009, 25, 8208–8216 CrossRef PubMed.
- H. Zhu, C. Zhang and Y. Yin, J. Cryst. Growth, 2004, 270, 722–728 CrossRef CAS.
- H. Kawasaki, Y. Kosaka, Y. Myoujin, T. Narushima, T. Yonezawa and R. Arakawa, Chem. Commun., 2011, 47, 7740 RSC.
- R. S. Aparna, S. S. Syamchand and S. George, J. Cluster Sci., 2017, 28, 2223–2238 CrossRef CAS.
- M. Brust, M. Walker, D. Bethell, D. J. Schiffrin and R. Whyman, J. Chem. Soc., Chem. Commun., 1994, 801–802 RSC.
- M. Brust, J. Fink, D. Bethell, D. J. Schiffrin and C. Kiely, J. Chem. Soc., Chem. Commun., 1995, 1655 RSC.
- P. J. G. Goulet and R. B. Lennox, J. Am. Chem. Soc., 2010, 132, 9582–9584 CrossRef CAS PubMed.
- Y. Li, O. Zaluzhna, B. Xu, Y. Gao, J. M. Modest and Y. J. Tong, J. Am. Chem. Soc., 2011, 133, 2092–2095 CrossRef CAS PubMed.
- W. Wei, Y. Lu, W. Chen and S. Chen, J. Am. Chem. Soc., 2011, 133, 2060–2063 CrossRef CAS PubMed.
- D. Li, Z. Chen, Z. Wan, T. Yang, H. Wang and X. Mei, RSC Adv., 2016, 6, 34090–34095 RSC.
- T. Zhou, Q. Yao, T. Zhao and X. Chen, Talanta, 2015, 141, 80–85 CrossRef CAS PubMed.
- M. Zhao, H. Feng, X. Zhang, H. Ao and Z. Qian, Analyst, 2017, 142, 4613–4617 RSC.
- J. Ma, P. Chen and H. Chang, Nanotechnology, 2014, 25, 195502 CrossRef PubMed.
- X. Jia, X. Yang, J. Li, D. Li and E. Wang, Chem. Commun., 2014, 50, 237–239 RSC.
- D. Li, B. Li and S. I. Yang, Anal. Methods, 2015, 7, 2278–2282 RSC.
- M. Cui, G. Song, C. Wang and Q. Song, Microchim. Acta, 2015, 182, 1371–1377 CrossRef CAS.
- Z. Li, S. Guo and C. Lu, Analyst, 2015, 140, 2719–2725 RSC.
- K. Basu, S. Paul, R. Jana, A. Datta and A. Banerjee, ACS Sustainable Chem. Eng., 2019, 7, 1998–2007 CrossRef CAS.
- S. Bhunia, S. Kumar and P. Purkayastha, ACS Omega, 2019, 4, 2523–2532 CrossRef CAS PubMed.
- S. K. Anand, M. R. Mathew and K. G. Kumar, J. Fluoresc., 2020, 30, 63–70 CrossRef CAS PubMed.
- X. Yang, Y. Feng, S. Zhu, Y. Luo, Y. Zhuo and Y. Dou, Anal. Chim. Acta, 2014, 847, 49–54 CrossRef CAS PubMed.
- U. Sivasankaran, J. Radecki, H. Radecka and K. Girish Kumar, Luminescence, 2019, 34, 243–248 CrossRef CAS PubMed.
- A. Dutta, U. Goswami and A. Chattopadhyay, ACS Appl. Mater. Interfaces, 2018, 10, 19459–19472 CrossRef CAS PubMed.
- Y.-J. Lin, P.-C. Chen, Z. Yuan, J.-Y. Ma and H.-T. Chang, Chem. Commun., 2015, 51, 11983–11986 RSC.
- X. Gao, C. Du, C. Zhang and W. Chen, ChemElectroChem, 2016, 3, 1266–1272 CrossRef CAS.
- B. Yin, R. R. Liu, H. L. Zhai, S. H. Lu and J. Y. Mi, Talanta, 2020, 213, 120838 CrossRef CAS PubMed.
- F. Qu, Q. Yang, B. Wang and J. You, Talanta, 2020, 207, 120289 CrossRef CAS PubMed.
- T. Luo, S. Zhang, Y. Wang, M. Wang, M. Liao and X. Kou, Luminescence, 2017, 32, 1092–1099 CrossRef CAS PubMed.
- B. Han, X. Hu, M. Yu, T. Peng, Y. Li and G. He, RSC Adv., 2018, 8, 22748–22754 RSC.
- C. Wang, L. Ling, Y. Yao and Q. Song, Nano Res., 2015, 8, 1975–1986 CrossRef CAS.
- C. Wang and Y. Huang, Nano, 2013, 08, 1350054 CrossRef.
- C. Wang, H. Cheng, Y. Sun, Q. Lin and C. Zhang, ChemNanoMat, 2015, 1, 27–31 CrossRef CAS.
- T. Zhou, W. Xu, Q. Yao, T. Zhao and X. Chen, Methods Appl. Fluoresc., 2015, 3, 044002 CrossRef PubMed.
- H. Huang, H. Li, A.-J. Wang, S.-X. Zhong, K.-M. Fang and J.-J. Feng, Analyst, 2014, 139, 6536–6541 RSC.
- N. K. Das, S. Ghosh, A. Priya, S. Datta and S. Mukherjee, J. Phys. Chem. C, 2015, 119, 24657–24664 CrossRef CAS.
- A. Baghdasaryan, R. Grillo, S. Roy Bhattacharya, M. Sharma, E. Reginato, H. Theraulaz, I. Dolamic, M. Dadras, S. Rudaz, E. Varesio and T. Burgi, ACS Appl. Nano Mater., 2018, 1, 4258–4267 CrossRef CAS.
- C. Wang, H. Cheng, Y. Huang, Z. Xu, H. Lin and C. Zhang, Analyst, 2015, 140, 5634–5639 RSC.
- Y. T. Tai, T. Simon, Y. Y. Chu and F. H. Ko, Sens. Bio-Sens. Res., 2020, 27, 100319 CrossRef.
- Z. Wu, J. Liu, Y. Gao, H. Liu, T. Li, H. Zou, Z. Wang, K. Zhang, Y. Wang, H. Zhang and B. Yang, J. Am. Chem. Soc., 2015, 137, 12906–12913 CrossRef CAS PubMed.
- L. Ai, W. Jiang, Z. Liu, J. Liu, Y. Gao, H. Zou, Z. Wu, Z. Wang, Y. Liu, H. Zhang and B. Yang, Nanoscale, 2017, 9, 12618–12627 RSC.
- M. Zhao, Z. Qian, M. Zhong, Z. Chen, H. Ao and H. Feng, ACS Appl. Mater. Interfaces, 2017, 9, 32887–32895 CrossRef CAS PubMed.
- Y. Du, J. Fang, H. Wang and Y. Yang, ACS Appl. Mater. Interfaces, 2017, 9, 11035–11044 CrossRef CAS PubMed.
- H. Rao, H. Ge, Z. Lu, W. Liu, Z. Chen, Z. Zhang, X. Wang, P. Zou, Y. Wang, H. He and X. Zeng, Microchim. Acta, 2016, 183, 1651–1657 CrossRef CAS.
- H. Cao, Z. Chen and Y. Huang, Talanta, 2015, 143, 450–456 CrossRef CAS PubMed.
- H. Cao, Z. Chen, H. Zheng and Y. Huang, Biosens. Bioelectron., 2014, 62, 189–195 CrossRef CAS PubMed.
- J.-S. Shen, Y.-L. Chen, Q.-P. Wang, T. Yu, X.-Y. Huang, Y. Yang and H.-W. Zhang, J. Mater. Chem. C, 2013, 1, 2092 RSC.
