Open Access Article
Open Access ArticleA method to identify small molecule/protein pairs susceptible to protein ubiquitination by the CRBN E3 ligase†
Pinwen
Cai
 ,
Chiara
Disraeli
,
Chiara
Disraeli
 ,
Basilius
Sauter
,
Basilius
Sauter
 ,
Saule
Zhanybekova
,
Saule
Zhanybekova
 and
Dennis
Gillingham
and
Dennis
Gillingham
 *
*
Department of Chemistry, University of Basel, 4056 Basel, Switzerland. E-mail: dennis.gillingham@unibas.ch
First published on 31st March 2025
Abstract
Although using DNA-encoded libraries (DELs) to find small molecule binders of target proteins is well-established, identifying molecules with functions beyond binding remains challenging in pooled screens. Here, we develop an approach for multiplexing functional screens that simultaneously evaluates encoded small molecules and encoded collections of protein targets in functional selections. We focus on ubiquitin (Ub) transfer with the cereblon-bound CRL4 E3 ligase because of its proven versatility in drug discovery. The functional selections recover small molecule/G-hairpin loop pairs based on their ability to promote Ub-transfer onto the G-hairpin loop. As Ub-transfer is the first step in tagging proteins for proteasomal destruction, finding small molecules capable of selectively reprogramming it is a significant challenge in contemporary drug development. Our work lays the foundation for functional DEL selections that match small molecule Ub-transfer catalysts with their optimal protein substrates.
Introduction
DNA-encoded libraries (DELs) are pooled collections of small molecules in which each compound's identity is encoded by a covalently linked DNA tag.1–5 Binders of target proteins are typically captured by affinity selection, followed by massive parallel sequencing to decode the identity of enriched molecules. While affinity-based bead selections for DEL screening have become a standard tool in early drug discovery, selecting DELs for a functional event remains far less advanced.6–10 Aside from the technical difficulty of a functional selection in pooled DEL format, an additional pitfall of functional selections is that they are often bespoke, i.e. specific to a function of the target protein. For DEL practitioners, a complex selection that is ultimately unique to a single target would usually not be worth the investment of resources. We present here an induced-proximity based DEL method that will be useful in identifying novel MGDs or PROTACs as well as their optimal protein substrates. The selection unites DEL technology with a widespread, robust, and largely commercially available in vitro biochemical technique – cullin ring ligase (CRL)-catalyzed Ub-transfer. Since we use DNA hybridization to pre-associate DEL ligands with potential protein substrates (also DNA encoded), the selection can reveal information on optimal ligand/substrate pairs in a single functional experiment. We chose the cereblon (CRBN)-bound CRL4 E3 ligase11–14 as a system to establish the technology because of its great potential in drug discovery. Many molecular glue degraders (MGDs) or proteolysis targeting chimeras (PROTACs) that exploit CRBN are either clinically approved or in clinical development.15,16 Nevertheless, recent results demonstrate that mining the neo-substrate repertoire of this ligase in response to CRBN-binders still regularly delivers surprises.17 Despite these exciting findings, the current state-of-the-art method is proteomic screening of compound libraries to search for CRBN-dependent protein disappearance, which is extremely cost and labor-intensive. Additionally, cellular proteomics screens, while having the advantage of directly delivering cell-active compounds, has the disadvantage that it would miss weakly active or cell impermeable compounds, as well as neosubstrates not expressed in a particular cell line. We present here foundational studies demonstrating that pooled functional screens could offer an alternative way to sample both compounds and Ub-substrates in a ternary complex screen with CRBN-directed E3 ligases.Small molecules that degrade proteins18 are exciting innovations because they enable drugs that mimic genetic techniques such as RNAi or CRISPR/Cas9.19–21 The ubiquitin proteasome pathway (UPP) is one of the primary natural mechanisms for the controlled degradation of proteins in eukaryotes.22 Within the UPP the key molecular signal for protein degradation is the sequential transfer of ubiquitin (Ub) proteins (typically K48-linked Ub 4–6-mers) onto client substrates.23 A challenge in modern medicinal chemistry is to find small molecules that can reprogram the UPP and stimulate Ub-transfer onto non-client disease-driving proteins. Early examples of such compounds were discovered through evolution or happenstance.24 Although new methods to discover molecular glue degraders (MGDs) are emerging,25 novel screening approaches remain in high demand. Once a binder to an E3-ligase substrate receptor is identified, a more rational approach to developing degraders is to create bispecific small molecules where one end binds the E3 ligase and the other end binds the target protein – a class of molecules now termed proteolysis targeting chimeras (PROTACs).15,16 Whether it is MGDs or PROTACs, identifying small molecules that promote degradative protein–protein interactions requires a different screening paradigm than classical inhibitor discovery. Against this backdrop, we considered how encoded methods might enable a more systematic, high-throughput approach to degrader discovery. An ideal screen would identify small molecule Ub-transfer catalysts and, conversely, reveal protein substrates susceptible to ubiquitination by a given small molecule. Several exciting recent works have explored the potential of DEL in discovering or optimizing E3 ligase recruiters, including through classical affinity selection26,27 as well as functional screens.6,26,28 The technology introduced here has a different application goal: characterizing a panel of Ub-transfer substrates29 across a library of encoded molecules in a single dataset.
