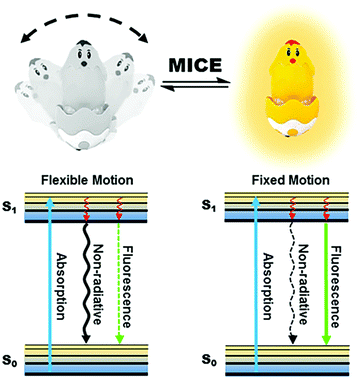
Motion-induced change in emission (MICE) for developing fluorescent probes
Dongdong
Su
 a,
Chai Lean
Teoh
a,
Lu
Wang
a,
Xiaogang
Liu
a,
Chai Lean
Teoh
a,
Lu
Wang
a,
Xiaogang
Liu
 b and
Young-Tae
Chang
b and
Young-Tae
Chang
 *ac
*ac
aLaboratory of Bioimaging Probe Development, Singapore Bioimaging Consortium, Agency for Science, Technology and Research (A*STAR), 11 Biopolis Way, Helios #02-02, 138667 Singapore
bSingapore University of Technology and Design, 8 Somapah Road, 487372 Singapore
cDepartment of Chemistry, Pohang University of Science and Technology, Pohang, 37673, Republic of Korea. E-mail: ytchang@postech.ac.kr
First published on 10th July 2017
Abstract
The need for detecting and labelling environmentally and biologically important analytes has driven considerable research efforts in developing fluorescent probes. During the sensing process, molecular motions (i.e., molecular rotations or vibrations) of a flexible fluorescent probe can be significantly altered by its embedding micro-environment or analyte, thereby leading to substantial changes in readout signals. Motion-induced change in emission (MICE) can be utilized as an effective sensing mechanism. However, in comparison to the well-understood sensing mechanisms, such as photo-induced electron transfer (PET), intramolecular charge transfer (ICT), aggregation-induced emission (AIE) and disaggregation-induced emission (DIE), MICE has not been systematically discussed to date. In this tutorial review, we will summarize the concept and mechanisms of MICE for developing single-molecular fluorescent probes, present unique advantages of MICE based sensors, demonstrate their various applications, and discuss technical challenges in this field. We expect that this review will promote a deeper understanding of MICE and facilitate the development of novel MICE based probes.
Key learning points(1) Motion-induced change in emission (MICE) as a sensing mechanism.(2) Structural and physical properties of MICE based fluorescent probes. (3) Representative applications of MICE probes. (4) Design considerations for developing dedicated MICE based probes. |
1. Introduction
Fluorescent molecular probes have emerged as indispensable tools in a broad range of bioimaging and biosensing applications.1 These probes offer a convenient approach to monitoring specific analytes with high sensitivity, great resolution, and vivid visibility. In recent years, along with a rapid advancement in microscopic imaging technologies, a great number of fluorescent probes have been developed, for labelling and detecting various environmentally and biologically important analytes. The main sensing strategy of these fluorescent probes relies on fluorescence changes before and after interactions with target species via a few known sensing mechanisms, such as photo-induced electron transfer (PET),2 intramolecular charge transfer (ICT),3 aggregation-induced emission (AIE),4 and disaggregation-induced emission (DIE).5 Most of these probes have a distinct chemical target, via a judicious choice of sensing recognizing motifs and signal reporting motifs. However, such clearly defined sensing motifs are not available in many micro-environmental sensing applications, i.e., in monitoring intracellular viscosity, temperature, and certain proteins. For these purposes, fluorescence changes can be achieved by modifying the motion changes in fluorescent probes in different microenvironments. It is noteworthy that such changes do not alter the chemical structure of a probe, but only adjust its molecular conformation. These physical changes are in stark contrast to reaction-based sensing mechanisms.6Motion changes in fluorescent probes are manifested by variations in their fluorescence intensity and lifetime, in response to the embedding environment or interacting analytes. These changes in readout signals have been utilized as an effective sensing strategy. Among many such sensing applications, aggregation-induced emission (AIE) has emerged in the past few decades, gained rapid development and been thoroughly reviewed.4 AIE based fluorescent molecules are non-emissive in monomers, but becomes highly emissive in molecular aggregates as intramolecular motions become restricted.7 The sensing process of AIE probes involves molecular aggregation of either the sensors themselves or with other objects including analytes and/or matrixes. Although the term AIE is useful and popular, it is difficult to classify many motion-based probes as AIE sensors. For example, many fluorescent sensors exhibit enhanced fluorescence upon one-to-one binding to target proteins, due to conformational rigidification. This one-to-one molecular recognition is probably better described as molecular complexation, rather than aggregation. Moreover, motion changes in fluorescent sensors in response to their embedding environment have permitted the development of abundant viscosity and temperature sensors that function in their monomer forms without molecular aggregation. To cover these less discussed situations in contrast to AIE, we raise Motion-Induced Change in Emission (MICE) as a descriptor for single-molecular sensors that function based on motion changes due to the surrounding environment or molecular complexation. MICE has not been fully appreciated as an independent concept or systematically discussed, in contrast to other well-established sensing mechanisms (e.g., PET, ICT, AIE & DIE). Yet, many existing fluorescent probes fall into the category of MICE. In this review, we aim to provide a rigorous but succinct overview of the MICE mechanism, MICE based molecular probes and their applications in viscosity, temperature and biomolecule recognition. We hope that this review offers a useful and timely introduction to MICE, and facilitates the development of high-performance MICE based probes for various applications.
2. Sensing mechanism of MICE
Motion changes in fluorescent probes, such as molecular rotations and vibrations of fluorophores, are results of their conformational flexibility (Fig. 1). These motion changes have important implications for the fluorescence properties of molecular probes. In the excited states of MICE based probes (the S1 state), the absorbed energy can be released primarily by non-radiative relaxation or by fluorescence emission. When the probe experiences flexible motions, non-radiative relaxation dominates energy transfer. In contrast, when the probe adopts a rigid molecular structure, energy is mostly released in the form of fluorescence. Consequently, the immediate surrounding environment modulates motion changes in the molecular probe, and adjusts the ratio of non-radiative relaxation against fluorescence. The coexistence and competition of these two kinds of energy release pathways make MICE an excellent sensing mechanism in developing fluorescent probes.The relatively simple concept of MICE holds great promises for designing functional probes. Firstly, as MICE based probes experience only physical changes (and no chemical reactions), their responses to variations in the surrounding environment are fast and highly reversible. Moreover, these characteristics of MICE probes permit dynamic and real-time monitoring of environmental changes, such as viscosity and temperature variations. Finally, the change of motion has a significant influence on the fluorescence output of MICE based probes, thus affording great sensitivity. Hence, MICE sensors are very useful in studying abnormal intracellular changes in microenvironments.