- X. J. Zhao and C. Z. Huang, New J. Chem., 2014, 38, 3673 RSC.
- H. Y. Zou, J. Lan and C. Z. Huang, RSC Adv., 2015, 5, 55832–55838 RSC.
- B. Han, T. Peng, Y. Li, M. Yu, X. Hu and G. He, Methods Appl. Fluoresc., 2018, 6, 035015 CrossRef CAS PubMed.
- X. Liu and D. Astruc, Coord. Chem. Rev., 2018, 359, 112–126 CrossRef CAS.
- X. H. Gao, S. J. He, C. M. Zhang, C. Du, X. Chen, W. Xing, S. L. Chen, A. Clayborne and W. Chen, Adv. Sci., 2016, 3, 1–7 Search PubMed.
- A. Eichhöfer, M. Kühn, S. Lebedkin, M. Kehry, M. M. Kappes and F. Weigend, Inorg. Chem., 2017, 56, 9330–9336 CrossRef PubMed.
- R. Langer, M. Yadav, B. Weinert, D. Fenske and O. Fuhr, Eur. J. Inorg. Chem., 2013, 3623–3631 CrossRef CAS.
- C. Latouche, S. Kahlal, Y. R. Lin, J. H. Liao, E. Furet, C. W. Liu and J. Y. Saillard, Inorg. Chem., 2013, 52, 13253–13262 CrossRef CAS PubMed.
- P. V. V. N. Kishore, D. R. Shi, J. H. Liao, A. K. Gupta and C. W. Liu, Inorg. Chim. Acta, 2019, 496, 119068 CrossRef CAS.
- P. K. Liao, C. S. Fang, A. J. Edwards, S. Kahlal, J. Y. Saillard and C. W. Liu, Inorg. Chem., 2012, 51, 6577–6591 CrossRef CAS PubMed.
- K. Nakamae, M. Tanaka, B. Kure, T. Nakajima, Y. Ura and T. Tanase, Chem. – Eur. J., 2017, 23, 9457–9461 CrossRef CAS PubMed.
- A. M. Polgar, A. Zhang, F. MacK, F. Weigend, S. Lebedkin, M. J. Stillman and J. F. Corrigan, Inorg. Chem., 2019, 58, 3338–3348 CrossRef CAS PubMed.
- F. Ke, Y. Song, H. Li, C. Zhou, Y. Du and M. Zhu, Dalton Trans., 2019, 48, 13921–13924 RSC.
- E. Dornsiepen, F. Weigend and S. Dehnen, Chem. – Eur. J., 2019, 25, 2486–2490 CrossRef CAS.
- H. Li, H. Zhai, C. Zhou, Y. Song, F. Ke, W. W. Xu and M. Zhu, J. Phys. Chem. Lett., 2020, 11, 4891–4896 CrossRef CAS.
- C. Latouche, J. H. Liao, Y. J. Li, R. Y. Shiu, V. Barone, S. Kahlal, C. W. Liu and J. Y. Saillard, Inorg. Chem., 2017, 56, 14135–14146 CrossRef CAS PubMed.
- R. P. Brocha Silalahi, G. R. Huang, J. H. Liao, T. H. Chiu, K. K. Chakrahari, X. Wang, J. Cartron, S. Kahlal, J. Y. Saillard and C. W. Liu, Inorg. Chem., 2020, 59, 2536–2547 CrossRef CAS PubMed.
- X. X. Yang, I. Issac, S. Lebedkin, M. Kühn, F. Weigend, D. Fenske, O. Fuhr and A. Eichhöfer, Chem. Commun., 2014, 50, 11043–11045 RSC.
- A. Eichhöfer, G. Buth, S. Lebedkin, M. Kühn and F. Weigend, Inorg. Chem., 2015, 54, 9413–9422 CrossRef PubMed.
- C. B. Khadka, B. K. Najafabadi, M. Hesari, M. S. Workentin and J. F. Corrigan, Inorg. Chem., 2013, 52, 6798–6805 CrossRef CAS PubMed.
- S. Dehnen, A. Schäfer, D. Fenske and R. Ahlrichs, Angew. Chem., Int. Ed. Engl., 1994, 33, 746–749 CrossRef.
- K. K. Chakrahari, J. H. Liao, S. Kahlal, Y. C. Liu, M. H. Chiang, J. Y. Saillard and C. W. Liu, Angew. Chem., Int. Ed., 2016, 55, 14704–14708 CrossRef CAS PubMed.
- K. K. Chakrahari, R. P. B. Silalahi, J. H. Liao, S. Kahlal, Y. C. Liu, J. F. Lee, M. H. Chiang, J. Y. Saillard and C. W. Liu, Chem. Sci., 2018, 9, 6785–6795 RSC.
- A. W. Cook, Z. R. Jones, G. Wu, S. J. Teat, S. L. Scott and T. W. Hayton, Inorg. Chem., 2019, 58, 8739–8749 CrossRef CAS PubMed.
- T. A. D. Nguyen, B. R. Goldsmith, H. T. Zaman, G. Wu, B. Peters and T. W. Hayton, Chem. – Eur. J., 2015, 21, 5341–5344 CrossRef CAS PubMed.
- Y. Li, J. Wang, P. Luo, X. Ma, X. Dong, Z. Wang, C. Du, S. Zang and T. C. W. Mak, Adv. Sci., 2019, 1900833, 1900833 CrossRef PubMed.
- M. Zhang, X. Dong, Z. Wang, H. Li, S. Li, X. Zhao and S. Zang, Angew. Chem., 2020, 132, 10138–10144 CrossRef.
- A. K. Gupta, P. V. V. N. Kishore, J. H. Liao, Y. J. Chen and C. W. Liu, Eur. J. Inorg. Chem., 2017, 2017, 1989–1993 CrossRef CAS.
- M. A. Huertos, I. Cano, N. A. G. Bandeira, J. Benet-Buchholz, C. Bo and P. W. N. M. van Leeuwen, Chem. – Eur. J., 2014, 20, 16121–16127 CrossRef CAS PubMed.
- A. Eichhöfer, J. Jiang, S. Lebedkin, D. Fenske, D. G. McDonald, J. F. Corrigan and C. Y. Su, Dalton Trans., 2012, 41, 3321–3327 RSC.
- A. W. Cook, Z. R. Jones, G. Wu, S. L. Scott and T. W. Hayton, J. Am. Chem. Soc., 2018, 140, 394–400 CrossRef CAS.
- R. S. Dhayal, J. H. Liao, Y. R. Lin, P. K. Liao, S. Kahlal, J. Y. Saillard and C. W. Liu, J. Am. Chem. Soc., 2013, 135, 4704–4707 CrossRef CAS.
- R. S. Dhayal, J.-H. Liao, X. Wang, Y.-C. Liu, M.-H. Chiang, S. Kahlal, J.-Y. Saillard and C. W. Liu, Angew. Chem., Int. Ed., 2015, 54, 13604–13608 CrossRef CAS PubMed.
- P. Y. Lin, D. Y. Li, F. H. Ho, J. H. Liao, S. K. Barik and C. W. Liu, J. Chin. Chem. Soc., 2019, 66, 988–995 CrossRef CAS.
- R. S. Dhayal, H. P. Chen, J. H. Liao, W. E. Van Zyl and C. W. Liu, ChemistrySelect, 2018, 3, 3603–3610 CrossRef CAS.
- C. Sun, N. Mammen, S. Kaappa, P. Yuan, G. Deng, C. Zhao, J. Yan, S. Malola, K. Honkala, H. Häkkinen, B. K. Teo and N. Zheng, ACS Nano, 2019, 13, 5975–5986 CrossRef CAS.