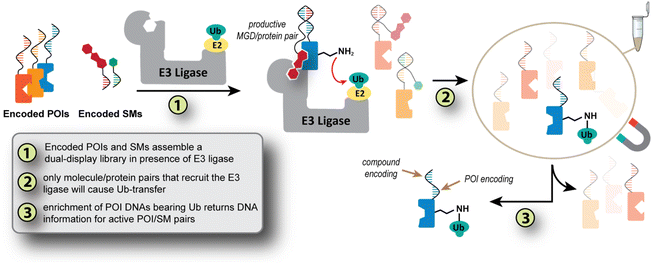
The concept is that a dual-display DEL3,30 will simultaneously probe SM/POI pairs for their ability to transfer Ub to the POI (Fig. 1). Specifically, a single-stranded small molecule DEL (SM_DEL, Fig. 1) is designed to hybridize with a collection of POIs encoded with DNAs (pooled POIs, Fig. 1), which also bear a DNA sequence with inverse complements to the codons from the SM_DEL. This hybridization-based encoding strategy (or templated encoding) is inspired by the pioneering work from Liu and Winssinger groups on DNA templated synthesis31–34 and DNA display35–37 respectively; indeed we use the Liu codon design system for the present work.34 The dual-display arrangement matches the SM_DEL's identity to its complementary POI-bound strand, while also inducing proximity of the small molecules to the POI. DNA hybridization serving dual encoding/proximity roles has previously been shown in interaction-dependent PCR.38 Once the POI and DEL are associated, a reconstituted E3 ligase system (Step 1, Fig. 1) is added. For SM/POI pairs able to form a catalytically active ternary complex with the E3 ligase, a Ub monomer is transferred to the POI (Step 2, Fig. 1). An anti-Ub bead purification (or another affinity capture method) then enriches only those DNA sequences connected to the Ub-modified POI (Step 3, Fig. 1). Finally, these enriched POI_DNA pools are sequenced to determine the active pairs (Step 4, Fig. 1). By creating a self-assembled protein/small molecule library, information on a pool of molecules and their Ub-transfer ability across a pool of POIs can be determined in a single experiment. The work herein establishes the proof-of-concept and first pooled selections for using Ub-transfer to select small molecules and their preferred protein partners in a molecular glue interaction.
Results
DNA tagged proteins and peptides
The selection system discussed in Fig. 1 requires protein or peptide molecules coupled with DNA encoding tags as the first pooled element. Fig. 2A outlines the synthetic approach to both the model DNA tagged proteins/peptides (later used for qPCR) as well as the pooling strategy of these individual coupling products. For proteins (top line of Fig. 2A) we selected the self-labelling enzyme SNAP-tag39,40 for attaching the DNA, while for peptides we used an azide handle that could be coupled to DNAs bearing strained alkynes (lower line Fig. 2A). The required DNAs could be purchased with amine-termini and coupled with an activated ester of DBCO to create DBCO_DNA, which could further be coupled with benzylguanine (BnG) to create BnG_DNA (Fig. 2B). These could be coupled with good yield to a variety of proteins (for BnG_DNA) or peptides (for DBCO_DNA), as exemplified by the example shown in Fig. 2C where the zing finger fragment of the Ikaros transcription factor (labelled IKZF1a, more on why we chose this later) is expressed as a fusion with SNAP-tag and then conjugated to a fluorescent DNA (see Fig. S2† for a complete list of proteins we have encoded). | ||
| Fig. 2 (A) DNA-linked proteins are encoded by expressing them as fusions with SNAP-tag and then incubating with BnG-bearing DNAs (top). DNA-linked peptides are encoded using the strain-promoted azide/alkyne cycloaddition (bottom). These are used individually or pooled for qPCR experiments. (B) Structure of BnG-linked DNAs and DBCO-linked DNAs (C). Representative example of DNA/protein coupling with IKZF1a protein (detailed structure and sequences given in the ESI†). BnG = benzylguanine, DBCO = dibenzocyclooctyne, POI = protein or peptide of interest, FAM = 6-carboxyfluorescein amide. | ||
Ubiquitin transfer with pomalidomide and its DNA-linked form
The CRL4CRBN E3 ligase (CRBN = cereblon) is ideal as a first test case because of its well-established ability to ubiquitinate C2H2 zinc finger motifs29 in the presence of arylglutarimides (also referred to as immunomodulatory agents, imids) such as pomalidomide (Pom, Fig. 3A).29,41 Aside from the well-established ground truths for bench-marking,29,42 an additional feature of this system is that new discoveries on either the small molecule or protein side are highly sought after in the community.17,43 Given the importance of the CRL4CRBN ligase in drug development, all components for in vitro reconstitution are commercially available. Our first step was to establish that both the protein and small molecule were tolerant to the changes required for implementing our functional selection, including adding a linker (Pom_N3, Fig. 3A), and connecting that linker to DNA (Pom_DNA, Fig. 3A). The precise protein sequence for our first test experiments was chosen based on a study demonstrating that the zinc finger 2 domain (residues 143–167) of IKZF1 (hereafter IKZF1a) is a potent and portable degron tag.41 Hence starting with the IKZF1a_DNA synthesized as shown in Fig. 2B and C above, we performed ubiquitination experiments with Pom, Pom_N3 and a Pom_DNA (Fig. 3B) first at 625 nM (Fig. 3C). Importantly IKZF1a was clearly ubiquitinated in the CRL4CRBN reaction using Pom as the molecular glue (Fig. 3C), but reactivity was reduced using the azide-bearing pomalidomide (Pom-N3, Fig. 3C, synthesized as shown in Scheme S3†). It should be noted that the preference for monoubiquitination is a result of using UBE2D3, which specializes in the pioneering round of Ub-transfer. If a processive E2 is added as well (UBE2G1), then multiple ubiquitination events are observed (Fig. S8†). The instances where several Ub-transfers seem to have occurred (see annotations in Fig. 3C) are likely still singular Ub-transfers that target different lysines. Although it was concerning that the azide linker in Pom_N3 seems to reduce Ub-transfer activity significantly, DNA-templating seems to more than compensate for this as Ub-conjugated bands are strong when Pom_DNA molecules are used. Across a series of Pom_DNAs from 6-mers to 20-mers we consistently see Ub-transfer, but surprisingly