3. Application of MICE in viscosity sensing
Intracellular viscosity has an important role in biological signalling processes. Abnormal changes in intracellular viscosity are shown to be associated with a variety of diseases. The intracellular viscosity varies according to different cell types and testing techniques, ranging from 1 cP to hundreds of cP (Fig. 2). For example, the viscosity values vary between 60 and 110 cP in mitochondria of living HeLa cells,8 and range from 50 to 90 cP in lysosomes of MCF-7 cells.9 Among various techniques for measuring viscosity, the use of fluorescent viscosity sensors has attracted considerable attention in bioimaging applications, as they are minimally invasive to live cells and this method affords higher spatial and temporal resolution than mechanical methods.10 Until now, most of the fluorescent viscosity sensors were developed based on the MICE mechanism.11 MICE based viscosity sensors, or molecular rotors, contain an intramolecular rotational moiety connected to or within a fluorophore scaffold. The freely rotating rotor in a non-viscous environment significantly increases the non-radiative decay rate, quenches fluorescence, and shortens the fluorescence lifetime. In contrast, restricting the rotation of the molecular rotor in a viscous environment greatly enhances the fluorescence intensity and lengthens the fluorescence lifetime.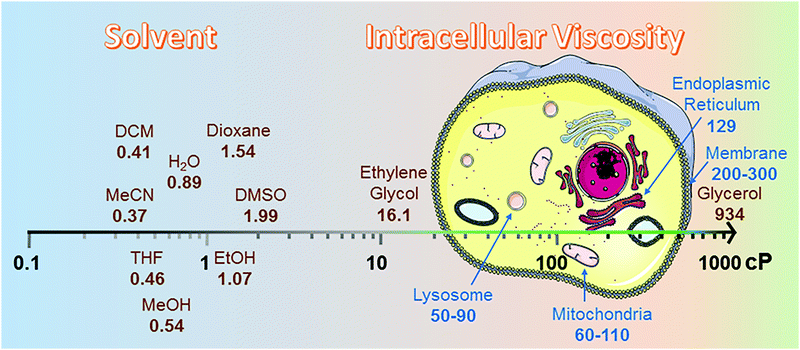 | ||
| Fig. 2 Viscosity data summary. The viscosities of different solvents were highlighted from 25 °C (298.15 K).12 cP is the abbreviation for centipoise, a standard unit of measurement for viscosity. The viscosities of different organelles, like lysosome,9 mitochondria,8 endoplasmic reticulum13 and membrane,14 were cited from research papers. | ||
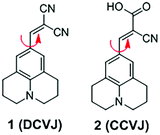
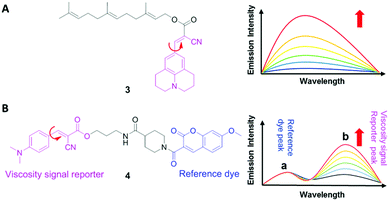
3.1 DCVJ family of viscosity sensors
9-(Dicyanovinyl)julolidine (DCVJ; 1) is a well-characterized molecular rotor (Fig. 3).15,16 Fluorescence quantum yields of DCVJ depend on environmental factors, such as solvent viscosity and temperature changes, because these factors affect intramolecular rotations of this fluorescent probe. Due to this attractive property, DCVJ was employed to study viscosities in microenvironments, such as cell membrane and phospholipid bilayers. However, DCVJ showed limited binding capacity. To overcome this shortcoming, different targeting motifs could be linked to a similar molecular rotor CCVJ (2) through a straightforward reaction with its carboxylic acid group (Fig. 3). In 2001, Theodorakis and coworkers reported a series of CCVJ-derived viscosity sensors for quantifying viscosities in cell membranes.17 Their molecular rotors carry aliphatic hydrocarbon chains of different lengths. The aliphatic hydrocarbon chains virtually do not affect viscosity-dependent fluorescence responses of CCVJ. Nevertheless, longer hydrocarbon chains showed increased membrane selectivity by reducing dye migration into the cytoplasm and other organelles. For example, a farnesyl-containing sensor 3 showed an increased affinity for cell membranes, which leads to >20 times improvement in sensitivity to membrane viscosities than that of DCVJ (Fig. 4A). Further designs based on a similar core structure have also been developed for mapping viscosity in live cells or in vivo.Although the fluorescence intensities of molecular rotors from the DCVJ family have a strong dependence on solvent viscosity, their emission intensities can also be affected by many other factors, such as variations in dye concentrations, changes in fluid optical properties in heterogeneous systems, and fluctuations in laser excitation power. To avoid these limitations and yield reliable viscosity information, Haidekker and co-workers constructed a ratiometric sensor 4via employing the FRET strategy (Fig. 4B).18 Ratiometric viscosity sensor 4 covalently links two distinct fluorescent dyes: an environment-insensitive fluorophore, MCCA (7-methoxycoumarin-3-carboxylic acid), acts as the energy donor and serves as a reference dye; an environment-sensitive CMAM fluorophore (2-cyano-3-(4-dimethylaminophenyl) acrylic acid methyl ester) was chosen as the energy acceptor and functions as a viscosity sensor. By exciting the donor fluorophore, energy transfers from the donor (the reference dye) to the acceptor (the viscosity sensor). In addition, the viscosity sensor shows a clear fluorescence enhancement, as solvent viscosity increases; in contrast, the fluorescence intensity of the reference fluorophore remains unchanged throughout the course. By taking ratios of emission intensities of the viscosity sensor and the reference dye, one can cancel out the effects of medium environments, dye concentrations and laser excitation power, and thus obtain accurate viscosity values.