- T.-A. D. Nguyen, Z. R. Jones, B. R. Goldsmith, W. R. Buratto, G. Wu, S. L. Scott and T. W. Hayton, J. Am. Chem. Soc., 2015, 137, 13319–13324 CrossRef CAS PubMed.
- A. J. Edwards, R. S. Dhayal, P. K. Liao, J. H. Liao, M. H. Chiang, R. O. Piltz, S. Kahlal, J. Y. Saillard and C. W. Liu, Angew. Chem., Int. Ed., 2014, 53, 7214–7218 CrossRef CAS PubMed.
- T. A. D. Nguyen, Z. R. Jones, D. F. Leto, G. Wu, S. L. Scott and T. W. Hayton, Chem. Mater., 2016, 28, 8385–8390 CrossRef CAS.
- S. K. Barik, S. Huo, C. Wu, T.-H. Chiu, J. Liao, X. Wang, S. Kahlal, J.-Y. Saillard and C. W. Liu, Chem. – Eur. J., 2020, 26, 10471–10479 CrossRef CAS PubMed.
- R. S. Dhayal, J. H. Liao, S. Kahlal, X. Wang, Y. C. Liu, M. H. Chiang, W. E. Van Zyl, J. Y. Saillard and C. W. Liu, Chem. – Eur. J., 2015, 21, 8369–8374 CrossRef CAS PubMed.
- Q. Tang, Y. Lee, D. Y. Li, W. Choi, C. W. Liu, D. Lee and D. E. Jiang, J. Am. Chem. Soc., 2017, 139, 9728–9736 CrossRef CAS PubMed.
- S. Lee, M. S. Bootharaju, G. Deng, S. Malola, W. Baek, H. Häkkinen, N. Zheng and T. Hyeon, J. Am. Chem. Soc., 2020, 142, 13974–13981 CrossRef CAS PubMed.
- L. M. Zhang and T. C. W. Mak, J. Am. Chem. Soc., 2016, 138, 2909–2912 CrossRef CAS PubMed.
- R. Langer, D. Fenske and O. Fuhr, Z. Naturforsch., B: J. Chem. Sci., 2013, 68, 575–580 CrossRef CAS.
- J. Weßing, C. Ganesamoorthy, S. Kahlal, R. Marchal, C. Gemel, O. Cador, A. C. H. Da Silva, J. L. F. Da Silva, J. Y. Saillard and R. A. Fischer, Angew. Chem., Int. Ed., 2018, 57, 14630–14634 CrossRef PubMed.
- S. Bestgen, O. Fuhr, P. W. Roesky and D. Fenske, Dalton Trans., 2016, 45, 14907–14910 RSC.
- P. Yuan, R. Chen, X. Zhang, F. Chen, J. Yan, C. Sun, D. Ou, J. Peng, S. Lin, Z. Tang, B. K. Teo, L. S. Zheng and N. Zheng, Angew. Chem., Int. Ed., 2019, 58, 835–839 CrossRef CAS PubMed.
- A. Baghdasaryan, C. Besnard, L. M. Lawson Daku, T. Delgado and T. Bürgi, Inorg. Chem., 2020, 59, 2200–2208 CrossRef CAS.
- R.-W. Huang, J. Yin, C. Dong, A. Ghosh, M. J. Alhilaly, X. Dong, M. N. Hedhili, E. Abou-Hamad, B. Alamer, S. Nematulloev, Y. Han, O. F. Mohammed and O. M. Bakr, J. Am. Chem. Soc., 2020, 142, 8696–8705 CrossRef PubMed.
- M.-L. Fu, I. Issac, D. Fenske and O. Fuhr, Angew. Chem., Int. Ed., 2010, 49, 6899–6903 CrossRef CAS PubMed.
- K. YAMAMOTO and S. KASHIDA, Solid State Ionics, 1991, 48, 241–248 CrossRef CAS.
- S. Chitsaz, D. Fenske and O. Fuhr, Angew. Chem., Int. Ed., 2006, 45, 8055–8059 CrossRef CAS PubMed.
- A. H. Pakiari and Z. Jamshidi, J. Phys. Chem. A, 2010, 114, 9212–9221 CrossRef CAS PubMed.
- A. Ganguly, I. Chakraborty, T. Udayabhaskararao and T. Pradeep, J. Nanopart. Res., 2013, 15, 1522 CrossRef.
- P. Ngene, A. Longo, L. Mooij, W. Bras and B. Dam, Nat. Commun., 2017, 8, 1846 CrossRef PubMed.
- Q. Lai, Y. Sun, T. Wang, P. Modi, C. Cazorla, U. B. Demirci, J. R. Ares Fernandez, F. Leardini and K. F. Aguey-Zinsou, Adv. Sustainable Syst., 2019, 3, 1–64 Search PubMed.
- R. S. Dhayal, W. E. Van Zyl and C. W. Liu, Acc. Chem. Res., 2016, 49, 86–95 CrossRef CAS.
- R. S. Dhayal, W. E. Van Zyl and C. W. Liu, Dalton Trans., 2019, 48, 3531–3538 RSC.
- A. J. Jordan, G. Lalic and J. P. Sadighi, Chem. Rev., 2016, 116, 8318–8372 CrossRef CAS PubMed.
- M. W. Heaven, A. Dass, P. S. White, K. M. Holt and R. W. Murray, J. Am. Chem. Soc., 2008, 130, 3754–3755 CrossRef CAS PubMed.
- R. Jin, Nanoscale, 2015, 7, 1549–1565 RSC.
- V. W.-W. Yam, C.-H. Lam, W. K.-M. Fung and K.-K. Cheung, Inorg. Chem., 2001, 40, 3435–3442 CrossRef CAS.
- W.-P. Leung, Y.-C. Chan and T. C. W. Mak, Dalton Trans., 2014, 43, 63–66 RSC.
- P. C. Ford and A. Vogler, Acc. Chem. Res., 1993, 26, 220–226 CrossRef CAS.
- Y. Guo, F. Cao, X. Lei, L. Mang, S. Cheng and J. Song, Nanoscale, 2016, 8, 4852–4863 RSC.
- Y.-S. Borghei, M. Hosseini, M. Khoobi and M. R. Ganjali, J. Fluoresc., 2017, 27, 529–536 CrossRef CAS PubMed.
- J. Yang, Z. Li and Q. Jia, Sens. Actuators, B, 2019, 297, 126807 CrossRef CAS.
- Y. Hong, J. W. Y. Lam and B. Z. Tang, Chem. Soc. Rev., 2011, 40, 5361 RSC.
- N. Goswami, Q. Yao, Z. Luo, J. Li, T. Chen and J. Xie, J. Phys. Chem. Lett., 2016, 7, 962–975 CrossRef CAS PubMed.
- K. T. Prakash, N. Singh and V. Venkatesh, Chem. Commun., 2019, 55, 322–325 RSC.
- T. Zhou, J. Zhu, L. Gong, L. Nong and J. Liu, J. Am. Chem. Soc., 2019, 141, 2852–2856 CrossRef CAS PubMed.
- X. Su and J. Liu, ACS Appl. Mater. Interfaces, 2017, 9, 3902–3910 CrossRef CAS PubMed.
- Y. Huang, H. Feng, W. Liu, Y. Zhou, C. Tang, H. Ao, M. Zhao, G. Chen, J. Chen and Z. Qian, Anal. Chem., 2016, 88, 11575–11583 CrossRef CAS PubMed.
- J. Yuan, L. Wang, Y. Wang and J. Hao, Chem. – Eur. J., 2020, 26, 3545–3554 CrossRef CAS PubMed.
- R. Jalili and A. Khataee, Microchim. Acta, 2019, 186, 29 CrossRef PubMed.
- M. Ye, Y. Yu, B. Lin, Y. Cai, Y. Cao, M. Guo and D. Zhu, Sens. Actuators, B, 2019, 284, 36–44 CrossRef CAS.