we see that the longer DNAs are less active. From 6-mers to 14-mers we see strong and similar activity, but beyond 16-mers the conversion seems to stop at ∼20% (additional longer oligos shown in Fig. S14 and 15†). We attribute this to shorter oligos being capable of ubiquitinating several protein substrates (i.e. catalysis)44–47 or that shorter oligos have more conformational flexibility once bound, better facilitating Ub-transfer. We currently prefer the later explanation since even high concentrations of longer oligos do not fully ubiquitinate the protein, despite the fact that they completely form stable duplexes (see ESI, Fig. S13†). As we show later, these results cannot be explained by untemplated reactivity since template-matched oligos are selectively enriched in pull-down experiments (quantified by qPCR). The poor conversion can be improved by running the reaction at higher DNA_Pom concentrations (Fig. 3D); nevertheless, so as to avoid several rounds of optimization we proceeded with these conditions to begin testing ubiquitination selectivity. Finally, before continuing we wanted to test whether the distance between protein and substrate was important for Ub-transfer. By varying the landing site for Watson–Crick base-pairing we could easily adapt the position between protein and small molecule. As experiments in Fig. S18† show, the shortest linker is best, but even over long distances (32 base-pairs) Ub-transfer is still observed. In summary, these test systems verify that in vitro ubiquitination of zinc fingers with CRLCRBN can be reconstituted using a DNA-linked version of common MGD scaffolds. | ||
| Fig. 3 (A) Structures of glutarimide derivatives and the corresponding DNA-linked version after bioconjugation. (B) In vitro ubiquitin transfer of Pom, Pom_N3, and Pom_DNA were tested and compared. (C) DNA templated Ub-transfer to IKZF1a with Pom, Pom-N3 and varying lengths of Pom_DNA (detailed conditions: ESI Section 5†). (D) The in vitro reconstituted CRL4CRBN E3 ligase targeting the IKZF1a, using Pom_DNA (10-mer) as the molecular glue (left) or Pom_N3 (right) at different concentrations (see ESI Section 7† for additional experiments on DNA length dependence, effect of linker and DNA positioning as well as a panel of controls for the in vitro Ub-transfer). | ||
Selective ubiquitination and enrichment
Next we wanted to explore the specificity of ubiquitination with different binders and how strongly we could enrich. To this end, we ran the reaction with Pom_DNA and DBCO_DNA (prepared this time as 55-mers to improve qPCR quantitation) and performed a ratiometric analysis of their relative enrichment after pull-down (according to qPCR of the DNA, Fig. 4A). The hypothesis would be that ubiquitination should predominate on the IKZF1a_DNA conjugate since it has complementarity to the Pom_DNA and it can form a ternary complex between IKZF1 and CRBN. Although only “T2” is drawn as a template DNA in Fig. 4A, the final enrichment data is shown with two different templates (T1 & T2 in Fig. 4B). The inclusion of a second template was to insure that no spurious effect from a specific oligonucleotide's behavior in qPCR would bias our signal. As the data in Fig. 4B indicates, we achieve robust enrichment above background after a single pulldown (∼6-fold), and this can be improved with a second (∼40-fold) and third (∼64-fold) pulldown (further pulldowns did not improve enrichments). It should be noted that although we were careful in making sure there were no differences between templates in these proof-of-concept studies, such concerns are not relevant in a proper DEL screen since every library member behaves nearly identically in PCR and libraries are tagged with a unique molecule identifier to detect PCR bias. In summary the data in Fig. 4B validate that DNA-templated ubiquitination can robustly identify active ternary complexes. | ||
| Fig. 4 Establishing the enrichment assay. (A) Pom_DNA and DBCO_DNA were independently tested for Ub-transfer to a partially complementary template DNA bearing IKZF1a (IKZF1a_DNA). (B) Fold enrichment of Pom_DNAversus DBCO DNA over three consecutive pulldowns using anti-Ub magnetic beads; different templates were tested (T1 and T2) to generate the data to insure that qPCR bias does not impact the enrichments (for absolute DNA quantitation by qPCR, as well as details on the pull-down conditions and optimization of the protocol, see ESI Section 8†). (C) Structures of Pom(5)_N3 and NVP, as well as synthesis of NVP_N3 and NVP_DNA. (D) In vitro ubiquitination of IKZFs with Pom_DNA and NVP_DNA expressed as fold enrichment (relative to DBCO-bearing DNA) after anti-Ub magnetic beads purification and qPCR. (E). In vitro ubiquitination of MGDs with and without linkers to examine the effect of linker on Ub-transfer efficiency. MGD = molecular glue degrader. | ||
With the feasibility of identifying small molecules established, we next set out quantify the sensitivity of the assay for identifying specific molecule/ZF combinations. As a test case we selected the recently published MGD (NVP-DKY709, hereafter NVP, Fig. 4C) that preferentially degrades Ikaros zinc finger 2 (IKZF2) over other zinc fingers, although an affinity to IKZF1 is retained.11 A variant of NVP was therefore synthesized bearing an azide linker to facilitate DNA conjugation (NVP_N3 and NVP_DNA, Fig. 4C). We planned ubiquitination reactions of Pom_DNA and NVP_DNA with both IKZF1a and IKZF2a (residues 138–162), as well as extended double-ZF versions (covering the ZF2 and ZF3 regions from amino acids 141–196, 136–191 respectively) that are known to have a higher affinity to CRBN in the presence of Imids.41,48 For clarity, the single SNAP-ZF fusions are labelled as ‘a’ and the double ZFs are labelled as ‘b’ (for example with IKZF2 we have IKZF2a and IKZF2b). We then tested the ubiquitination and enrichment of both of these ZF sets with both Pom_DNA and NVP_DNA, according to the conditions established in Fig. 3.