3.2 BODIPY-based viscosity sensors
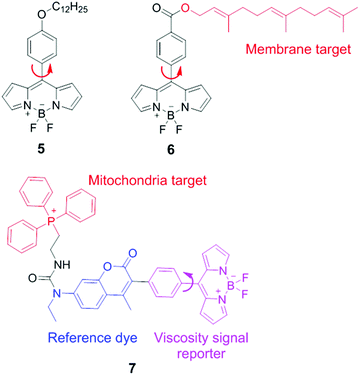
BODIPY (boron difluoride dipyrromethene) derivatives are another type of molecular rotors for measuring viscosities. In these molecular rotors, rotations of the meso phenyl ring with respect to the dipyrromethene core play a critical role in determining the fluorescence output. In a low-viscosity medium, free rotations of the meso phenyl ring accelerate non-radiative decays, and lead to weak fluorescence. In a high-viscosity medium, the rotation of the phenyl group is restricted and these molecular rotors emit strong fluorescence. The viscosity response of these rotors can be further tuned via chemical substitution either on the BODIPY scaffold or on the meso phenyl ring.In 2008, Kuimova and Suhling developed a BODIPY molecular rotor 5 to map the viscosity of live cells via fluorescence lifetime imaging (FLIM; Fig. 5).19 They synthesized a series of BODIPY-based fluorescent dyes with different chemical substituents and carefully compared their fluorescence properties. The results show that with a substituent in the ortho-positions of the phenyl ring or positions 1 and 7 in the BODIPY core structure, the free rotation of the meso phenyl ring around a single bond is sterically restricted. Such dyes exhibit bright fluorescence but lose viscosity sensitivity. In contrast, sensor 5, which contains a long tail in the para position of the phenyl ring, demonstrates a dramatic enhancement in both fluorescence quantum yield and fluorescence lifetime as solvent viscosity increases. The fluorescence lifetime of 5 increased from 0.7 ± 0.05 to 3.8 ± 0.1 ns as the viscosity increases from 28 to 950 cP. Similar to ratiometric fluorescence imaging, lifetime based measurements eliminate dependence on dye concentrations and laser excitation power, etc., thus leading to an accurate determination of solvent viscosity. Subsequently, FLIM experiments with molecular rotor 5 have been performed and provided detailed cellular viscosity information with good spatial resolution.
Suhling and co-workers also reported another BODIPY-based fluorescent rotor 6 with a long and hydrophobic tail. This tail locates 6 in membrane regions of intracellular organelles (Fig. 5).20 Moreover, both fluorescence intensity and lifetime of 6 increase substantially with increasing viscosities. Consequently, this molecular rotor has been applied in FLIM experiments to determine the viscosity of organelle membranes and study their dynamic changes in various biological environments.
By combining the ratiometric sensing strategy with a organelle-specific chemical group, Kim and co-workers developed a self-calibrating bipartite sensor 7 (Fig. 5).8 The molecular structure of 7 consists of three parts: a viscosity insensitive coumarin dye (the reference dye), a viscosity-sensitive BODIPY rotor, and a phosphonium cation which serves as a mitochondria-targeting unit. Consequently, this probe monitors the viscosity changes in mitochondria, via either ratiometric fluorescence intensity or ratiometric fluorescence lifetime analysis. Kim et al. found that the average mitochondrial viscosity of living cells was ∼62 cP. Furthermore, treatment of mitochondria with monensin and nystatin led to an increase in mitochondrial viscosity to ∼110 cP, due to drug-induced swelling and structural changes.
3.3 CHO-based viscosity sensors
In 2011, Peng and co-workers reported a fluorescent viscosity sensor based on the MICE mechanism.21 By connecting an aldehyde group (CHO) to the meso position of Cy5, they synthesized a viscosity sensor, 8 (Fig. 6). Sensor 8 shows good viscosity responses in live cells via both ratiometric fluorescence imaging and FLIM, as a result of the motion changes in the CHO group. In a non-viscous media or a high-temperature media, the free rotation of the CHO group activates significant non-radiative decays; in a viscous or a low-temperature media, restricting rotations of the CHO group increase the fluorescence intensity and lifetime of 8. Moreover, two emission peaks (de-excitations from the S1 and S2 states, respectively) make 8 a ratiometric fluorescent sensor with a large pseudo-Stokes shift. Viscosities derived from the ratiometric imaging have a good agreement with those determined from FLIM experiments in NIH-3T3 cells, indicating good measurement reliabilities. The same group also reported a BODIPY viscosity sensor, 9, based on a similar design (Fig. 6).22 Motion changes in the CHO group of sensor 9 induce a considerable variation in fluorescence intensity as solvent viscosity changes. Consequently, sensor 9 maps viscosity information in live cells and demonstrates the increase of cytoplasmic viscosities during apoptosis in real time.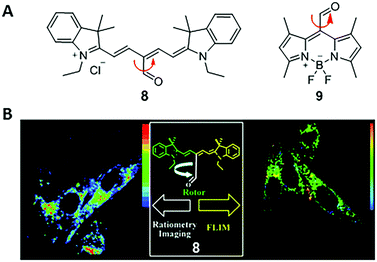 | ||
| Fig. 6 (A) Chemical structures of CHO-based viscosity sensors 8 and 9; (B) dual mode cell imaging of sensor 8. Reprinted with permission from ref. 21. Copyright 2011 American Chemical Society. | ||
3.4 Dimer-based viscosity sensor
In 2009, Kuimova and co-workers developed a new type of ratiometric fluorescent viscosity sensor 10 (Fig. 7).23 Conjugated porphyrin dimer 10 displays dual emissions, as a result of its two conformations: planar and twisted porphyrin units around the linear diyne linker. In a non-viscous environment, the emission of the dimer was dominated by the planar conformation, with an emission peaked at 780 nm. In a highly viscous environment, the less stable and twisted conformers are preserved due to the restriction of free rotations, thus enabling emissions peaked at 710 nm.24 The ratiometric response of dimer 10 permits accurate determination of viscosities. Also, dimer 10 has high cell permeability and photostability and can be used in photodynamic therapy (PDT) to treat cancers. Experimental results show that cellular viscosity significantly increases upon photo-induced cell death. During this process, sensor 10 provides a useful tool for monitoring the dynamics of diffusion in cells.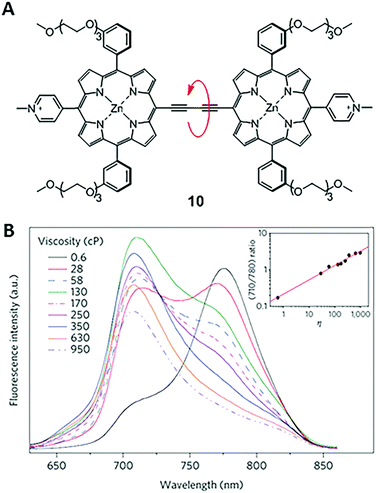 | ||
| Fig. 7 (A) Structure of a dimer-based fluorescent ratiometric viscosity sensor 10; (B) fluorescence spectra of 10 in solvent mixtures of different viscosities. The inset is the intensity ratio at 710 and 780 nm versus viscosities of solvent mixtures. Reprinted with permission from ref. 23. Copyright 2009 Nature Publishing Group. | ||
4. Application of MICE in temperature sensing
Apart from applications in monitoring intracellular viscosity, MICE based fluorescent probes were also utilized to sense intracellular temperatures. Sustaining and regulating body temperatures are of fundamental importance in endothermic animals. Monitoring intracellular temperatures using fluorescent probes has thus attracted increasing interest in the fields of chemistry and biology. In the last 20 years, different types of thermometers have been developed, including nanoparticle-, protein- and fluorophore-based thermometers.25 Among these reported thermometers, MICE based fluorescent probes display many advantages, such as simplicities in operations and chemical modifications, fast responses, and organelle targeting capabilities.As described in the MICE mechanism, faster molecular rotations and vibrations at higher temperatures cause higher non-radiative decay rates in fluorophores, thus decreasing their fluorescence intensities and shortening their fluorescence lifetimes.26 This correlation between temperatures and fluorescence intensities/lifetimes permits the development of fluorescent thermometers. Moreover, due to rapid diffusion of fluorophores into the extracellular environment in single cell analysis, it is often preferred to locate the probe in specific organelles to dynamically monitor subcellular temperature changes. Such temperature information is critical in understanding biological activities in these organelles (Fig. 8A). To this end, several MICE based fluorescent thermometers have been developed.