- F. Geng, C. Zou, J. Liu, Q. Zhang, X. Guo, Y. Fan, H. Yu, S. Yang, Z. Liu and L. Li, Anal. Chim. Acta, 2019, 1076, 131–137 CrossRef CAS.
- L. Lin, Y. Hu, L. Zhang, Y. Huang and S. Zhao, Biosens. Bioelectron., 2017, 94, 523–529 CrossRef CAS PubMed.
- D. Li, Y. Zhao, Z. Chen, X. Mei and X. Qiu, Mater. Sci. Eng., C, 2017, 78, 653–657 CrossRef CAS PubMed.
- Y. Huang, H. Feng, W. Liu, S. Zhang, C. Tang, J. Chen and Z. Qian, J. Mater. Chem. B, 2017, 5, 5120–5127 RSC.
- Y. Liu, D. Yao and H. Zhang, ACS Appl. Mater. Interfaces, 2018, 10, 12071–12080 CrossRef CAS PubMed.
- A. Han, L. Xiong, S. Hao, Y. Yang, X. Li, G. Fang, J. Liu, Y. Pei and S. Wang, Anal. Chem., 2018, 90, 9060–9067 CrossRef CAS PubMed.
- Z. Wu, H. Liu, T. Li, J. Liu, J. Yin, O. F. Mohammed, O. M. Bakr, Y. Liu, B. Yang and H. Zhang, J. Am. Chem. Soc., 2017, 139, 4318–4321 CrossRef CAS PubMed.
- T. G. Schaaff and R. L. Whetten, J. Phys. Chem. B, 2000, 104, 2630–2641 CrossRef CAS.
- C. Noguez and I. L. Garzón, Chem. Soc. Rev., 2009, 38, 757 RSC.
- S. Knoppe and T. Bürgi, Acc. Chem. Res., 2014, 47, 1318–1326 CrossRef CAS PubMed.
- O. Lopez-Acevedo, H. Tsunoyama, T. Tsukuda, H. Häkkinen and C. M. Aikens, J. Am. Chem. Soc., 2010, 132, 8210–8218 CrossRef CAS PubMed.
- I. Dolamic, S. Knoppe, A. Dass and T. Bürgi, Nat. Commun., 2012, 3, 798 CrossRef PubMed.
- S. Knoppe, O. A. Wong, S. Malola, H. Häkkinen, T. Bürgi, T. Verbiest and C. J. Ackerson, J. Am. Chem. Soc., 2014, 136, 4129–4132 CrossRef CAS PubMed.
- M.-R. Goldsmith, C. B. George, G. Zuber, R. Naaman, D. H. Waldeck, P. Wipf and D. N. Beratan, Phys. Chem. Chem. Phys., 2006, 8, 63–67 RSC.
- V. Humblot, S. Haq, C. Muryn, W. A. Hofer and R. Raval, J. Am. Chem. Soc., 2002, 124, 503–510 CrossRef CAS PubMed.
- X. Hu, W. Wang and Y. Huang, Talanta, 2016, 154, 409–415 CrossRef CAS PubMed.
- J. Li, W. Fu, J. Bao, Z. Wang and Z. Dai, ACS Appl. Mater. Interfaces, 2018, 10, 6965–6971 CrossRef CAS PubMed.
- J. R. Bhamore, B. Deshmukh, V. Haran, S. Jha, R. K. Singhal, N. Lenka, S. K. Kailasa and Z. V. P. Murthy, New J. Chem., 2018, 42, 1510–1520 RSC.
- R. Liu, L. Zuo, X. Huang, S. Liu, G. Yang, S. Li and C. Lv, Microchim. Acta, 2019, 186, 250 CrossRef.
- H. Zhang, Y. Guan, X. Li, L. Lian, X. Wang, W. Gao, B. Zhu, X. Liu and D. Lou, Anal. Sci., 2018, 34, 1155–1161 CrossRef CAS PubMed.
- X. Feng, J. Zhang, J. Wang, A. Han, G. Fang, J. Liu and S. Wang, Anal. Methods, 2020, 12, 3130–3136 RSC.
- H. Huang, H. Li, J.-J. Feng, H. Feng, A.-J. Wang and Z. Qian, Sens. Actuators, B, 2017, 241, 292–297 CrossRef CAS.
- T. Tang, J. Ouyang, L. Hu, L. Guo, M. Yang and X. Chen, Microchim. Acta, 2016, 183, 2831–2836 CrossRef CAS.
- D. K. Sahu, D. Singha and K. Sahu, Sens. Bio-Sens. Res., 2019, 22, 100250 CrossRef.
- Y. Huang, H. Zhang, X. Xu, J. Zhou, F. Lu, Z. Zhang, Z. Hu and J. Luo, Spectrochim. Acta, Part A, 2018, 202, 65–69 CrossRef CAS PubMed.
- S. M. Lin, S. Geng, N. Li, S. G. Liu, N. B. Li and H. Q. Luo, Sens. Actuators, B, 2017, 252, 912–918 CrossRef CAS.
- Z.-C. Liu, J.-W. Qi, C. Hu, L. Zhang, W. Song, R.-P. Liang and J.-D. Qiu, Anal. Chim. Acta, 2015, 895, 95–103 CrossRef CAS PubMed.
- Z. Qing, L. Zhu, S. Yang, Z. Cao, X. He, K. Wang and R. Yang, Biosens. Bioelectron., 2016, 78, 471–476 CrossRef CAS PubMed.
- Y. Wang, T. Chen, Z. Zhang and Y. Ni, RSC Adv., 2018, 8, 9057–9062 RSC.
- Z. Shen, C. Zhang, X. Yu, J. Li, B. Liu and Z. Zhang, Microchem. J., 2019, 145, 517–522 CrossRef CAS.
- J. Yang, N. Song, X. Lv and Q. Jia, Sens. Actuators, B, 2018, 259, 226–232 CrossRef CAS.
- L. J. Ou, X. Y. Li, H. W. Liu, L. J. Li and X. Chu, Anal. Sci., 2014, 30, 723–727 CrossRef CAS.
- Z. Liu, X. Jing, S. Zhang and Y. Tian, Anal. Chem., 2019, 91, 2488–2497 CrossRef CAS.
- B. Han, R. Xiang, X. Hou, M. Yu, T. Peng, Y. Li and G. He, Anal. Methods, 2017, 9, 2590–2595 RSC.
- R. S. Aparna, J. S. Anjali Devi, R. R. Anjana, J. Nebu and S. George, Sens. Actuators, B, 2019, 291, 298–305 CrossRef CAS.
- X. Gao, X. Zhuang, C. Tian, H. Liu, W. F. Lai, Z. Wang, X. Yang, L. Chen and A. L. Rogach, Sens. Actuators, B, 2020, 307, 127626 CrossRef CAS.
- A. Nain, Y.-T. Tseng, Y.-S. Lin, S.-C. Wei, R. P. Mandal, B. Unnikrishnan, C.-C. Huang, F.-G. Tseng and H.-T. Chang, Sens. Actuators, B, 2020, 321, 128539 CrossRef CAS.
- A. Singh, S. Kaur, A. Kaur, T. Aree, N. Kaur, N. Singh and M. S. Bakshi, ACS Sustainable Chem. Eng., 2014, 2, 982–990 CrossRef CAS.
- C. Chen, F. Geng, Y. Wang, H. Yu, L. Li, S. Yang, J. Liu and W. Huang, Talanta, 2019, 205, 120132 CrossRef CAS PubMed.
- X. Hu, X. Mao, X. Zhang and Y. Huang, Sens. Actuators, B, 2017, 247, 312–318 CrossRef CAS.
- C. Boonmee, V. Promarak, T. Tuntulani and W. Ngeontae, Talanta, 2018, 178, 796–804 CrossRef CAS PubMed.
- J. Pang, Y. Lu, X. Gao, L. He, J. Sun, F. Yang, Z. Hao and Y. Liu, Microchim. Acta, 2019, 186, 1–9 CrossRef CAS PubMed.
- L. Jin, Z. Zhang, A. Tang, C. Li and Y. Shen, Biosens. Bioelectron., 2016, 79, 108–113 CrossRef CAS PubMed.
- Z. Wen, S. Song, T. Hu, C. Wang, F. Qu, P. Wang and M. Yang, Microchim. Acta, 2019, 186, 258 CrossRef PubMed.
- H. Liao, L. Hu, Y. Zhang, X. Yu, Y. Liu and R. Li, Microchim. Acta, 2018, 185, 143 CrossRef PubMed.
- G. Zhang, R. Wang, L. Shi, C. Zhang, Y. Zhang, Y. Zhou, C. Dong, G. Li and S. Shuang, Sens. Actuators, B, 2019, 279, 361–368 CrossRef CAS.
- X. Cao, X. Li, F. Liu, Y. Luo and L. Yu, Luminescence, 2018, 33, 981–985 CrossRef CAS PubMed.
- Z. Chen, Y. Niu, G. Cheng, L. Tong, G. Zhang, F. Cai, T. Chen, B. Liu and B. Tang, Analyst, 2017, 142, 2781–2785 RSC.
- F. Pena-Pereira, N. Capón, I. de la Calle, I. Lavilla and C. Bendicho, Sens. Actuators, B, 2019, 299, 126979 CrossRef CAS.
- J. Cang, C.-W. Wang, P.-C. Chen, Y.-J. Lin, Y.-C. Li and H.-T. Chang, Anal. Methods, 2017, 9, 5254–5259 RSC.
- S. Han and X. Chen, Spectrochim. Acta, Part A, 2019, 210, 315–320 CrossRef CAS PubMed.
- X.-J. Zheng, R.-P. Liang, Z.-J. Li, L. Zhang and J.-D. Qiu, Sens. Actuators, B, 2016, 230, 314–319 CrossRef CAS.
- Q. Du, X. Zhang, H. Cao and Y. Huang, Microchim. Acta, 2020, 187, 357 CrossRef CAS PubMed.
- H. B. Wang, H. D. Zhang, Y. Chen, Y. Li and T. Gan, RSC Adv., 2015, 5, 77906–77912 RSC.
- Y. Zhong, C. Deng, Y. He, Y. Ge and G. Song, Microchim. Acta, 2016, 183, 2823–2830 CrossRef CAS.
- H. Li, Y. Lu, J. Pang, J. Sun, F. Yang, Z. Wang and Y. Liu, Microchim. Acta, 2019, 186, 862 CrossRef CAS PubMed.
- S. Gou, Y. Shi, P. Li, H. Wang, T. Li, X. Zhuang, W. Li and Z. Wang, ACS Appl. Mater. Interfaces, 2019, 11, 6561–6567 CrossRef CAS PubMed.
- C. Ma, M. Chen, H. Liu, K. Wu, H. He and K. Wang, Chin. Chem. Lett., 2018, 29, 136–138 CrossRef CAS.
- R. Patel, S. Bothra, R. Kumar, G. Crisponi and S. K. Sahoo, Biosens. Bioelectron., 2018, 102, 196–203 CrossRef CAS PubMed.
- H. Li, J. Chang, T. Hou, L. Ge and F. Li, Talanta, 2016, 160, 475–480 CrossRef CAS PubMed.
- Z. Bao, K. Zhang, J. Jian, Z. Hu, K. Yuan, H. Shao, K. Peng, Z. Jiang, J. A. Zapien, Y. Yan, C. Zhang and H. Zhou, Microchim. Acta, 2018, 185, 507 CrossRef PubMed.
- W. J. Zhang, S. G. Liu, L. Han, Y. Ling, L. L. Liao, S. Mo, H. Q. Luo and N. B. Li, Anal. Methods, 2018, 10, 4251–4256 RSC.
- K. Shanmugaraj and S. A. John, New J. Chem., 2018, 42, 7223–7229 RSC.
- X. Li, X. Wu, F. Zhang, B. Zhao and Y. Li, Talanta, 2019, 195, 372–380 CrossRef CAS PubMed.
- R. S. Aparna, J. S. Anjali Devi, P. Sachidanandan and S. George, Sens. Actuators, B, 2018, 254, 811–819 CrossRef CAS.
- G. Hambarde, S. Bothra, Y. Upadhyay, R. K. Bera and S. K. Sahoo, Microchem. J., 2019, 147, 899–904 CrossRef CAS.
- Q. Li, Y. Li, H. Li, X. Yan, G. Han, F. Chen, Z. Song, J. Zhang, W. Fan, C. Yi, Z. Xu, B. Tan and W. Yan, Nanomaterials, 2020, 10, 1531 CrossRef CAS PubMed.
- C. Song, W. Hong, X. Zhang and Y. Lu, Analyst, 2018, 143, 1829–1834 RSC.
- Y. He, F. Tian, J. Zhou and B. Jiao, Microchim. Acta, 2019, 186, 199 CrossRef PubMed.
- Y. Xiong, B. Gao, K. Wu, Y. Wu, Y. Chai, X. Huang and Y. Xiong, Nanoscale, 2018, 10, 19890–19897 RSC.
- Y. Zhang, Z. Zhu, X. Teng, Y. Lai, S. Pu, P. Pang, H. Wang, C. Yang, C. J. Barrow and W. Yang, Talanta, 2019, 202, 279–284 CrossRef CAS PubMed.
- Y. Zhang, Y. Lai, X. Teng, S. Pu, Z. Yang, P. Pang, H. Wang, C. Yang, W. Yang and C. J. Barrow, Anal. Methods, 2020, 12, 1752–1758 RSC.
- Y. Ma, Y. Yu, B. Lin, L. Zhang, Y. Cao and M. Guo, Microchim. Acta, 2019, 186, 206 CrossRef PubMed.
- Z. Cai, X. Yin, J. Fang, J. Zhao, T. Wu, E. Hao, F. Tian and C. Zhang, Nano, 2020, 15, 2050063 CrossRef CAS.
- K. Fan, W. Kang, S. Qu, L. Li, B. Qu and L. Lu, Talanta, 2019, 197, 645–652 CrossRef CAS.
- Y. Yang, Q. Wei, T. Zou, Y. Kong, L. Su, D. Ma and Y. Wang, Sens. Actuators, B, 2020, 321, 128534 CrossRef CAS.
- S. Chen, Y. Wang and L. Feng, Talanta, 2020, 210, 120627 CrossRef CAS.
- R. Rajamanikandan and M. Ilanchelian, Mater. Sci. Eng., C, 2019, 98, 1064–1072 CrossRef CAS.
- H.-B. Wang, H.-D. Zhang, Y. Chen, K.-J. Huang and Y.-M. Liu, Sens. Actuators, B, 2015, 220, 146–153 CrossRef CAS.
- Z. Miao, W. Hou, M. Liu, Y. Zhang and S. Yao, New J. Chem., 2018, 42, 1446–1456 RSC.
- W. He, R. Gui, H. Jin, B. Wang, X. Bu and Y. Fu, Talanta, 2018, 178, 109–115 CrossRef CAS PubMed.
- X. Zhang, Q. Liu, Z.-W. Wang, H. Xu, F.-P. An, Q. Huang, H.-B. Song and Y.-W. Wang, Microchim. Acta, 2020, 187, 329 CrossRef CAS PubMed.
- X.-M. Wu, F. Zhang and Y. Li, J. Mater. Chem. B, 2018, 6, 5466–5475 RSC.
- R. S. Aparna, J. S. Anjali Devi, R. R. Anjana, J. Nebu and S. George, Analyst, 2019, 144, 1799–1808 RSC.
- A. Hatefi, E. Rahimpour, M. Khoubnasabjafari, M. Edalat, V. Jouyban-Gharamaleki, S. Alvani-Alamdari, A. Nokhodchi, M. H. Pournaghi-Azar and A. Jouyban, Microchim. Acta, 2019, 186, 194 CrossRef.
- H. Bagheri, A. Afkhami, H. Khoshsafar, A. Hajian and A. Shahriyari, Biosens. Bioelectron., 2017, 89, 829–836 CrossRef CAS PubMed.
- Z. Wang, C. C. Zhang, J. Gao and Q. Wang, J. Lumin., 2017, 190, 115–122 CrossRef CAS.
- Y. Luo, H. Miao and X. Yang, Talanta, 2015, 144, 488–495 CrossRef CAS.
- K. Yang, Y. Wang, C. Lu and X. Yang, J. Lumin., 2018, 196, 181–186 CrossRef CAS.
- Y. Wang, T. Chen, Q. Zhuang and Y. Ni, Talanta, 2018, 179, 409–413 CrossRef CAS.
- C. Ma, W. Ren, J. Tang, X. Wang, D. Ji, R. Meng, C. Zhang and Q. Wang, Luminescence, 2019, 34, 767–773 CrossRef CAS PubMed.
- X. Bu, Y. Fu, H. Jin and R. Gui, New J. Chem., 2018, 42, 17323–17330 RSC.
- S. Xu, K. Zhou, D. Fang and L. Ma, Molecules, 2018, 24, 95 CrossRef PubMed.
- Z. Cai, H. Li, J. Wu, L. Zhu, X. Ma and C. Zhang, RSC Adv., 2020, 10, 8989–8993 RSC.
- Z. Cai, R. Zhu, C. Zhang, E. Hao, J. Zhao and T. Wu, Chem. Pap., 2020, 74, 3839–3846 CrossRef CAS.
- X. Bu, Y. Fu, X. Jiang, H. Jin and R. Gui, Microchim. Acta, 2020, 187, 154 CrossRef CAS PubMed.
- A. Yousefzadeh, J. Hassanzadeh, S. M. J. Mousavi and M. Yousefzadeh, Sens. Actuators, B, 2019, 286, 154–162 CrossRef CAS.
- C. Muñoz-Bustos, A. Tirado-Guízar, F. Paraguay-Delgado and G. Pina-Luis, Sens. Actuators, B, 2017, 244, 922–927 CrossRef.
- R. Rajamanikandan and M. Ilanchelian, Anal. Methods, 2018, 10, 3666–3674 RSC.
- T. Li, Z. Wang, D. Jiang, H. Wang, W. F. Lai, Y. Lv and Y. Zhai, Sens. Actuators, B, 2019, 290, 535–543 CrossRef CAS.
- Y. Shi, F. Han, L. Xie, C. Zhang, T. Li, H. Wang, W.-F. Lai, S. Luo, W. Wei, Z. Wang and Y. Huang, Microchim. Acta, 2020, 187, 38 CrossRef CAS PubMed.
- L. Zhang, Q.-Y. Cai, J. Li, J. Ge, J.-Y. Wang, Z.-Z. Dong and Z.-H. Li, Biosens. Bioelectron., 2015, 69, 77–82 CrossRef CAS PubMed.
- X. Ouyang, M. Wang, L. Guo, C. Cui, T. Liu, Y. Ren, Y. Zhao, Z. Ge, X. Guo, G. Xie, J. Li, C. Fan and L. Wang, Angew. Chem., 2020, 132, 11934–11942 CrossRef.
- M. Zhang, J. Qiao, S. Zhang and L. Qi, Talanta, 2018, 182, 595–599 CrossRef CAS PubMed.
- Y. S. Borghei, M. Hosseini, M. Khoobi and M. R. Ganjali, Luminescence, 2017, 32, 1045–1050 CrossRef CAS PubMed.
- Y. Wang, T. Chen, Q. Zhuang and Y. Ni, ACS Appl. Mater. Interfaces, 2017, 9, 32135–32141 CrossRef CAS PubMed.
- Q. Song, R. Wang, F. Sun, H. Chen, Z. Wang, N. Na and J. Ouyang, Biosens. Bioelectron., 2017, 87, 760–763 CrossRef CAS PubMed.
- Y. M. Wang, J. W. Liu, L. Y. Duan, S. J. Liu and J. H. Jiang, Microchim. Acta, 2017, 184, 4183–4188 CrossRef CAS.
- S. S. Zhou, L. Zhang, Q. Y. Cai, Z. Z. Dong, X. Geng, J. Ge and Z. H. Li, Anal. Bioanal. Chem., 2016, 408, 6711–6717 CrossRef CAS PubMed.
- X. Gao, J. Liu, X. Zhuang, C. Tian, F. Luan, H. Liu and Y. Xiong, Sens. Actuators, B, 2020, 308, 127720 CrossRef CAS.
- J. Ge, Z. Z. Dong, L. Zhang, Q. Y. Cai, D. M. Bai and Z. H. Li, RSC Adv., 2016, 6, 91077–91082 RSC.
- Y. Wang, H. Cui, Z. Cao, C. Lau and J. Lu, Talanta, 2016, 154, 574–580 CrossRef CAS PubMed.
- F. Qu, B. Wang, K. Li, J. You and W. Han, Microchim. Acta, 2020, 187, 457 CrossRef CAS PubMed.
- J. Cao, W. Wang, B. Bo, X. Mao, K. Wang and X. Zhu, Biosens. Bioelectron., 2017, 90, 534–541 CrossRef CAS PubMed.
- H. Liao, Y. Zhou, Y. Chai and R. Yuan, Biosens. Bioelectron., 2018, 114, 10–14 CrossRef CAS.
- Y. Li, D. Tang, L. Zhu, J. Cai, C. Chu, J. Wang, M. Xia, Z. Cao and H. Zhu, Anal. Biochem., 2019, 585, 113346 CrossRef CAS PubMed.
- Y. Wang, X. Zhang, L. Zhao, T. Bao, W. Wen, X. Zhang and S. Wang, Biosens. Bioelectron., 2017, 98, 386–391 CrossRef CAS.
- Y. Tang, M. Liu, Z. Zhao, Q. Li, X. Liang, J. Tian and S. Zhao, Microchim. Acta, 2019, 186, 133 CrossRef PubMed.
- Y.-S. Borghei, M. Hosseini and M. R. Ganjali, Clin. Chim. Acta, 2018, 483, 119–125 CrossRef CAS PubMed.
- Y. S. Borghei, M. Hosseini and M. R. Ganjali, Microchim. Acta, 2017, 184, 2671–2677 CrossRef CAS.
- Y. Zhou, H. Wang, H. Zhang, Y. Chai and R. Yuan, Anal. Chem., 2018, 90, 3543–3549 CrossRef CAS PubMed.
- Y. Wang, W. Meng, X. Chen and Y. Zhang, Microchim. Acta, 2020, 187, 4 CrossRef CAS.
- J. Ge, Z. Z. Dong, D. M. Bai, L. Zhang, Y. L. Hu, D. Y. Ji and Z. H. Li, New J. Chem., 2017, 41, 9718–9723 RSC.
- H. Zhao, J. Dong, F. Zhou and B. Li, Sens. Actuators, B, 2017, 238, 828–833 CrossRef CAS.
- W. Zhuang, Y. Li, J. Chen, W. Liu and H. Huang, Anal. Methods, 2019, 11, 2597–2604 RSC.
- J. Yin, F. Liu, T. Fan, Y. Ren and Y. Jiang, Sens. Actuators, B, 2018, 276, 499–506 CrossRef CAS.
- D. Gao, H. Zhang, Y. Xu, Y. Liu, H. Xu and J. Cui, Anal. Biochem., 2018, 559, 5–10 CrossRef CAS PubMed.
- T. Qing, C. Long, X. Wang, K. Zhang, P. Zhang and B. Feng, Microchim. Acta, 2019, 186, 248 CrossRef PubMed.
- Q. Liu, Q. Lai, N. Li and X. Su, Microchim. Acta, 2018, 185, 182 CrossRef PubMed.
- Z. Qing, X. He, T. Qing, K. Wang, H. Shi, D. He, Z. Zou, L. Yan, F. Xu, X. Ye and Z. Mao, Anal. Chem., 2013, 85, 12138–12143 CrossRef CAS PubMed.
- X. Peng, S.-Y. Chen, L.-J. Ou, F.-W. Luo, S.-W. Qin and A. Sun, Analyst, 2018, 143, 415–419 RSC.
- H.-B. Wang, Y. Li, Y. Chen, Z.-P. Zhang, T. Gan and Y.-M. Liu, Microchim. Acta, 2018, 185, 102 CrossRef PubMed.
- Y. Hu, Y. He, Y. Han, Y. Ge, G. Song and J. Zhou, Microchim. Acta, 2019, 186, 5 CrossRef.
- R. Li, Q. Liu, Y. Jin and B. Li, Sens. Actuators, B, 2019, 281, 28–33 CrossRef CAS.
- M. Cao, Y. Jin and B. Li, Anal. Methods, 2016, 8, 4319–4323 RSC.
- Y. Ling, J. Zhou, X. F. Zhang, X. H. Wang, N. B. Li and H. Q. Luo, Sens. Actuators, B, 2019, 286, 46–51 CrossRef CAS.
- G. Liu, W. He and C. Liu, Talanta, 2019, 195, 320–326 CrossRef CAS PubMed.
- Z.-Z. Dong, L. Zhang, M. Qiao, J. Ge, A.-L. Liu and Z.-H. Li, Talanta, 2016, 146, 253–258 CrossRef CAS PubMed.
- J. Ge, L. Zhang, Z. Z. Dong, Q. Y. Cai and Z. H. Li, Anal. Methods, 2016, 8, 2831–2836 RSC.
- X. Zhang, Q. Liu, Y. Jin and B. Li, Microchim. Acta, 2019, 186, 1–9 CrossRef PubMed.
- L. Zhang, J. Zhao, H. Zhang, J. Jiang and R. Yu, Biosens. Bioelectron., 2013, 44, 6–9 CrossRef CAS PubMed.
- M. Wang, L. Liu, X. Xie, X. Zhou, Z. Lin and X. Su, Sens. Actuators, B, 2020, 313, 128023 CrossRef CAS.
- P. Ni, Y. Sun, S. Jiang, W. Lu, Y. Wang, Z. Li and Z. Li, Sens. Actuators, B, 2017, 240, 651–656 CrossRef CAS.
- F. Zhou, X. Cui, A. Shang, J. Lian, L. Yang, Y. Jin and B. Li, Microchim. Acta, 2017, 184, 773–779 CrossRef CAS.
- K. Zhang, X. Wang, C. Long, J. Xu, Z. Jiang, B. Feng, P. Zhang, J. Fei and T. Qing, Sens. Actuators, B, 2020, 316, 128072 CrossRef CAS.
- J. Yang, Z. Li and Q. Jia, Chem. Commun., 2020, 56, 3081–3084 RSC.
- X. Huang, H. Zhao, W. Qiu, J. Wang, L. Guo, Z. Lin, W. Pan, Y. Wu and B. Qiu, Microchim. Acta, 2020, 187, 240 CrossRef CAS PubMed.
- T. Ye, Y. Peng, M. Yuan, H. Cao, J. Yu, Y. Li and F. Xu, Microchim. Acta, 2019, 186, 760 CrossRef CAS PubMed.
- J. Yang, N. Song and Q. Jia, Nanoscale, 2019, 11, 21927–21933 RSC.
- S. Chen, Z. Huang and Q. Jia, Sens. Actuators, B, 2020, 319, 128305 CrossRef CAS.
- M. Wang, L. Wang, Q. Liu and X. Su, Sens. Actuators, B, 2018, 256, 691–698 CrossRef CAS.
- H.-B. Wang, H.-D. Zhang, Y. Chen and Y.-M. Liu, Biosens. Bioelectron., 2015, 74, 581–586 CrossRef CAS PubMed.
- Y. He and B. Jiao, Sens. Actuators, B, 2018, 265, 387–393 CrossRef CAS.
- X.-G. Li, F. Zhang, Y. Gao, Q.-M. Zhou, Y. Zhao, Y. Li, J.-Z. Huo and X.-J. Zhao, Biosens. Bioelectron., 2016, 86, 270–276 CrossRef CAS.
- Q. Song, L. Yang, H. Chen, R. Zhang, N. Na and J. Ouyang, Talanta, 2017, 174, 444–447 CrossRef CAS.
- L. J. Ou, X. Y. Li, L. J. Li, H. W. Liu, A. M. Sun and K. J. Liu, Analyst, 2015, 140, 1871–1875 RSC.
- S. Zhang, C. Chen, X. Qin, Q. Zhang, J. Liu, J. Zhu, Y. Gao, L. Li and W. Huang, Talanta, 2018, 189, 92–99 CrossRef CAS PubMed.
- L. Wang, F. Shi, Y. Li and X. Su, Sens. Actuators, B, 2016, 222, 945–951 CrossRef CAS.
- Y. Hu, Y. He, Y. Han, Y. Ge, G. Song and J. Zhou, Microchim. Acta, 2018, 185, 383 CrossRef PubMed.
- F. M. Moghadam and M. Rahaie, Biosens. Bioelectron., 2019, 132, 186–195 CrossRef CAS.
- M. Chen, F. Yeasmin Khusbu, C. Ma, K. Wu, H. Zhao, H. Chen and K. Wang, New J. Chem., 2018, 42, 13702–13707 RSC.
- L. Zhao and Z. Ma, Sens. Actuators, B, 2017, 241, 849–854 CrossRef CAS.
- J. Ye, X. Dong, H. Jiang and X. Wang, J. Mater. Chem. B, 2017, 5, 691–696 RSC.
- C. Wang, Y. Yao and Q. Song, Colloids Surf., B, 2016, 140, 373–381 CrossRef CAS.
- Y. Qiao, T. Xu, Y. Zhang, C. Zhang, L. Shi, G. Zhang, S. Shuang and C. Dong, Sens. Actuators, B, 2015, 220, 1064–1069 CrossRef CAS.
- Z. Cai, C. Zhang and K. Jia, Chem. Pap., 2020, 74, 1831–1838 CrossRef CAS.
- X. Mao, S. Liu, C. Yang, F. Liu, K. Wang and G. Chen, Anal. Chim. Acta, 2016, 909, 101–108 CrossRef CAS.
- E. Shokri, M. Hosseini, A. A. Sadeghan, A. Bahmani, N. Nasiri and S. Hosseinkhani, Sens. Actuators, B, 2020, 321, 128634 CrossRef CAS.
- Y. Zhao, X. Wang, J. Mi, Y. Jiang and C. Wang, Part. Part. Syst. Charact., 2019, 36, 1–19 Search PubMed.
- T. Qing, K. Zhang, Z. Qing, X. Wang, C. Long, P. Zhang, H. Hu and B. Feng, Microchim. Acta, 2019, 186, 670 CrossRef CAS.
- Y. An, Y. Ren, M. Bick, A. Dudek, E. Hong-Wang Waworuntu, J. Tang, J. Chen and B. Chang, Biosens. Bioelectron., 2020, 154, 112078 CrossRef CAS.
- X. Hu, T. Liu, Y. Zhuang, W. Wang, Y. Li, W. Fan and Y. Huang, TrAC, Trends Anal. Chem., 2016, 77, 66–75 CrossRef CAS.
- Z. Qing, A. Bai, S. Xing, Z. Zou, X. He, K. Wang and R. Yang, Biosens. Bioelectron., 2019, 137, 96–109 CrossRef CAS.
- Q. Cao, J. Li and E. Wang, Biosens. Bioelectron., 2019, 132, 333–342 CrossRef CAS.
- WHO, W. H. O., 2004, 1–31 Search PubMed.
- WHO, W. H. O., 2019, 1–6 Search PubMed.
- H. Sun, J. Brocato and M. Costa, Curr. Environ. Health Rep., 2015, 2, 295–303 CrossRef CAS PubMed.
- WHO, W. H. O., 2003, 1–10 Search PubMed.
- WHO, W. H. O., 2003, 1–41 Search PubMed.
- WHO, W. H. O., 2011, 1–31 Search PubMed.
- H. Komaba and M. Fukagawa, Kidney Int., 2016, 90, 753–763 CrossRef CAS PubMed.
- WHO, W. H. O., 2018, 1–5 Search PubMed.
- WHO, W. H. O., 2008, 1–7 Search PubMed.
- J.-M. Xia, X. Wei, X.-W. Chen, Y. Shu and J.-H. Wang, Microchim. Acta, 2018, 185, 205 CrossRef PubMed.
- Y. Wang, Y. Cui, R. Liu, Y. Wei, X. Jiang, H. Zhu, L. Gao, Y. Zhao, Z. Chai and X. Gao, Chem. Commun., 2013, 49, 10724–10726 RSC.
- M. J. Barthel, I. Angeloni, A. Petrelli, T. Avellini, A. Scarpellini, G. Bertoni, A. Armirotti, I. Moreels and T. Pellegrino, ACS Nano, 2015, 9, 11886–11897 CrossRef CAS PubMed.
- G. S. Heo, Y. Zhao, D. Sultan, X. Zhang, L. Detering, H. P. Luehmann, X. Zhang, R. Li, A. Choksi, S. Sharp, S. Levingston, T. Primeau, D. E. Reichert, G. Sun, B. Razani, S. Li, K. N. Weilbaecher, F. Dehdashti, K. L. Wooley and Y. Liu, ACS Appl. Mater. Interfaces, 2019, 11, 19669–19678 CrossRef CAS PubMed.
- W. F. Lai, W. T. Wong and A. L. Rogach, Adv. Mater., 2020, 1906872, 1–21 Search PubMed.
- J. Xia, W. Wang, X. Hai, S. E, Y. Shu and J. Wang, Chin. Chem. Lett., 2019, 30, 421–424 CrossRef CAS.
- A. T. Simon, D. Dutta, A. Chattopadhyay and S. S. Ghosh, ACS Omega, 2019, 4, 4697–4706 CrossRef CAS PubMed.
- U. Goswami, A. Dutta, A. Raza, R. Kandimalla, S. Kalita, S. S. Ghosh and A. Chattopadhyay, ACS Appl. Mater. Interfaces, 2018, 10, 3282–3294 CrossRef CAS PubMed.
- Y. Liu, J. Liang, Q. Wang, Y. He and Y. Chen, J. Appl. Toxicol., 2016, 36, 454–463 CrossRef CAS PubMed.
- A. Halder, L. A. Curtiss, A. Fortunelli and S. Vajda, J. Chem. Phys., 2018, 148, 110901 CrossRef PubMed.
- B. Yang, C. Liu, A. Halder, E. C. Tyo, A. B. F. Martinson, S. Seifert, P. Zapol, L. A. Curtiss and S. Vajda, J. Phys. Chem. C, 2017, 121, 10406–10412 CrossRef CAS.
- C. Liu, B. Yang, E. Tyo, S. Seifert, J. Debartolo, B. Von Issendorff, P. Zapol, S. Vajda and L. A. Curtiss, J. Am. Chem. Soc., 2015, 137, 8676–8679 CrossRef CAS PubMed.
- C. Cheng, X. Zhang, Z. Yang and Z. Zhou, ACS Appl. Mater. Interfaces, 2018, 10, 32903–32912 CrossRef CAS PubMed.
- E. Fernandez, M. Boronat and A. Corma, J. Phys. Chem. C, 2020, 124, 21549–21558 CrossRef CAS.
- P. López-Caballero, A. W. Hauser and M. Pilar De Lara-Castells, J. Phys. Chem. C, 2019, 123, 23064–23074 CrossRef PubMed.
- L. Liu and A. Corma, Chem. Rev., 2018, 118, 4981–5079 CrossRef CAS PubMed.
- N. Vilar-Vidal, J. Rivas and M. A. López-Quintela, ACS Catal., 2012, 2, 1693–1697 CrossRef CAS.
- C. Xie, X. Wen, C. Xiao, S. Wei, X. Wu, S. Liu and J. Cao, Water, Air, Soil Pollut., 2020, 231, 280 CrossRef CAS.
- M. B. Gawande, A. Goswami, F.-X. Felpin, T. Asefa, X. Huang, R. Silva, X. Zou, R. Zboril and R. S. Varma, Chem. Rev., 2016, 116, 3722–3811 CrossRef CAS PubMed.
- F. Alonso, Y. Moglie and G. Radivoy, Acc. Chem. Res., 2015, 48, 2516–2528 CrossRef CAS PubMed.
- F. Fu, A. Martinez, C. Wang, R. Ciganda, L. Yate, A. Escobar, S. Moya, E. Fouquet, J. Ruiz and D. Astruc, Chem. Commun., 2017, 53, 5384–5387 RSC.
- Z. Wang, A. S. Susha, B. Chen, C. Reckmeier, O. Tomanec, R. Zboril, H. Zhong and A. L. Rogach, Nanoscale, 2016, 8, 7197–7202 RSC.
- Z. Wang, B. Chen, A. S. Susha, W. Wang, C. J. Reckmeier, R. Chen, H. Zhong and A. L. Rogach, Adv. Sci., 2016, 3, 1600182 CrossRef PubMed.
- Z. Wang, Y. Shi, X. Yang, Y. Xiong, Y. Li, B. Chen, W.-F. Lai and A. L. Rogach, Adv. Funct. Mater., 2018, 28, 1802848 CrossRef.
- D. Bain, S. Maity and A. Patra, Phys. Chem. Chem. Phys., 2019, 21, 5863–5881 RSC.
- E. P. W. Ward, I. Arslan, P. A. Midgley, A. Bleloch and J. M. Thomas, Chem. Commun., 2005, 5805–5807 RSC.
- J. Liu, N. Jian, I. Ornelas, A. J. Pattison, T. Lahtinen, K. Salorinne, H. Häkkinen and R. E. Palmer, Ultramicroscopy, 2017, 176, 146–150 CrossRef CAS.
- Z. W. Wang, O. Toikkanen, B. M. Quinn and R. E. Palmer, Small, 2011, 7, 1542–1545 CrossRef CAS PubMed.
- F. Yin, Z. W. Wang and R. E. Palmer, J. Exp. Nanosci., 2012, 7, 703–710 CrossRef CAS.
- P. Chakraborty and T. Pradeep, NPG Asia Mater., 2019, 11, 48 CrossRef.
- Y. Lu and W. Chen, Anal. Chem., 2015, 87, 10659–10667 CrossRef CAS PubMed.
- L. Pitkänen and A. M. Striegel, TrAC, Trends Anal. Chem., 2016, 80, 311–320 CrossRef PubMed.
- F. Tagliaro, G. Manetto, F. Crivellente and F. Smith, Forensic Sci. Int., 1998, 92, 75–88 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2021 |