As seen in Fig. 4D the results with DNA-linked NVP compounds fully recapitulate the expected selectivity profile,11 giving stronger ubiquitin enrichments relative to Pom when IKZF2a or IKZF2b are the substrates. Surprisingly, Pom enrichments with IKZF1, although strong, were not superior to NVP, which runs counter to what literature data would suggest. We wondered whether the presence of the linker reduces Ub-transfer efficiency of Pom derivatives, but not NVP. Indeed, we had already seen hints of this in the initial assay development (compare Pom and Pom-N3 in Fig. 3D). The PROTAC field uses Pom-like ligands regularly with great success to target CRBN,49–51 but the impact of molecular alterations outside of the CRBN-binding motif (glutarimide) on the repertoire of G-hairpin loop structures is less well understood. Recent reports have begun to quantitatively examine this issue, finding that while modified pomalidomide derivatives remain strong CRBN binders, once linkers at C4 or C5 are introduced, their ability to form effective ternary complexes with ZFs are often attenuated.11,42 We therefore performed in vitro Ub-transfer tests with compounds bearing linkers, but that were not coupled to DNA. Our results are consistent with the in vitro ubiquitination data of DNA-linked substrates. In particular we see that NVP is an excellent Ub-transfer catalyst both with and without the linker (lanes 5 & 6 in lower gel of Fig. 4E), and that it is a better catalyst for IKZF2 than IKZF1 – as the literature would suggest (compare upper and lower gels in Fig. 4E). Pom strongly prefers IKZF1, but the presence of a linker seems to significantly reduce Ub-transfer ability (compare lanes 2 & 3 with lane 4 in Fig. 4E). Taken together, the above experiments suggest that in vitro ubiquitination benefits from induced proximity driven by DNA hybridization, but that the linker to DNA can impact relative efficiency of Ub-transfer. To achieve the ultimate goal of using a dual display library to match optimal pairs, the remaining challenge was to verify that the selection operated in pooled format for both small molecules and proteins.
Ub transfer in a mini-pool of DNA-linked molecules
We first tested the selection against IKZF2b with a handful of DNA-linked molecules (Fig. 5A) to see if we could identify the best binder while holding the protein constant. IKZF2b was conjugated individually with 5 different templates and equally mixed. In principle, if DNA template-based recruitment of the small molecules was not essential to dictate Ub-transfer efficiency, then each of these POI-template conjugates would be equally ubiquitinated and indistinguishable from background since the only difference between them is their DNA sequence. NVP_DNA, Pom_DNA, lenalidomide (Len_DNA), glutarimide (Gluta_DNA) and amine_DNA conjugates – each with complementarity to one of the POI-template sequences – were then added along with all other components of the in vitro ubiquitination mixture (see the ESI Section 5.1 for precise chemical structures of Len_DNA, Gluta_DNA). A negative control with all the small molecules replaced by amine was also performed (shown in Fig. S29†). Based on both absolute DNA quantitation (ESI Fig. S27 and 28†) and relative enrichment values (Fig. 5A) we see that binders are clearly distinguished from background (amine_DNA). Importantly, all binders were correctly identified and the best binder to IKZF2 (NVP) always gave the highest amount of DNA. To summarize, the results in Fig. 5A indicate that the functional selection system could rank Ub-transfer efficiency of MGDs to a specific POI.Ub transfer in a mini-pool of protein targets
After the validation of the selection from a mixture of DNA-linked molecules, we next needed to see whether MGDs could be selected out of a mixture of protein targets. NVP was chosen as the test molecule and was coupled with 5 different DNA strands complementary to 5 DNA templates. The DNA templates were linked to four different ZFs as well as one control protein (SNAP). The NVP_DNA conjugates were added to the POI-templates mixture, along with the in vitro ubiquitination system. A negative control with all the NVP replaced by amine was also performed (shown in Fig. S30 and 31†). The four ZF-bearing proteins in the mixture were expected to be ubiquitinated by NVP in the ubiquitination condition, while the dummy protein (SNAP) should not. Gratifyingly, the ZF-linked templates all show enrichments relative to the template bearing no protein. Although the assay gave only expected Ub-transfer events, in contrast to small molecule variation (where the expected winner was clearly identified, i.e.Fig. 5A), the ranking of optimal protein substrates is less efficient. Although ZFs are clearly identifiable over background, the superiority of IKZF2 over IKZF1 for NVP is likely too small to distinguish in this case (Fig. 5B). Nevertheless, these results validate that MGDs could be identified and ranked based on their Ub-transfer ability on a pool of protein substrates.Exploring the size restriction for a pooled dual display screen
The potential library size we could explore in a dual display MGD/substrate screen will be set by the screen's ability to robustly detect individual library members at higher and higher dilution. As such, to define an upper limit for library size we mimicked increasing library size by diluting active MGD-bound DNAs with inactive background DNA (Fig. 5C). We set a threshold of 10-fold enrichment over background as the minimum value we should maintain (brown dotted line in Fig. 5C) to give robust data. To explore assay sensitivity we performed the DNA-templated NVP/IKZF2 ubiquitination using 10 pmol of NVP_DNA and 10 pmol of amine-bearing DNA while adding varying amounts of partially double-stranded background DNA (first four columns of Fig. 5C, the amount of background DNA is converted to a “virtual library size” to simplify the mental math for the reader). To our delight, the signal of the template complementary to NVP_DNA was much higher than the signal of the template complementary to amine_DNA, suggesting large amounts of background DNA (as is present in a real encoded library setup) are not detrimental to sensitivity. To further explore the limits of library size we then reduced the amount of input DNA for the template-matched system. Specifically, we kept the total library input (i.e.NVP_DNA + amine_DNA + dummy DNA) as 10 nmol, while incrementally decreasing (10 pmol, 5 pmol, 2.5 pmol, 1 pmol, 0.1 pmol) the copy number of the NVP_DNA (see columns 5–8 in Fig. 5C). Even at the very lowest copy number of 0.1 pmol of the NVP_DNA we still see robust enrichment of the positive signal over background (∼16-fold, last column in Fig. 5C). While it seems there would still be room for additional dilution, these conditions would support a library size of 105 members – which is already more than we plan to synthesize in the MGD DEL. For calibration, in a dual display setup, screening 10 proteins with this size of MGD library would give Ub-transfer data for a million potential interactions.Conclusion
In summary, our research has demonstrated that DNA hybridization-induced proximity can accelerate selective Ub-transfer, and hence be used to select winners in pooled selections. The method continues to deliver strong enrichment signal even with simulated libraries of up to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 members—a size sufficient for large-scale library selections. While DEL selections are now standard in early drug discovery, conducting functional selections in a pooled format continues to pose challenges. Recent innovative efforts underscore the significant commitment to establishing functional DEL selections.6,26,28,52,53 Our work introduces a novel functional selection applicable to pooled format, setting the stage for a full screen that determines optimal pairs of MGDs and protein substrates for Ub-transfer reactions. We focus on Ub-transfer due to the challenge of discovering novel MGDs, but in principle similar functional assays could be developed for any enrichable tag that can be biochemically reconstituted (SUMOylation, phosphorylation). A drawback of our approach is the need for custom library synthesis for each selection and that the effect of the linker will be built into the selection data. Hence the application needs to be carefully chosen such that the insights gained from the resulting dataset justify the effort in library construction and that linker incorporation is not problematic. The current application, which connects MGDs with potential substrates using the CRBN E3 ligase, is particularly well-suited for this approach. Since the mechanism of action of CRBN-directed therapeutics has been defined,14,54,55 enormous efforts have been made across numerous research labs to define both active MGDs and their optimal protein degrome for the CRBN E3 ligase.13,17,29,56 The approach we developed here has the potential to generate unprecedentedly large datasets exploring this problem, while simultaneously reducing cost and effort – we are currently synthesizing a CRBN-focused DEL to generate this dataset.
000 members—a size sufficient for large-scale library selections. While DEL selections are now standard in early drug discovery, conducting functional selections in a pooled format continues to pose challenges. Recent innovative efforts underscore the significant commitment to establishing functional DEL selections.6,26,28,52,53 Our work introduces a novel functional selection applicable to pooled format, setting the stage for a full screen that determines optimal pairs of MGDs and protein substrates for Ub-transfer reactions. We focus on Ub-transfer due to the challenge of discovering novel MGDs, but in principle similar functional assays could be developed for any enrichable tag that can be biochemically reconstituted (SUMOylation, phosphorylation). A drawback of our approach is the need for custom library synthesis for each selection and that the effect of the linker will be built into the selection data. Hence the application needs to be carefully chosen such that the insights gained from the resulting dataset justify the effort in library construction and that linker incorporation is not problematic. The current application, which connects MGDs with potential substrates using the CRBN E3 ligase, is particularly well-suited for this approach. Since the mechanism of action of CRBN-directed therapeutics has been defined,14,54,55 enormous efforts have been made across numerous research labs to define both active MGDs and their optimal protein degrome for the CRBN E3 ligase.13,17,29,56 The approach we developed here has the potential to generate unprecedentedly large datasets exploring this problem, while simultaneously reducing cost and effort – we are currently synthesizing a CRBN-focused DEL to generate this dataset.
Data availability
The data supporting the article SC-EDG-02-2025-001251 have been included as part of the ESI.†Author contributions
Pinwen Cai helped concenptualize the study, ran most of the experiments, performed data analysis, and helped with preparing the draft of the manuscript. Chiara Disraeli generated data contributing to Fig. 2A, C and 4C–E, helped with data analysis and visualization, performed solid-phase peptide synthesis, and proof-read the paper. Basilius Sauter supervised the scientific work, helped in data analysis, helped with data visualization and participated in manuscript writing. Saule Zhanybekova helped in supervision for developing biochemical assays and cloning, and also helped with protein production. Dennis Gillingham conceptualized the study, supervised the experiments, and wrote the first draft of the manuscript with input from all authors.Conflicts of interest
The authors report no conflict of interest.Acknowledgements
We would like to thank Koder Dagher for the help of protein production. We would also like to acknowledge the Swiss National Science Foundation for funding (Grant: 200020_215758).References
- S. Brenner and R. A. Lerner, Encoded combinatorial chemistry, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 5381–5383 CAS.
- N. Favalli, S. Biendl, M. Hartmann, J. Piazzi, F. Sladojevich, S. Gräslund, P. J. Brown, K. Näreoja, H. Schüler, J. Scheuermann, R. Franzini and D. Neri, A DNA-Encoded Library of Chemical Compounds Based on Common Scaffolding Structures Reveals the Impact of Ligand Geometry on Protein Recognition, ChemMedChem, 2018, 13, 1303–1307 CAS.
- S. Melkko, J. Scheuermann, C. E. Dumelin and D. Neri, Encoded self-assembling chemical libraries, Nat. Biotechnol., 2004, 22, 568–574 CAS.
- A. Gironda-Martínez, E. J. Donckele, F. Samain and D. Neri, DNA-Encoded Chemical Libraries: A Comprehensive Review with Successful Stories and Future Challenges, ACS Pharmacol. Transl. Sci., 2021, 4, 1265–1279 Search PubMed.
- A. A. Peterson and D. R. Liu, Small-molecule discovery through DNA-encoded libraries, Nat. Rev. Drug Discovery, 2023, 22, 699–722 CAS.
- W. G. Cochrane, M. L. Malone, V. Q. Dang, V. Cavett, A. L. Satz and B. M. Paegel, Activity-Based DNA-Encoded Library Screening, ACS Comb. Sci., 2019, 21, 425–435 CAS.
- T. Kodadek, N. G. Paciaroni, M. Balzarini and P. Dickson, Beyond protein binding: recent advances in screening DNA-encoded libraries, Chem. Commun., 2019, 55, 13330–13341 CAS.
- N. Favalli, G. Bassi, J. Scheuermann and D. Neri, DNA-encoded chemical libraries – achievements and remaining challenges, FEBS Lett., 2018, 592, 2168–2180 CrossRef CAS PubMed.
- Y. Huang, Y. Li and X. Li, Strategies for developing DNA-encoded libraries beyond binding assays, Nat. Chem., 2022, 14, 129–140 CAS.
- L. A. Schneider, B. Sauter, K. Dagher and D. Gillingham, Recording binding information directly into DNA-Encoded libraries using terminal deoxynucleotidyl transferase, J. Am. Chem. Soc., 2023, 145, 20874–20882 CrossRef CAS PubMed.
- S. Bonazzi, E. d'Hennezel, R. E. J. Beckwith, L. Xu, A. Fazal, A. Magracheva, R. Ramesh, A. Cernijenko, B. Antonakos, H.-e. C. Bhang, R. G. Caro, J. S. Cobb, E. Ornelas, X. Ma, C. A. Wartchow, M. C. Clifton, R. R. Forseth, B. H. Fortnam, H. Lu, A. Csibi, J. Tullai, S. Carbonneau, N. M. Thomsen, J. Larrow, B. Chie-Leon, D. Hainzl, Y. Gu, D. Lu, M. J. Meyer, D. Alexander, J. Kinyamu-Akunda, C. A. Sabatos-Peyton, N. A. Dales, F. J. Zécri, R. K. Jain, J. Shulok, Y. K. Wang, K. Briner, J. A. Porter, J. A. Tallarico, J. A. Engelman, G. Dranoff, J. E. Bradner, M. Visser and J. M. Solomon, Discovery and characterization of a selective IKZF2 glue degrader for cancer immunotherapy, Cell Chem. Biol., 2023, 30, 235–247 CrossRef CAS PubMed.
- G. Lu, R. E. Middleton, H. Sun, M. Naniong, C. J. Ott, C. S. Mitsiades, K.-K. Wong, J. E. Bradner and W. G. Kaelin, The Myeloma Drug Lenalidomide Promotes the Cereblon-Dependent Destruction of Ikaros Proteins, Science, 2014, 343, 305–309 CrossRef CAS PubMed.
- M. E. Matyskiela, G. Lu, T. Ito, B. Pagarigan, C.-C. Lu, K. Miller, W. Fang, N.-Y. Wang, D. Nguyen, J. Houston, G. Carmel, T. Tran, M. Riley, L. A. Nosaka, G. C. Lander, S. Gaidarova, S. Xu, A. L. Ruchelman, H. Handa, J. Carmichael, T. O. Daniel, B. E. Cathers, A. Lopez-Girona and P. P. Chamberlain, A novel cereblon modulator recruits GSPT1 to the CRL4CRBN ubiquitin ligase, Nature, 2016, 535, 252–257 CrossRef CAS PubMed.
- T. Ito, H. Ando, T. Suzuki, T. Ogura, K. Hotta, Y. Imamura, Y. Yamaguchi and H. Handa, Identification of a Primary Target of Thalidomide Teratogenicity, Science, 2010, 327, 1345–1350 CrossRef CAS PubMed.
- M. Békés, D. R. Langley and C. M. Crews, PROTAC targeted protein degraders: the past is prologue, Nat. Rev. Drug Discovery, 2022, 21, 181–200 CrossRef PubMed.
- D. A. Nalawansha and C. M. Crews, PROTACs: An Emerging Therapeutic Modality in Precision Medicine, Cell Chem. Biol., 2020, 27, 998–1014 CrossRef CAS PubMed.
- G. Petzold, P. Gainza, S. Annunziato, I. Lamberto, P. Trenh, L. McAllister, B. Demarco, L. Schwander, R. D. Bunker, M. Zlotosch, R. SriRamaratnam, S. Gilberto, G. Langousis, E. J. Donckele, C. Quan, V. Strande, G. M. D. Donatis, S. B. Alabi, J. Alers, M. Matysik, C. Staehly, A. Dubois, A. Osmont, M. Garskovas, D. Lyon, L. Wiedmer, V. Oleinikovas, R. Lieberherr, N. T. Rubin, D. T. Lam, N. I. Widlund, A. Ritzén, R. M. Caceres, D. Vigil, J. Tsai, O. Wallace, M. Peluso, A. Sadok, A. M. Paterson, V. Zarayskiy, B. Fasching, D. Bonenfant, M. Warmuth, J. Castle and S. A. Townson, Mining the CRBN Target Space Redefines Rules for Molecular Glue-induced Neosubstrate Recognition, bioRxiv, 2024, preprint, DOI:10.1101/2024.10.07.616933, 2024.2010.2007.616933.
- D. L. Buckley and C. M. Crews, Small-Molecule Control of Intracellular Protein Levels through Modulation of the Ubiquitin Proteasome System, Angew. Chem., Int. Ed., 2014, 53, 2312–2330 CrossRef CAS PubMed.
- G. M. Burslem and C. M. Crews, Small-Molecule Modulation of Protein Homeostasis, Chem. Rev., 2017, 117, 11269–11301 CrossRef CAS PubMed.
- A. C. Runcie, K.-H. Chan, M. Zengerle and A. Ciulli, Chemical genetics approaches for selective intervention in epigenetics, Curr. Opin. Chem. Biol., 2016, 33, 186–194 CrossRef CAS PubMed.
- C. P. Tinworth, H. Lithgow and I. Churcher, Small molecule-mediated protein knockdown as a new approach to drug discovery, MedChemComm, 2016, 7(12), 2206–2216 RSC.
- S. H. Lecker, A. L. Goldberg and W. E. Mitch, Protein Degradation by the Ubiquitin–Proteasome Pathway in Normal and Disease States, J. Am. Soc. Nephrol., 2006, 17, 1807–1819 CrossRef CAS PubMed.
- C. Behrends and J. W. Harper, Constructing and decoding unconventional ubiquitin chains, Nat. Struct. Mol. Biol., 2011, 18, 520–528 CrossRef CAS PubMed.
- S. L. Schreiber, The Rise of Molecular Glues, Cell, 2021, 184, 3–9 CrossRef CAS PubMed.
- C. Mayor-Ruiz, S. Bauer, M. Brand, Z. Kozicka, M. Siklos, H. Imrichova, I. H. Kaltheuner, E. Hahn, K. Seiler, A. Koren, G. Petzold, M. Fellner, C. Bock, A. C. Müller, J. Zuber, M. Geyer, N. H. Thomä, S. Kubicek and G. E. Winter, Rational discovery of molecular glue degraders via scalable chemical profiling, Nat. Chem. Biol., 2020, 16(11), 1199–1207 CrossRef CAS PubMed.
- J. W. Mason, Y. T. Chow, L. Hudson, A. Tutter, G. Michaud, M. V. Westphal, W. Shu, X. Ma, Z. Y. Tan, C. W. Coley, P. A. Clemons, S. Bonazzi, F. Berst, K. Briner, S. Liu, F. J. Zécri and S. L. Schreiber, DNA-encoded library-enabled discovery of proximity-inducing small molecules, Nat. Chem. Biol., 2024, 20, 170–179 CrossRef CAS PubMed.
- Q. Chen, C. Liu, W. Wang, X. Meng, X. Cheng, X. Li, L. Cai, L. Luo, X. He, H. Qu, J. Luo, H. Wei, S. Gao, G. Liu, J. Wan, D. I. Israel, J. Li and D. Dou, Optimization of PROTAC Ternary Complex Using DNA Encoded Library Approach, ACS Chem. Biol., 2023, 18, 25–33 CrossRef CAS PubMed.
- B. Cai, A. B. Mhetre and C. J. Krusemark, Selection methods for proximity-dependent enrichment of ligands from DNA-encoded libraries using enzymatic fusion proteins, Chem. Sci., 2023, 14, 245–250 RSC.
- Q. L. Sievers, G. Petzold, R. D. Bunker, A. Renneville, M. Słabicki, B. J. Liddicoat, W. Abdulrahman, T. Mikkelsen, B. L. Ebert and N. H. Thomä, Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN, Science, 2018, 362, eaat0572 CrossRef PubMed.
- M. Wichert, N. Krall, W. Decurtins, R. M. Franzini, F. Pretto, P. Schneider, D. Neri and J. Scheuermann, Dual-display of small molecules enables the discovery of ligand pairs and facilitates affinity maturation, Nat. Chem., 2015, 7, 241–249 CrossRef CAS PubMed.
- Z. J. Gartner and D. R. Liu, The Generality of DNA-Templated Synthesis as a Basis for Evolving Non-Natural Small Molecules, J. Am. Chem. Soc., 2001, 123, 6961–6963 CrossRef CAS PubMed.
- Z. J. Gartner, B. N. Tse, R. Grubina, J. B. Doyon, T. M. Snyder and D. R. Liu, DNA-Templated Organic Synthesis and Selection of a Library of Macrocycles, Science, 2004, 305, 1601–1605 CrossRef CAS PubMed.
- X. Li and D. R. Liu, DNA-templated organic synthesis: Nature's strategy for controlling chemical reactivity applied to synthetic molecules, Angew. Chem., Int. Ed., 2004, 43, 4848–4870 CAS.
- D. L. Usanov, A. I. Chan, J. P. Maianti and D. R. Liu, Second-generation DNA-templated macrocycle libraries for the discovery of bioactive small molecules, Nat. Chem., 2018, 10, 704–714 CrossRef CAS PubMed.
- B. R. Vummidi, L. Farrera-Soler, J.-P. Daguer, M. Dockerill, S. Barluenga and N. Winssinger, A mating mechanism to generate diversity for the Darwinian selection of DNA-encoded synthetic molecules, Nat. Chem., 2022, 14, 141–152 CAS.
- J. P. Daguer, C. Zambaldo, M. Ciobanu, P. Morieux, S. Barluenga and N. Winssinger, DNA display of fragment pairs as a tool for the discovery of novel biologically active small molecules, Chem. Sci., 2015, 6, 739–744 RSC.
- F. Debaene, L. Mejias, J. L. Harris and N. Winssinger, Synthesis of a PNA-encoded cysteine protease inhibitor library, Tetrahedron, 2004, 60, 8677–8690 CrossRef CAS.
- L. M. McGregor, D. J. Gorin, C. E. Dumelin and D. R. Liu, Interaction-Dependent PCR: Identification of Ligand−Target Pairs from Libraries of Ligands and Libraries of Targets in a Single Solution-Phase Experiment, J. Am. Chem. Soc., 2010, 132, 15522–15524 CrossRef CAS PubMed.
- A. Gautier, A. Juillerat, C. Heinis, I. R. Corrêa Jr, M. Kindermann, F. Beaufils and K. Johnsson, An Engineered Protein Tag for Multiprotein Labeling in Living Cells, Chem. Biol., 2008, 15, 128–136 CrossRef CAS PubMed.
- A. Keppler, M. Kindermann, S. Gendreizig, H. Pick, H. Vogel and K. Johnsson, Labeling of fusion proteins of O6-alkylguanine-DNA alkyltransferase with small molecules in vivo and in vitro, Methods, 2004, 32, 437–444 CrossRef CAS PubMed.
- V. Koduri, S. K. McBrayer, E. Liberzon, A. C. Wang, K. J. Briggs, H. Cho and W. G. Kaelin, Peptidic degron for IMiD-induced degradation of heterologous proteins, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 2539–2544 CrossRef CAS PubMed.
- T. M. Nguyen, V. Sreekanth, A. Deb, P. Kokkonda, P. K. Tiwari, K. A. Donovan, V. Shoba, S. K. Chaudhary, J. A. M. Mercer, S. Lai, A. Sadagopan, M. Jan, E. S. Fischer, D. R. Liu, B. L. Ebert and A. Choudhary, Proteolysis-targeting chimeras with reduced off-targets, Nat. Chem., 2024, 16, 218–228 CrossRef CAS PubMed.
- V. Oleinikovas, P. Gainza, T. Ryckmans, B. Fasching and N. H. Thomä, From Thalidomide to Rational Molecular Glue Design for Targeted Protein Degradation, Annu. Rev. Pharmacol. Toxicol., 2024, 64, 291–312 CrossRef CAS PubMed.
- K. K. Sadhu and N. Winssinger, Detection of miRNA in Live Cells by Using Templated RuII-Catalyzed Unmasking of a Fluorophore, Chem.–Eur. J., 2013, 19, 8182–8189 CrossRef CAS PubMed.
- M. Röthlingshöfer, K. Gorska and N. Winssinger, Nucleic Acid Templated Uncaging of Fluorophores Using Ru-Catalyzed Photoreduction with Visible Light, Org. Lett., 2011, 14(2), 482–485 CrossRef.
- Z. Pianowski, K. Gorska, L. Oswald, C. A. Merten and N. Winssinger, Imaging of mRNA in Live Cells Using Nucleic Acid-Templated Reduction of Azidorhodamine Probes, J. Am. Chem. Soc., 2009, 131, 6492–6497 CrossRef CAS PubMed.
- Z. L. Pianowski and N. Winssinger, Fluorescence-based detection of single nucleotide permutation in DNA via catalytically templated reaction, Chem. Commun., 2007, 3820–3822 RSC.
- Q. L. Sievers, J. A. Gasser, G. S. Cowley, E. S. Fischer and B. L. Ebert, Genome-wide screen identifies cullin-RING ligase machinery required for lenalidomide-dependent CRL4CRBN activity, Blood, 2018, 132, 1293–1303 CrossRef CAS PubMed.
- W. T. Jauslin, M. Schild, T. Schaefer, C. Borsari, C. Orbegozo, L. Bissegger, S. Zhanybekova, D. Ritz, A. Schmidt, M. Wymann and D. Gillingham, A high affinity pan-PI3K binding module supports selective targeted protein degradation of PI3Kα, Chem. Sci., 2024, 15, 683–691 RSC.
- J. Lu, Y. Qian, M. Altieri, H. Dong, J. Wang, K. Raina, J. Hines, J. D. Winkler, A. P. Crew, K. Coleman and C. M. Crews, Hijacking the E3 Ubiquitin Ligase Cereblon to Efficiently Target BRD4, Chem. Biol., 2015, 22, 755–763 CrossRef CAS PubMed.
- G. E. Winter, D. L. Buckley, J. Paulk, J. M. Roberts, A. Souza, S. Dhe-Paganon and J. E. Bradner, Phthalimide conjugation as a strategy for in vivo target protein degradation, Science, 2015, 348, 1376–1381 CrossRef CAS PubMed.
- B. Cai, D. Kim, S. Akhand, Y. Sun, R. J. Cassell, A. Alpsoy, E. C. Dykhuizen, R. M. Van Rijn, M. K. Wendt and C. J. Krusemark, Selection of DNA-Encoded Libraries to Protein Targets within and on Living Cells, J. Am. Chem. Soc., 2019, 141, 17057–17061 CrossRef CAS PubMed.
- Y. Huang, L. Meng, Q. Nie, Y. Zhou, L. Chen, S. Yang, Y. M. E. Fung, X. Li, C. Huang, Y. Cao, Y. Li and X. Li, Selection of DNA-encoded chemical libraries against endogenous membrane proteins on live cells, Nat. Chem., 2021, 13, 77–88 CrossRef CAS PubMed.
- M. E. Matyskiela, S. Couto, X. Zheng, G. Lu, J. Hui, K. Stamp, C. Drew, Y. Ren, M. Wang, A. Carpenter, C.-W. Lee, T. Clayton, W. Fang, C.-C. Lu, M. Riley, P. Abdubek, K. Blease, J. Hartke, G. Kumar, R. Vessey, M. Rolfe, L. G. Hamann and P. P. Chamberlain, SALL4 mediates teratogenicity as a thalidomide-dependent cereblon substrate, Nat. Chem. Biol., 2018, 14, 981–987 CrossRef CAS PubMed.
- G. Petzold, E. S. Fischer and N. H. Thomä, Structural basis of lenalidomide-induced CK1α degradation by the CRL4CRBN ubiquitin ligase, Nature, 2016, 532, 127 CrossRef CAS PubMed.
- M. E. Matyskiela, W. Zhang, H.-W. Man, G. Muller, G. Khambatta, F. Baculi, M. Hickman, L. LeBrun, B. Pagarigan, G. Carmel, C.-C. Lu, G. Lu, M. Riley, Y. Satoh, P. Schafer, T. O. Daniel, J. Carmichael, B. E. Cathers and P. P. Chamberlain, A Cereblon Modulator (CC-220) with Improved Degradation of Ikaros and Aiolos, J. Med. Chem., 2017, 61, 535–542 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5sc01251a |
| This journal is © The Royal Society of Chemistry 2025 |