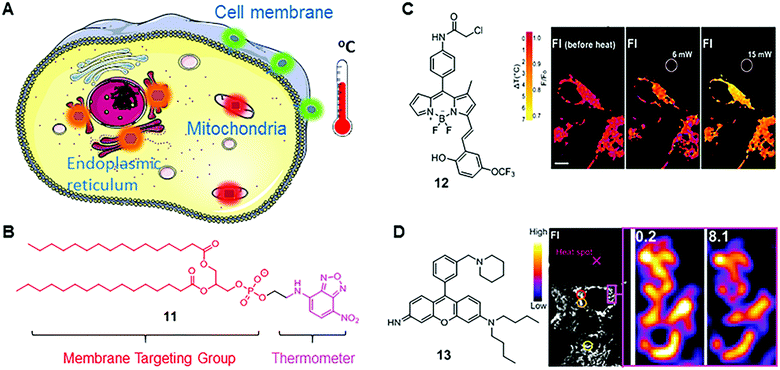 | ||
| Fig. 8 (A) Scheme of organelle-targeting fluorescent thermometers; (B) chemical structure of fluorescent thermometer 11; (C) chemical structure of fluorescent thermometer 12 and the temperature mapping of ER thermo yellow at different laser powers. Reprinted with permission from ref. 28. Copyright 2014 Nature Publishing Group; (D) chemical structure of fluorescent thermometer 13 and confocal fluorescence images in 3T3 cells stained with Mito thermo yellow at a single mitochondrion level. Reprinted with permission from ref. 29 Copyright 2015 The Royal Society of Chemistry. | ||
Nitrobenzoxadiazole (NDB) is a commonly used fluorophore with green to yellow fluorescence. It functions as a fluorescent thermometer when conjugated with phospholipids or fatty acids. In 1995, such a membrane-bound NBD-conjugated thermometer 11 was developed by Tromberg's group (Fig. 8B).27 Given the fact that the temperature sensitivity of NBD dyes is mainly from the rapid molecular vibrations of alkylated amine, Tromberg and co-workers conjugated an NBD fluorophore with phospholipids, such as 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine, to enhance the temperature sensitivity and achieve membrane specificity. Later, they applied 11 in microscopic imaging and suspension measurements of cells and phospholipid vesicles, demonstrating a reasonable temperature resolution (∼2 °C) over a broad temperature range from 15 to 70 °C.
BODIPY-based fluorescent thermometers have also been developed. In 2014, Chang's group reported a BODIPY-based thermometer 12 for real-time monitoring of temperature dynamics in the endoplasmic reticulum (Fig. 8B).28 After screening the BODIPY library with various molecular structures, thermometer 12 was selected for its significant temperature responses. Chang and co-workers demonstrated that the fluorescence intensity of 12 decreases by 1.7% °C−1 in HEPES buffer solution between 32 and 37 °C, and by 3.9% °C−1 in live and fixed HeLa cells. Chang and co-workers also confirmed the capability of 12 in monitoring heat production, induced by changing intracellular Ca2+ levels. Different from the molecular structure of fluorescent viscosity sensor 5, sensor 12 has one methyl group in position 1 and an extended π-conjugated structure in position 3 of the BODIPY scaffold. These structural differences probably induce different rotational and vibrational patterns in 5 and 12, thus endowing them with distinct sensitivities towards viscosity and temperature, respectively.
Besides BODIPY-based fluorophores, rhodamine- and rosamine-based fluorophores also display considerable fluorescence intensity variations as a function of temperature. Recently, Chang's group reported a rosamine-based mitochondria-targeting fluorescent thermometer, 13, for sensing intracellular temperature fluctuations (Fig. 8B).29 It is known that the alkylated amine and the meso phenyl group play a major role in the MICE mechanism. Therefore, Chang and co-workers modified the substituent on the phenyl group and found that attaching one additional piperidine ring greatly enhances the temperature sensitivity of 13. In cell imaging experiments, they demonstrated that 13 could monitor temperature changes in real time in live 3T3 cells, and notably, with a substantial fluorescence intensity variation (2.5–2.8% °C−1). Besides, fluorescent thermometer 13 was also used to detect intracellular temperatures in multi-cellular spheroidal HeLa cells, which simulates the properties of tumor cells in vivo.
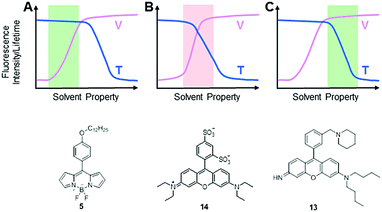
It is noteworthy that an intracellular environment is very complex. Many factors, such as viscosity, temperature, pH and ionic strength, coexist and collectively regulate cellular activities. Theoretically speaking, both intracellular viscosity and temperature changes affect molecular rotations and vibrations of MICE based fluorescent probes and thereby influence their fluorescence responses. It is thus not surprising that some fluorescent viscosity and temperature probes share similar molecular structures. For example, BODIPY fluorophores have been applied in detecting both viscosity and temperature changes. Nevertheless, it is still possible to develop MICE based probes that are dedicated to only one parameter, either viscosity or temperature in a cellular environment (Fig. 9). Indeed, while viscosity or temperature variations in cells/organelles are usually confined within a certain range, one could adjust the sensitivity and dynamic range of a probe in response to these environmental factors via careful molecular structure modifications. Consequently, some fluorophores may greatly respond to both viscosity and temperature variations in a cellular environment (the red area in Fig. 9B). However, when only the dynamic range of viscosity responses complies with that in cells, this probe serves as a dedicated viscosity sensor (the green area in Fig. 9A). Similarly, when only the dynamic range of temperature responses matches that in cells, this probe functions as a dedicated temperature sensor (the green area in Fig. 9C). Three such examples are discussed below.
In 2015, Kuimova's group studied the effects of temperature on viscosity sensors.30 They compared the response of a BODIPY-based sensor, 5, and a sulforhodamine-based sensor, 14, to both viscosity and temperature changes. These two MICE based probes were previously reported as a viscosity sensor (5) and a thermometer (14), respectively. While examining fluorescence lifetime responses of 14 in water and glycerol mixtures between 20 and 60 °C, Kuimova and co-workers showed that 14 was responsive to both temperature and viscosity changes. In contrast, similar experiments showed that the fluorescence lifetime and quantum yield of 5 depend only on viscosity from 5 to 61 °C. Therefore, sensor 5 functions as a dedicated intracellular viscosity sensor. The third example is from the rosamine-based mitochondria-targeting fluorescent thermometer 13. Probe 13 demonstrates a good response to temperature changes, while its fluorescence intensity only slightly changes from 60 cP to 264 cP. These properties make it a useful tool to monitor temperature fluctuations inside mitochondria. In sum, it is possible to apply rational molecular designs and develop fluorescent probes for monitoring only viscosity or temperature changes in the relatively stable intracellular environment.
5. Application of MICE in molecular recognitions
5.1 Protein characterization
MICE based probes have been widely used in numerous protein characterizations, such as for studying protein conformational dynamics and molecular assemblies (Fig. 10). Among these applications, thioflavin T (ThT; 15) is one representative dye. ThT was first introduced as a histochemical dye to stain amyloid deposits in tissues. Later, it became popular as an extrinsic fluorescent sensor to probe the formation of amyloid fibrils in the presence of precursor proteins. Amyloid fibrils are formed when misfolded proteins aggregate into highly ordered structures with fibre morphologies (Fig. 10A). Although proteins in amyloid fibrils do not share any sequence or structural homology, amyloid fibrils possess several common properties, such as a characteristic cross-beta structural motif. To date, more than 25 types of proteins have been known to form fibrils. These fibrils are associated with several debilitating conditions, such as Alzheimer's disease.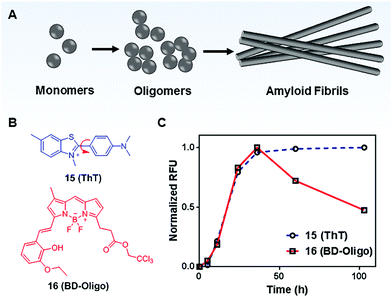 | ||
| Fig. 10 (A) Schematic representation of the amyloid fibril formation process; (B) chemical structures of Thioflavin T (ThT; 15) and BD-Oligo (16); (C) time-dependent amyloid fibril formation was monitored by ThT, whereas BD-Oligo detects on-fibril pathway oligomers. Reprinted with permission from ref. 31. Copyright 2015 American Chemical Society. | ||
ThT is the gold standard for kinetics measurement during the amyloid fibril formation process in vitro. ThT by itself is almost non-emissive in water. However, upon its association with amyloid fibrils, ThT shows a dramatic increase in emission intensity (Fig. 10C).31 This unique fluorescence response of ThT is related to molecular rotations. In a low viscosity solvent, the benzylamine and benzathiole rings of ThT rotate around the carbon–carbon bond and form a ∼90° twisted conformation upon photoexcitation. This twisted conformation corresponds to the twisted intramolecular charge transfer (TICT) state, which is typically non-emissive.32 Consequently, a low fluorescence signal is seen in unbound ThT. In contrast, amyloid fibrils present a binding site that prevents the TICT rotation in fibril-bound ThT dye, thereby enhancing ThT fluorescence. Indeed, ThT emits strong fluorescence in viscous solvents and confined environments.33 These facts support the TICT model and are in good agreement with the MICE mechanism.
Recent emerging evidence, however, suggests that oligomeric intermediates that formed during the fibrillization process (Fig. 10A) are principally responsible for the pathogenesis of amyloid diseases. As amyloid pathology is poorly correlated with clinical presentation, it is now believed that the levels of oligomers play a more important role in disease progression. To this end, Teoh et al. reported a BODIPY-based probe, BD-Oligo (16), that preferentially binds to Aβ oligomeric assemblies over monomers or fibrils.31 The oligomer selectivity is due to π–π stacking interactions and H-bonding between the dye and the exposed hydrophobic patches of oligomers. In contrast, such hydrophobic patches are not exposed in either Aβ fibrils or monomers. Consequently, BD-Oligo demonstrates dynamic oligomer-monitoring ability during Aβ peptide fibrillogenesis. With good permeability through the blood–brain-barrier, BD-Oligo is also able to stain Aβ oligomers in vivo. Like ThT, BD-Oligo alone in aqueous solution emits little fluorescence, due to its intramolecular rotation upon photoexcitation. When BD-Oligo is bound in oligomers via π–π stacking, the rotation of BD-Oligo is restricted, and the fluorescence is recovered.
Molecular rotors have also been applied to monitor protein polymerization. For example, DCVJ was deployed to follow the polymerization of tubulin.34 As tubulin assembles into a variety of polymorphic structures (such as microtubules and Zn2+ induced sheets), the fluorescence of DCVJ increases. This increase is a consequence of structural perturbations in the dye binding site of tubulin, which restricts the rotational freedom of DCVJ.
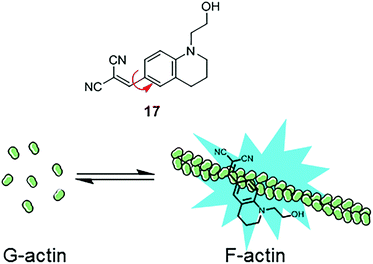
Similarly, another MICE based probe 1-(2-hydroxyethyl)-6-[(2,2-dicyano)vinyl]-2,3,4-trihydroquinoline (DCQ; 17) becomes fluorescent as actin proteins transform from the monomeric G state to the filamentous F state (Fig. 11).35 The fluorescence intensification of DCQ is mainly caused by restricting intramolecular rotation around the single bond between dialkylaminophenyl and dicyanovinyl groups when embedded in F-actins.
Besides biological research, emerging attention on protein aggregation during manufacturing, storage, and handling of biopharmaceuticals demands a sensitive protein characterization method. For example, in the formulations of therapeutic proteins, polysorbates are commonly used as a surfactant to prevent surface-induced denaturation and aggregation of proteins. However, polysorbates may undergo various degradation reactions and thus negatively affect long-term stabilities of proteins, especially under quiescent conditions. To sensitively detect aggregated proteins, viscosity sensitive MICE dyes, DCVJ (1) and CCVJ (2), have been applied in formulation screening of polysorbate-containing IgG products.36 Both probes displayed enhanced emission intensities for aggregated IgG formulations. CCVJ was also able to detect thermally induced protein aggregations in several commercial polysorbate-containing products.
In summary, non-covalent interactions of MICE based probes with hydrophobic pockets in various proteins result in confined intramolecular rotations in these probes, thereby leading to significant fluorescence enhancement. These phenomena are very useful for detecting the presence of proteins. In many cases, MICE based probes even permit the study of protein assemblies and structural dynamics.
5.2 Protein labeling
In addition to investigating structural changes and assemblies of proteins, MICE based sensors have been applied in detecting specific biomolecular interactions, such as peptide–protein interactions (Fig. 12A). Using a well characterized p53–Mdm2 interaction as a model system, Goh et al. conjugated CCVJ to a p53 peptide reporter (18; Fig. 12B). Once 18 interacts with its partner protein Mdm2, molecular rotations in CCVJ become immobilized, and its fluorescence turns on.37 With this construct, Goh and co-workers could screen and discover drugs that inhibit the p53–Mdm2 interaction. Their work represents an important therapeutic strategy, as the p53 protein is a critical tumor suppressor and Mdm2 regulates and targets the p53 protein for proteasomal degradation.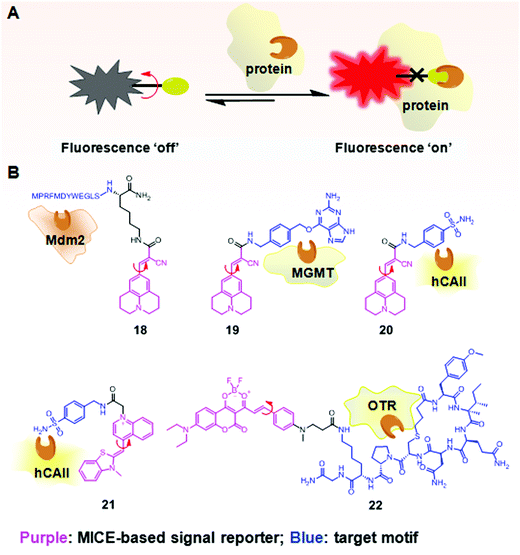 | ||
| Fig. 12 (A) Schematic illustration of the fluorescence switchable probes based on MICE for selective detection of proteins; (B) examples of MICE-based sensors 18–22 in protein sensing. | ||
In a similar way, CCVJ has also been conjugated with protein-specific ligands for selective detection of proteins. For example, in Sensor 19, CCVJ was conjugated with O6-benzylguanine (O6-BG) to detect and track O6-methyl-guanine-methyltransferase (MGMT) in real time in live cells (Fig. 12B).38 MGMT is a suicide enzyme. Upon reacting with O6-BG in an enzyme active site, the alkylated-MGMT degrades rapidly in cells. As a result, the presence of MGMT first triggers strong fluorescence in 19, as the CCVJ fragment loses its rotational freedom. Subsequently, the alkylated-MGMT protein degrades in living cells, and the bond rotation in CCVJ becomes unrestricted. The fluorescence of CCVJ thus greatly decreases.
A CCVJ derived reporter probe (20) has also been constructed to detect human carbonic anhydrase II (hCAII). hCAII is a monomeric soluble protein and consists of a benzene-sulfonamide binding site. The binding between hCAII and the benzene-sulfonamide involves a non-enzymatic process. Consequently, the conjugation of CCVJ with sulfonamide benzylamine forms a good hCAII probe 20 (Fig. 12B).38 Similarly, thiazole orange (TO) has also been used in selective hCAII detections when conjugated with sulfonamide benzylamine (21; Fig. 12B).39 Both sensors 20 and 21 emit weak fluorescence in a buffer. However, strong fluorescence turns on in the presence of hCAII, as molecular rotations of CCVJ and TO become restricted upon ligand–protein binding.
A new class of far-red probes (22) based on dioxaborine (DXBRed) has also been used to monitor ligand–receptor binding (Fig. 12B).40 Sensor 22 is conjugated with carbetocin. This molecular structure allows the detection and visualization of the oxytocin G protein-coupled receptor (OTR) expressed on cell surfaces.
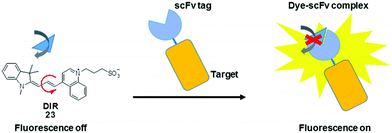
In addition to their conjugation with a peptide or protein specific ligand, MICE probes (23) have also been constructed in an alternative protein labeling strategy, which utilizes single-chain variable fragment (scFv) antibodies to create dye–protein pairs (namely fluoromodules).41,42 It is reported that scFv activates fluorescence from TO and related merocyanine compounds, which cover much of the visible and near-infrared spectra. These exogenous dyes are inherently non-fluorescent when free in solution. However, upon complexation with scFv, their bond rotations become restricted, resulting in significant fluorescence enhancement. This strategy allows visualization of various target proteins tagged with an scFv fluoromodule, in a way synonymous to genetically encoded fluorescent proteins (Fig. 13).
Taken together, the switchable fluorescence properties of MICE based probes enable dynamic monitoring of the binding events between a peptide/ligand to the target protein.
5.3 RNA tagging
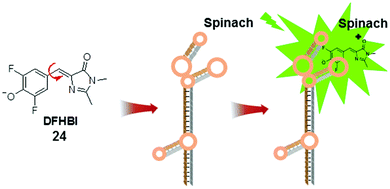
The development of fluorescent RNA tags is largely inspired by the green fluorescent protein (GFP). The fluorescent unit in GFP, 4-hydroxybenzylidene imidazolinone (HBI), is formed when three amino acid residues undergo autocatalytic intramolecular cyclization and become encased within the protein.43 The protein cavity in GFP constrains the intramolecular rotational freedom of HBI and renders it fluorescent. In contrast, chemically synthesized HBI is non-fluorescent, in accordance with the MICE mechanism.Like proteins, RNA has important roles in cellular functions. However, a comparable approach to label RNA (like GFP in labelling proteins) is still lacking, due to several challenging technical requirements. First, the fluorescent RNA tag should be emissive only when bound to RNA. In addition, the small fluorescent molecules should selectively bind to specific RNA sequences. To address this challenge, a GFP mimic aptamer, namely Spinach, was developed (Fig. 14).44 This 97 nucleotide RNA aptamer was designed to bind to DFHBI (24) in vitro. DFHBI is an HBI derivative, and non-fluorescent in aqueous solution. The crystal structure of DFHBI-bound Spinach revealed that Spinach has an elongated conformation, with two helical segments next to a G-quadruplex motif. This hydrophobic stacking platform supports the binding of DFHBI and immobilizes its molecular rotation.45 Consequently, binding to Spinach activates the fluorescence of DFHBI.
The development of fluorescent RNA tags is largely inspired by the green fluorescent protein (GFP). The fluorescent unit in GFP, 4-hydroxybenzylidene imidazolinone (HBI), is formed when three amino acid residues undergo autocatalytic intramolecular cyclization and become encased within the protein.43 The protein cavity in GFP constrains the intramolecular rotational freedom of HBI and renders it fluorescent. In contrast, chemically synthesized HBI is non-fluorescent, in accordance with the MICE mechanism.
Like proteins, RNA has important roles in cellular functions. However, a comparable approach to label RNA (like GFP in labelling proteins) is still lacking, due to several challenging technical requirements. First, the fluorescent RNA tag should be emissive only when bound to RNA. In addition, the small fluorescent molecules should selectively bind to specific RNA sequences. To address this challenge, a GFP mimic aptamer, namely Spinach, was developed (Fig. 14).44 This 97 nucleotide RNA aptamer was designed to bind to DFHBI (24) in vitro. DFHBI is an HBI derivative, and non-fluorescent in aqueous solution. The crystal structure of DFHBI-bound Spinach revealed that Spinach has an elongated conformation, with two helical segments next to a G-quadruplex motif. This hydrophobic stacking platform supports the binding of DFHBI and immobilizes its molecular rotation.45 Consequently, binding to Spinach activates the fluorescence of DFHBI.
6. Conclusions and outlook
MICE based single-molecular probes are non-fluorescent or weakly emissive when their molecular structures experience significant motions upon photoexcitation (i.e., molecular rotations and vibrations). When these motions are restricted by the surrounding environment, MICE based probes exhibit enhanced fluorescence intensities and lengthened fluorescence lifetimes. This modulation of fluorescence output involves no chemical reactions, but only conformational variations in MICE based probes. As a result, MICE based probes demonstrate fast responses, high reversibility and great sensitivities to micro-environmental changes. They have been successfully applied in a broad range of applications, such as viscosity, temperature, and molecular recognitions. By presenting a collection of representative MICE sensors and their applications, this tutorial review provides a useful introduction to the MICE mechanism and outlines technical challenges in designing dedicated MICE based probes. We expect that a good understanding of the MICE mechanism will enable the rapid development of novel and highly efficient fluorescent probes for biological research and development.Acknowledgements
This study was supported by an intramural funding from the A*STAR (Agency for Science, Technology and Research, Singapore) Biomedical Research Council and the National Medical Research Council grant NMRC/TCR/016-NNI/2016. X.L. acknowledges a Start-up Research Grant from the Singapore University of Technology and Design.Notes and references
- H. Kobayashi, M. Ogawa, R. Alford, P. L. Choyke and Y. Urano, Chem. Rev., 2010, 110, 2620–2640 CrossRef CAS PubMed.
- B. Daly, J. Ling and A. P. de Silva, Chem. Soc. Rev., 2015, 44, 4203–4211 RSC.
- A. P. de Silva, H. Q. Gunaratne, T. Gunnlaugsson, A. J. Huxley, C. P. McCoy, J. T. Rademacher and T. E. Rice, Chem. Rev., 1997, 97, 1515–1566 CrossRef CAS PubMed.
- J. Mei, N. L. Leung, R. T. Kwok, J. W. Lam and B. Z. Tang, Chem. Rev., 2015, 115, 11718–11940 CrossRef CAS PubMed.
- D. Zhai, W. Xu, L. Zhang and Y. T. Chang, Chem. Soc. Rev., 2014, 43, 2402–2411 RSC.
- H. N. Kim, M. H. Lee, H. J. Kim, J. S. Kim and J. Yoon, Chem. Soc. Rev., 2008, 37, 1465–1472 RSC.
- D. Ding, K. Li, B. Liu and B. Z. Tang, Acc. Chem. Res., 2013, 46, 2441–2453 CrossRef CAS PubMed.
- Z. Yang, Y. He, J. H. Lee, N. Park, M. Suh, W. S. Chae, J. Cao, X. Peng, H. Jung, C. Kang and J. S. Kim, J. Am. Chem. Soc., 2013, 135, 9181–9185 CrossRef CAS PubMed.
- L. Wang, Y. Xiao, W. Tian and L. Deng, J. Am. Chem. Soc., 2013, 135, 2903–2906 CrossRef CAS PubMed.
- M. K. Kuimova, Phys. Chem. Chem. Phys., 2012, 14, 12671–12686 RSC.
- G. S. Kottas, L. I. Clarke, D. Horinek and J. Michl, Chem. Rev., 2005, 105, 1281–1376 CrossRef CAS PubMed.
- CRC Handbook of Chemistry and Physics: A Ready-Reference Book of Chemical and Physical Data, ed. D. R. Lide, 85th edn, CRC Press, Boca Raton, FL, 2004 Search PubMed.
- Z. Yang, Y. He, J. H. Lee, W. S. Chae, W. X. Ren, J. H. Lee, C. Kang and J. S. Kim, Chem. Commun., 2014, 50, 11672–11675 RSC.
- I. Lopez-Duarte, T. T. Vu, M. A. Izquierdo, J. A. Bull and M. K. Kuimova, Chem. Commun., 2014, 50, 5282–5284 RSC.
- M. A. Haidekker, N. L'Heureux and J. A. Frangos, Am. J. Physiol.: Heart Circ. Physiol., 2000, 278, H1401–H1406 CAS.
- M. A. Haidekker and E. A. Theodorakis, Org. Biomol. Chem., 2007, 5, 1669–1678 CAS.
- M. A. Haidekker, T. Ling, M. Anglo, H. Y. Stevens, J. A. Frangos and E. A. Theodorakis, Chem. Biol., 2001, 8, 123–131 CrossRef CAS PubMed.
- M. A. Haidekker, T. P. Brady, D. Lichlyter and E. A. Theodorakis, J. Am. Chem. Soc., 2006, 128, 398–399 CrossRef PubMed.
- M. K. Kuimova, G. Yahioglu, J. A. Levitt and K. Suhling, J. Am. Chem. Soc., 2008, 130, 6672–6673 CrossRef CAS PubMed.
- J. A. Levitt, M. K. Kuimova, G. Yahioglu, P.-H. Chung, K. Suhling and D. Phillips, J. Phys. Chem. C, 2009, 113, 11634–11642 CAS.
- X. Peng, Z. Yang, J. Wang, J. Fan, Y. He, F. Song, B. Wang, S. Sun, J. Qu, J. Qi and M. Yan, J. Am. Chem. Soc., 2011, 133, 6626–6635 CrossRef CAS PubMed.
- H. Zhu, J. Fan, M. Li, J. Cao, J. Wang and X. Peng, Chemistry, 2014, 20, 4691–4696 CrossRef CAS PubMed.
- M. K. Kuimova, S. W. Botchway, A. W. Parker, M. Balaz, H. A. Collins, H. L. Anderson, K. Suhling and P. R. Ogilby, Nat. Chem., 2009, 1, 69–73 CrossRef CAS PubMed.
- M. U. Winters, J. Kärnbratt, M. Eng, C. J. Wilson, H. L. Anderson and B. Albinsson, J. Phys. Chem. C, 2007, 111, 7192–7199 CAS.
- X. D. Wang, O. S. Wolfbeis and R. J. Meier, Chem. Soc. Rev., 2013, 42, 7834–7869 RSC.
- T. T. Vu, R. Meallet-Renault, G. Clavier, B. A. Trofimov and M. K. Kuimova, J. Mater. Chem. C, 2016, 4, 2828–2833 RSC.
- C. F. Chapman, Y. Liu, G. J. Sonek and B. J. Tromberg, Photochem. Photobiol., 1995, 62, 416–425 CrossRef CAS PubMed.
- S. Arai, S. C. Lee, D. Zhai, M. Suzuki and Y. T. Chang, Sci. Rep., 2014, 4, 6701 CrossRef CAS PubMed.
- S. Arai, M. Suzuki, S. J. Park, J. S. Yoo, L. Wang, N. Y. Kang, H. H. Ha and Y. T. Chang, Chem. Commun., 2015, 51, 8044–8047 RSC.
- A. Vysniauskas, M. Qurashi, N. Gallop, M. Balaz, H. L. Anderson and M. K. Kuimova, Chem. Sci., 2015, 6, 5773–5778 RSC.
- C. L. Teoh, D. Su, S. Sahu, S. W. Yun, E. Drummond, F. Prelli, S. Lim, S. Cho, S. Ham, T. Wisniewski and Y. T. Chang, J. Am. Chem. Soc., 2015, 137, 13503–13509 CrossRef CAS PubMed.
- Z. R. Grabowski, K. Rotkiewicz and W. Rettig, Chem. Rev., 2003, 103 Search PubMed.
- N. Amdursky, Y. Erez and D. Huppert, Acc. Chem. Res., 2012, 45, 1548–1557 CrossRef CAS PubMed.
- C. E. Kung and J. K. Reed, Biochemistry, 1989, 28, 6678–6686 CrossRef CAS PubMed.
- T. Iio, S. Takahashi and S. Sawada, J. Biochem., 1993, 113, 196–199 CrossRef CAS PubMed.
- A. Hawe, V. Filipe and W. Jiskoot, Pharm. Res., 2010, 27, 314–326 CrossRef CAS PubMed.
- W. L. Goh, M. Y. Lee, T. L. Joseph, S. T. Quah, C. J. Brown, C. Verma, S. Brenner, F. J. Ghadessy and Y. N. Teo, J. Am. Chem. Soc., 2014, 136, 6159–6162 CrossRef CAS PubMed.
- W.-T. Yu, T.-W. Wu, C.-L. Huang, I. C. Chen and K.-T. Tan, Chem. Sci., 2016, 7, 301–307 RSC.
- H. P. Lai, R. C. Gao, C. L. Huang, I. C. Chen and K. T. Tan, Chem. Commun., 2015, 51, 16197–16200 RSC.
- I. A. Karpenko, Y. Niko, V. P. Yakubovskyi, A. O. Gerasov, D. Bonnet, Y. P. Kovtun and A. S. Klymchenko, J. Mater. Chem. C, 2016, 4, 3002–3009 RSC.
- H. Ozhalici-Unal, C. L. Pow, S. A. Marks, L. D. Jesper, G. L. Silva, N. I. Shank, E. W. Jones, J. M. Burnette 3rd, P. B. Berget and B. A. Armitage, J. Am. Chem. Soc., 2008, 130, 12620–12621 CrossRef CAS PubMed.
- N. I. Shank, K. J. Zanotti, F. Lanni, P. B. Berget and B. A. Armitage, J. Am. Chem. Soc., 2009, 131, 12960–12969 CrossRef CAS PubMed.
- H. Niwa, S. Inouye, T. Hirano, T. Matsuno, S. Kojima, M. Kubota, M. Ohashi and F. I. Tsuji, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 13617–13622 CrossRef CAS.
- J. S. Paige, K. Y. Wu and S. R. Jaffrey, Science, 2011, 333, 642–646 CrossRef CAS PubMed.
- H. Huang, N. B. Suslov, N. S. Li, S. A. Shelke, M. E. Evans, Y. Koldobskaya, P. A. Rice and J. A. Piccirilli, Nat. Chem. Biol., 2014, 10, 686–691 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |