 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The material–microorganism interface in microbial hybrid electrocatalysis systems
Jiyao
Li†
abc,
Hexing
Han†
c,
Yanhong
Chang
*ab and
Bin
Wang
 *c
*c
aDepartment of Environmental Science and Engineering, University of Science and Technology Beijing, Beijing 100083, China. E-mail: yhchang@ustb.edu.cn
bBeijing Key Laboratory of Resource-oriented Treatment of Industrial Pollutants, Beijing 100083, China
cCAS Key Laboratory of Nanosystem and Hierarchical Fabrication, National Center for Nanoscience and Technology (NCNST), Beijing 100190, China. E-mail: wangb@nanoctr.cn
First published on 27th February 2023
Abstract
This review presents a comprehensive summary of the material–microorganism interface in microbial hybrid electrocatalysis systems. Microbial hybrid electrocatalysis has been developed to combine the advantages of inorganic electrocatalysis and microbial catalysis. However, electron transfer at the interfaces between microorganisms and materials is a very critical issue that affects the efficiency of the system. Therefore, this review focuses on the electron transfer at the material–microorganism interface and the strategies for building efficient microorganism and material interfaces. We begin with a brief introduction of the electron transfer mechanism in both the bioanode and biocathode of bioelectrochemical systems to understand the material–microorganism interface. Next, we summarise the strategies for constructing efficient material–microorganism interfaces including material design and modification and bacterial engineering. We also discuss emerging studies on the bio-inorganic hybrid electrocatalysis system. Understanding the interface between electrode/active materials and the microorganisms, especially the electron transfer processes, could help to drive the evolution of material–microorganism hybrid electrocatalysis systems towards maturity.
1. Introduction
The increasing energy demands and impending climate change have been driving the development of sustainable pathways to produce fuels and chemicals. Inorganic electrocatalysis can electrochemically convert water, carbon dioxide, or nitrogen into value-added fuels and chemicals.1–5 This will play a key role in energy conversion technologies of the future. However, inorganic electrocatalysis also presents some problems. Some critical reactions are often kinetically sluggish and operate in the oxidation/reduction potential windows and cause significant overpotential and catalyst stability issues.6 For example, there are high energy demands and costs in the water electrolysis process,7–9 and relatively low selectivity, slow production speed and unsatisfactory energy conversion efficiency in the carbon dioxide (CO2) electrolysis process.10–12 Microorganisms are probably capable of tackling these synthetic challenges through their intracellular metabolic pathways. Microbial cells contain an entire system of metabolic pathways and can produce metabolites with high specificity. They are capable of decomposing complex organic substrates into small molecules but also synthesize complex products from the simplest and most stable feedstocks (e.g., H2O, CO2, N2, etc.).13 Besides, microorganisms can self-replicate and self-repair; thus, the cost of microbial catalysts is comparatively low. Therefore, the combination of inorganic electrocatalysis and microbial catalysis has been attracting significant attention.Over the past decades, microbial catalysis has been combined with electrocatalysis to construct bioelectrochemical system (BES). BES uses microorganisms as the catalysts14 to realize the conversion of matter and energy, such as the conversion from chemical energy to electricity in microbial fuel cell (MFC),15 the generation of hydrogen in microbial electrolytic cell (MEC),16 and CO2 reduction in microbial electrosynthesis (MES).17 However, BES is still far from real applications, because of the low efficacy and output of the scaled-up systems, which are not economically feasible for commercial applications.18 There are still many challenges due to thermal limitations, cost, and technical barriers, which hinder the scale-up and application of BES. Apart from the economic factors, the practical application of BES is mainly restricted by low power and product generation, and the limited electron transfer efficiency is usually one of the major bottlenecks.
In recent years, bio-inorganic hybrid systems that couple living microorganisms with inorganic nanomaterials have received much attention.19–22 The bio-inorganic hybrid system combines the advantages of inorganic catalysis, especially artificial photosynthesis, and microbial catalysis to overcome the intrinsic limitations of inorganic catalysis and promote and even change the microbial metabolisms.13,23 In a bio-inorganic hybrid electrocatalysis system, nanomaterials are closely attached/coated on the outer membrane of the microbial cells or even inside the cells, not just as a separate traditional electrode to participate in the electrochemical reactions.
In both BES and bio-inorganic hybrid electrocatalysis systems, electrons and matter are transferred and converted between microorganisms and materials. Therefore, in order to improve the efficiency of the combination of microbial catalysis and inorganic electrocatalysis, it is necessary to understand what happens at the material–microorganism interface and how to construct efficient microorganism and material interfaces, but a specific discussion on this issue is missing.
This review discusses the electron transfer mechanism at the material–microorganism interface and the strategies for building efficient microorganism and material interfaces. By summarizing and understanding the solved problems and unsolved challenges on this topic, we hope this review will attract more researchers to pay attention to this fast-growing field and thereby assist in moving such microbial hybrid electrocatalysis systems towards practical uses.
2. The electron transfer process at the material–microorganism interface
Most microorganisms cannot exchange electrons with extracellular electrodes or materials because their outer layers are usually known to consist of non-conductive lipid membranes, peptidoglycans and lipopolysaccharides.24,25 In contrast, electroactive microorganisms can exchange electrons with their extracellular environment across their cell membranes. Exoelectrogens can transfer electrons generated from the organic substrates to extracellular insoluble electron acceptors, such as electrodes or iron oxides, via the extracellular electron transfer (EET) pathways.26,27 Electrotrophs can accept electrons from extracellular electron donors via extracellular electron uptake (EEU) processes. Due to the electron exchange ability, electrons can be transferred at the interface between microorganisms and materials. There are already many studies on the electron exchange pathways; here, we focus on and summarize the mechanisms of electron transfer at the material–microorganism interfaces.2.1. The electron transfer from the microorganism to the anode
In the bioanode of BES, exoelectrogens transfer intracellular electrons mainly by direct electron transfer (DET) through redox-active proteins28–30 on the outer cell surface and conductive pili and microbial nanowires,31–33 or mediated electron transfer (MET) mediated by soluble electron mediators secreted by microorganisms34–36 or present in the natural environment.37,38 At present, mechanisms on EET mainly focus on some model exoelectrogens.The EET pathways of two typical Gram-negative exoelectrogens, Shewanella oneidensis MR-1 and Geobacter sulfurreducens, have been well studied.39 They transfer electrons across the outer membrane through porin–cytochrome complexes. The EET pathway of S. oneidensis MR-1 is called the metal-reducing (Mtr) pathway (Fig. 1A), which involves six c-type cytochromes (c-Cyts): CymA, Fcc3, MtrA, MtrC, OmcA, small tetrahaem cytochrome (STC) and the porin-like outer membrane protein MtrB.28,40–43 CymA acquires electrons from the quinone pool and transfers the electrons to Fcc3, STC and then to MtrA, MtrB and MtrC.44–54 On the cell surface, MtrC and OmcA transfer electrons directly to the extracellular acceptors.55–61S. oneidensis MR-1 also has the structure of microbial nanowires containing MtrC and OmcA, which are extensions of the outer membrane that can physically connect with neighbouring cells,32 mediating electron transfer through the multistep hopping mechanism.62 Besides, S. oneidensis MR-1 can transfer electrons by releasing flavin extracellularly.63–65 Through the above processes, S. oneidensis MR-1 can transfer electrons to extracellular electron receptors. The EET pathway in G. sulfurreducens is called the porin-cytochrome (Pcc) pathway (Fig. 1B). Inner membrane cytochromes consisting of quinol oxidases ImcH and CbcL connect the electron flow between intracellular electron transport chains and periplasmic cytochromes.66,67 Periplasmic cytochrome homologs PpcA, PpcB, PpcC, PpcD, and PpcE link the electron transfer between inner and outer membranes.68,69 Outer membrane c-Cyts are the final step in the direct EET consisting of porin-cytochrome complexes. The complexes consist of porin-like outer membrane proteins OmbB and OmbC, periplasmic c-Cyts OmaB and OmaC, and outer membrane c-Cyts OmcB and OmcC.29,70–72G. sulfurreducens also has electrically conductive pili (e-pili) comprised of c-Cyts OmcS.73 These c-Cyts and porin-like proteins transfer electrons from quinone and the quinone pool and across the extracellular membrane to the extracellular electron acceptors. Through the above EET pathways, S. oneidensis MR-1 and G. sulfurreducens transfer intracellular electrons to extracellular electron acceptors.
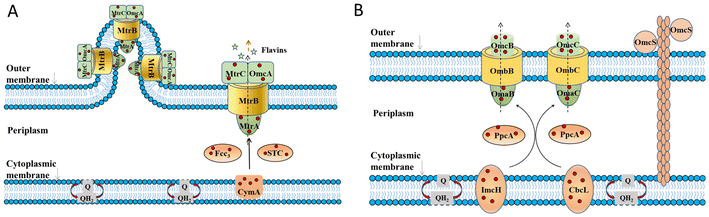 | ||
| Fig. 1 The proposed extracellular electron transfer pathways in Gram-negative exoelectrogens. (A) The metal-reducing (Mtr) pathway in S. oneidensis MR-1 and (B) porin–cytochrome (Pcc) pathway in G. sulfurreducens. Adapted from ref. 39 with permission from Nature Publishing Group, Copyright 2016. | ||
In the last decade, several Gram-positive bacteria have been found to perform extracellular electron transfer, though they lack outer membranes and contain a thick cell wall that is thought to prevent electron transfer.74–76 The studies on EET mechanisms of Gram-positive bacteria were only carried out for a few bacteria. Genomic analysis of Thermincola potens suggested that multiheme c-type cytochromes could be involved in the EET process.77 A model for the EET in T. potens has been presented that a series of a multiheme c-type cytochromes, TherJR_1117, TherJR_0764–0765, TherJR_0333, TherJR_1122 and TherJR_2595, transfer electrons through the cell wall to the extracellular electron acceptor.78Listeria monocytogenes has been reported to use a flavin-based EET mechanism to deliver electrons to iron or an electrode.79 An eight-gene locus was identified as being responsible for EET. Ndh2 transfers electrons from NAD to DMK. Electrons are then transferred from DMK to FMN groups on PplA or free flavin shuttles EetA and EetB, and ultimately to a terminal electron acceptor.79 Further studies are needed to clarify the EET mechanisms of Gram-positive bacteria.
To date, phylogenetically diverse exoelectrogens have been found, but the full diversity of exoelectrogens is still poorly understood.80 Extensive and in-depth studies are still needed to clearly and thoroughly understand the mechanisms of EET.
2.2. The electron transfer from the cathode to the microorganisms
The mechanisms of EEU are proposed, based on the electron transfer pathways in the anode.81 EEU also includes the DET and MET routes. In a direct EEU, microorganisms attach to solid cathode surfaces and take up electrons from them through direct contact, or the nanowires. During indirect EEU, microorganisms acquire electrons from the electrode through mediators, by-products, or extracellular polymeric substances.82 So far, EEU processes have been found in several microorganisms including metal-oxidizing bacteria, sulfur-reducing bacteria (SRB) and sulfur-oxidizing bacteria (SOB).Some microorganisms take up electrons directly from electrodes. For instance, the phototrophic Fe(II)-oxidizing bacterium Rhodopseudomonas palustris TIE-1 has been reported to accept electrons from a poised electrode, with CO2 as the sole carbon source/electron acceptor.83R. palustris TIE-1 has phototrophic iron oxidation (Pio) operon encoding three proteins: a multiheme cytochrome c (PioA), an outer membrane porin (PioB), and a periplasmic high-potential iron–sulfur protein (PioC).84 Electrons can enter the photosynthetic electron transport chain through the EEU and are then linked to the CO2 fixation process via the Calvin–Benson–Bassham cycle.85 Another Fe(II)-oxidizing bacterium Sideroxydans lithotrophicus ES-1 was hypothesized to oxidize iron via its Mto gene cluster. MtoA is a decaheme c-type cytochrome that can oxidize soluble Fe(II),86 while MtoD acts as an electron shuttle to transfer electrons from MtoA to CymAES-1 in the cytoplasmic membrane.87 CymAES-1 then reduces quinone in the cytoplasmic membrane to quinol. The sulfur-reducing bacteria (SRB) and sulfur-oxidizing bacteria (SOB) were also reported to use electrons through DET from a cathode. The electron uptake of SRB Desulfovibrio ferrophilus IS5 from the cathode is dependent on direct cell contact via a biofilm on the cathode surface rather than through secreted intermediates.88 The SOB Thioclava electrotropha ElOx9 was also demonstrated to perform electron uptake from cathodes through a direct-contact mechanism with a formal potential of −94 mV vs. the standard hydrogen electrode (SHE), rather than soluble intermediate electron carriers.89 Two typical exoelectrogens S. oneidensis MR-1 and G. sulfurreducens have been found to uptake the electrons from the cathode. Current studies suggest that S. oneidensis MR-1 can receives electrons from the cathode in a reverse Mtr pathway. It proposes that electrons from the electrode surface are transferred to MtrC. MtrC then transfers electrons to MtrA by interacting through MtrB. Then electrons are passed to CymA and through the menaquinone pool to a second CymA interacting with FccA.81 However, G. sulfurreducens was considered to have mechanisms for accepting electrons from the cathode that are substantially different from those for transferring electrons to the anode. A gene (GSU3274) encoding a putative monoheme, c-type cytochrome is vital to EEU. The deletion of genes for outer-surface proteins essential for EET had no impact on EEU from electrodes. However, the deletion of GSU3274 completely inhibited EEU but had no impact on EET.90
Microorganisms can also use artificial mediators, by-products, or extracellular polymeric substances and other substances to take electrons from the electrode through indirect electron transfer.91 For example, hydrogen and formate can act as special electron shuttles. Most acetogens, methanogens and SRB can use hydrogen as an electron donor.92 Electrochemically generated formate has been demonstrated to shuttle electrons from the cathode to Ralstonia eutropha H16.93 Methyl viologen and flavin have also been shown to mediate electron transfer from electrodes to microorganisms during the reduction of acetate and O2.94,95 Overall, the ability of a microorganism to take up electrons from cathode materials has been explored in MES, however, a detailed understanding of how electrons are transferred from the cathode to the microbes is still needed, especially for the DET process. The mechanism and direct observation of electron transfer from the cathode to the microorganism still require further investigation.
In addition to the extracellular electron uptake, the reduction reaction by the enzymes of microorganisms after obtaining electrons is also a key concern. Hydrogen production and CO2 reduction are the two main reactions in MES biocathodes, which are closely associated with the material–microorganism interfaces. Hydrogen production is mainly catalyzed by hydrogenases in a microbial system.96Desulfovibrio species are known for their ability to catalyze hydrogen production, where the hydrogenases including [FeFe], [NiFe] and [NiFeSe] are involved in the species. Desulfovibrio paquesii attached to a graphite electrode was found to catalyze hydrogen production at a potential of less than −900 mV vs. SHE.97Desulfovibrio vulgaris was found to catalyze the reaction in the presence of methyl viologen as a redox mediator,98 although Desulfovibrio was reported to handle direct EET as studies with purified Desulfovibrio enzymes have shown direct electron transfer from a cathode through cytochrome c3 to a [NiFe]-hydrogenase to produce hydrogen.99 For the Desulfovibrio genus, c-type cytochromes seem to be required for electron transfer to and from hydrogenases. The genome sequence of D. vulgaris (Hildenborough) reveald the presence of a pool of c-type cytochromes, likely providing the electrical wiring for connecting multiple periplasmic enzymes, including hydrogenases.100 Two typical exoelectrogens S. oneidensis MR-1 and G. sulfurreducens also encode several hydrogenases. Four [NiFe]-hydrogenases are encoded in the G. sulfurreducens genome: two periplasmically oriented, membrane-bound hydrogenases Hya and Hyb, and two cytoplasmic hydrogenases, Mvh and Hox.101G. sulfurreducens produces hydrogen when it acts as the biocatalyst at the cathode.102S. oneidensis MR-1 encodes a [NiFe]- and an [FeFe]-hydrogenase,103 which was found to catalyze cathodic hydrogen evolution at a potential of −758 mV vs. SHE.104 Several microorganisms have been reported to obtain electrons from the cathode, and then transfer the electrons to the intracellular hydrogenases to generate hydrogen.
Microorganisms known as electroautotrophs are reported to use CO2 as the sole carbon source and electrochemical-reducing power as the energy source. Electroautotrophs have been discovered in several groups of microorganisms, including iron-oxidizing bacteria, iron-reducing bacteria, nitrate-reducing bacteria, acetogens, methanogens and sulfate-reducing bacteria.92 Methanogens are one of the important biocatalysts to reduce CO2 in the MES. The ability of microorganisms to produce methane directly from CO2 and electrons was reported in 2009. At a set potential of less than −700 mV (vs. Ag/AgCl), CO2 was reduced to methane with Methanobacterium palustre as the biocatalyst using the electrons from the cathode.105Methanococcus vannielii, Methanococcus maripaludis, Methanolacinia petrolearia, Methanobacterium congolense and Methanoculleus submarinus were also able to produce methane at −700 mV vs. SHE.106 There are two possible mechanisms for methane formation including acetotrophic and hydrogenotrophic methanogens based on the substrates. The observed methane production was via the electron transfer and interspecies hydrogen transfer between H2-producing and H2-utilizing methanogens.107,108 Acetogenic bacteria have also been used for the production of commodity chemicals from CO2 and electricity in MES. The Gram-negative bacteria Sporomusa silvacetica and Sporomusa sphaeroides can reduce CO2 to acetate. In addition, Gram-positive bacteria Clostridium ljungdahlii, Clostridium aceticum and Moorella thermoacetica were reported to capture electrons from the cathode for acetic acid synthesis. Among them, Sporomusa ovata is the biocatalyst with the highest acetate production rate as described in reports109 because it has a unique autotrophic metabolism that uses CO2 as an electron acceptor and generates acetyl–CoA via the Wood-Ljungdahl pathway for efficient reduction.110
Overall, several microorganisms have been reported to obtain electrons from the cathode, and then transfer the electrons to the intracellular hydrogenases to generate hydrogen. However, the underlying mechanisms of the relationship between the electron-transferring components (e.g. c-type cytochromes, other oxidoreductases or mediators) and the hydrogenase activity remains unsettled. CO2 can also be reduced to various products through the metabolic systems of various microorganisms at the biocathode. In an actual BES, the biocathodes are often composed of mixed bacteria, also including the microorganisms that do not use and/or convert CO2. The reactions on biocathodes are complex and often involve multiple different reactions. More attention is required to fully clarify the underlying mechanisms through experiments and simulations to drive the development of this field.
3. The strategies for constructing efficient material–microorganism interfaces
High interfacial electron transfer is essential for electrocatalysis systems. The reported electron transfer efficiency is still very low, limited by the sluggish transmembrane and EET processes. To date, many methods have been used to build efficient material–microorganism interfaces, including electrode materials design and modification, microorganisms engineering, and the bio-inorganic hybrid system that has attracted much attention. Next, we summarize and discuss the three strategies for building efficient material–microorganism interfaces.3.1. The material design and modification
At the material–microorganism interface, the extracellular membrane components of the microorganism can interact with the electrode surface, which is where the electron exchange takes place. Due to the diverse physical and chemical properties of electrode materials, different material–microbe interfaces are formed, which affect the electron transfer at the interface. It is beneficial to form efficient interfaces for electron transfer by optimizing the design and modification of electrode materials.Materials with high electrical conductivity have been used to optimize the material–microorganism interface to facilitate electron transfer.111,112 Various carbon materials in diverse morphologies such as graphite, carbon cloth, carbon paper, active carbon, carbon black, carbon nanoparticles, carbon nanotubes and graphene have been used as electrode materials due to high conductivity and good biocompatibility.113,114 For example, graphitized mesophase pitch-based carbon foam was used as an MFC anode, and the GMCF–MFC exhibited an excellent power density of 1800 mW m−2.115 Nitrogen-doped carbon dots (CDs) were decorated onto the carbon paper surface to prepare the MFC anode, which was reported to be beneficial to the formation of a highly hydrophilic surface. A hydrophilic surface is usually considered a key parameter to enable microbial immobilization. The electrode showed a small charge transfer barrier, which enhanced the interaction between microbes and the anodes as stated by the researchers, producing power 1.1 times higher than that of the raw carbon paper anode with the same measurement area.116 Carbon nanotubes (CNT) have been employed as the electrode in MFC due to their unique mechanical and electrical properties.117 Nitrogen-doped carbon nanotubes with a bamboo-like nanostructure significantly reduced the internal resistance of the anode and improved the performance of an MFC in current production and power output.118 Graphene is the most famous 2D carbon material and it has low charge transfer resistance.119,120 Graphene and its derivatives such as graphene oxide (GO), reduced graphene oxide (r-GO), and functionalized graphene have been used as electrodes. For instance, the crumpled graphene particles showed the highest maximum power density (3.6 W m−3) of MFC.121 Metal-based materials have much higher conductivity than carbon-based materials, but they are easily corroded and of high cost, which make them difficult to use in large-scale systems, therefore, metal materials are often used to decorate carbon materials. For example, gold nanoparticles (Au NPs) were sputtered on carbon paper as electrodes of MFCs. The highest power density was obtained by depositing carbon paper with an Au thickness of 50 nm and 100 nm on each side. The modification of Au NPs can reduce the electrode resistance.122 Conductive polymers were also involved in the anode applications due to their intrinsic conductivity and biological compatibility. For instance, the cationic polythiophene derivative poly(3-(3′-N,N,N-triethylamino-1′-propyloxy)-4-methyl-2,5-thiophene hydrochloride) (PMNT) could enhance bacterial biofilm formation, improve the bacterial viability, decrease the resistance value and accelerate the EET at the material–microorganism interface, and as a result, the maximum current density and power density respectively increased 4.6 and 5.5 times.123 A cathode with electropolymerizing conductive poly (3,4-ethylenedioxythiophene) (PEDOT) on GO film-modified carbon cloth was used in MES to convert CO2. The GO/PEDOT modified electrode was applied in the MES of CO2 reduction to CH4, and a maximum CH4 production rate of 315.3 ± 13.2 mM m−2 d−1 was achieved with a Faraday efficiency >92% at 900 mV (vs. Ag/AgCl).124 On the whole, selecting materials with high conductivity or improving the conductivity of materials through modification can reduce internal resistance, stimulate the formation of biofilm, and promote electron transfer at the material–microorganism interface.
Large surface area and porosity are good for microbial attachment, biofilm formation and electron transfer. Nanostructures, especially three-dimensional (3D) structures, have been widely used to increase specific surface area.125 For example, a 3D porous inverse opal-indium tin oxide (IO-ITO) electrode hosted a large population of Geobacter and attained a current density of 3 mA cm−2 stemming from bacterial respiration (Fig. 2A–C). Geobacter was considered to express more electron-relaying proteins when interfaced with electrodes.126 A 3D CNTs-coated carbon-textile electrode provided a macroporous structure, which created an open space for efficient feed transport and internal bacteria immobilization.127 A 3D rGO–Ni foam-based anode was synthesized by controlling the deposition of rGO sheets onto a current collector (nickel foams). This 3D rGO–Ni anode not only has a highly accessible surface area for bacterial immobilization, but also a conductive macro-porous structure for efficient electron transfer and mass diffusion. The rGO–Ni anode produced a remarkable volumetric power density of 661 W m−3 (27 W m−3) at a stable power generation, calculated based on the volume of the anode material (based on the anode chamber volume).128 Similarly, a hierarchical porous nitrogen-doped carbon nanotube (N-CNTs)/rGO composite possessed a 3D hierarchically porous structure for rich microbial biofilm growth (Fig. 2D). The improved bio-electrocatalysis could be attributed to the enhanced adsorption of flavins on the N-doped carbon surface and the high density of biofilm adhesion for fast interfacial electron transfer. The maximum power density reached 1137 mW m−2, 8.9 times that of the carbon cloth anode, and higher than that of N–CNTs, N–rGO and the CNTs/rGO composite without nitrogen doping.129 A MES with 3D CNTs on reticulated vitreous carbon achieved acetate production rates of 1.3 mM cm−2 d−1 from CO2. The improved performance was attributed to the nanostructure of the electrode.130 Generally, 3D materials have a large surface area, open macroporous structure and suitable surface properties for bacteria attachment and electron transfer and can facilitate biofilm formation.131
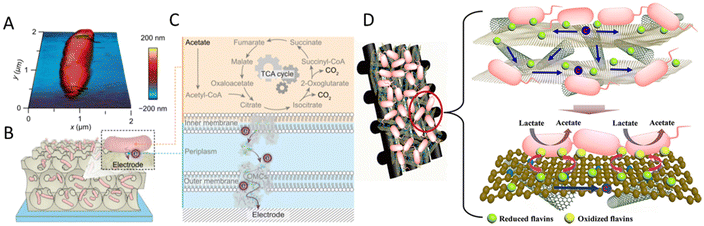 | ||
| Fig. 2 (A) Atomic force microscopy (AFM) image of G. sulfurreducens on a silicon wafer. (B) Schematic representation of a biohybrid electrode where G. sulfurreducens colonized the IO-ITO scaffold. (C) Extracellular electron transfer at the interface between G. sulfurreducens and an electrode. Acetate is metabolized into CO2via the TCA cycle and excess electrons are discharged to an external electrode via OMCs. (A–C) Reproduced from ref. 126 with permission from the National Academy of Sciences of the United States of America, Copyright 2020. (D) Mechanism of synchronous enhanced biofilm extracellular electron transport and electronic mediator electrochemistry of a hierarchically porous nitrogen-doped carbon nanotubes (N–CNTs)/reduced graphene oxide (rGO) composite anode. (D) Reproduced from ref. 129 with permission from the American Chemical Society, Copyright 2018. | ||
Special structures such as microelectrodes and nanowires at the microbial interface are reported to enhance the interaction between bacteria and electrodes. The microbial current generated by Shewanella loihica PV-4 could be greatly improved 115-fold by adding antimony-doped tin oxide (ATO) nanoparticles because the ATO nanoparticles acted as microelectrodes to facilitate the formation of cells/ATO composite biofilm. These ATO nanoparticles stimulated the reduction of the outer membrane c-Cyts.132 A Sn-doped In2O3 nanowires array with a flat F-doped In2O3 (FTO) electrode was reported to promote EET via outer membrane c-type cytochromes (OMCs) by physical contact with microbes, and the Sn-doped In2O3 nanowires were considered to provide a suitable energy level to facilitate the EET via the OMC-flavins cofactor, rather than the certain redox state of OMCs at a given potential (Fig. 3A–C). The composite electrode can highly boost EET by over 60 times at a certain potential of 0.2 V.133 In addition, the long nanowire structure endows materials with good binding capacity with the microbial cell walls, which also assists the interfacial electron transfer.134,135 Multi-walled carbon nanotubes (MWCNTs) could be directly grown in the radial direction from the wires of stainless steel meshes for use. MWCNTs can capture microbes like tentacles, resulting in good charge transfer characteristics between CNTs and the pili of microbes. The long-term performance showed a maximum power density of 3360 mW m−2, 7.4 times higher than that of carbon cloth.136 A MWCNTs network was also used to modify the anodic surface to form a highly conductive, interconnected, biocompatible, and microbial cell wall-attached network in the anolyte. The carbonaceous network is reported to transfer electrons away from suspended microorganisms, which greatly improves the electron transfer efficiency.137 Besides, unique interfaces can be formed to produce special effects to facilitate electron transfer. A suction effect was demonstrated in a bioinspired active anode by constructing polypyrrole nanotubular arrays on carbon textiles. A vacuum in the nanotubular space was formed due to the depletion of the oxygen in the inner space of the nanosucker, which then activated the electrode to draw the microorganism by suction to improve the EET.138 A cathode by using nickel nanowires anchored to graphite was used in the MES for Sporomusa-catalyzed reduction of CO2 to acetate. This porous nickel-nanowire-network-coated graphite electrode was reported to increase the interfacial area and interfacial interactions between the cathode surface and the microbial biofilm. Around 282 mM m−2 d−1 of acetate was produced with 82 ± 14% of the electrons consumed being recovered in acetate.
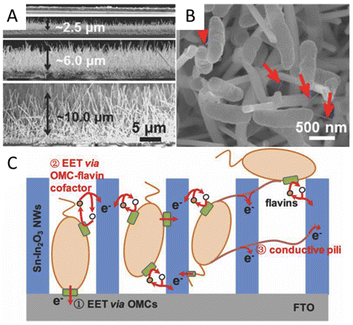 | ||
| Fig. 3 (A) Cross-section SEM images of the flat FTO film and SINW/FTO with the nanowire lengths of 2.47, 5.80, and 9.85 μm. (B) SEM images of SINW/FTO electrodes integrated with microbes of S. loihica PV-4 after 25 h electrochemical culture at 0.2 V. The red arrows mark the pili grown among cells. (C) Cartoon illustrating the active EET pathways on the SiNW/FTO electrode. Reproduced from ref. 133 with permission from Wiley Online Library, Copyright 2018. | ||
Fe oxides were employed as electrode modifiers to improve bacterial electron transfer because of their high biocompatibility and great specific affinity to the bacterial OM c-Cyts.139,140 For instance, a study showed that S. loihica PV-4 had the ability to self-organize an electrically conductive network using outer-membrane proteins OM c-Cyts and semiconductive minerals α-Fe2O3 as a long-distance electron transfer conduit.141 An α-Fe2O3 nanorod and chitosan were used to modify the ITO as the anode. The MFC with modified anode produced a higher quantity of electricity output with 320% enhancement as compared with the bare anode.142 A carbon-coated Fe2O3 electrode MFC exhibited the maximum biocurrent density of 0.22 ± 0.01 mA cm−2, which was nearly 6-times higher than that of a bare carbon cloth electrode.140 Similarly, Fe3O4, Fe3O4/CNT and graphite belt hosted bentonite–Fe have also been applied in the systems.143,144
In addition to the morphologies and type of electrode materials, other properties affect the material–microorganism interface. The surface charge of the electrode material was reported to influence microbial attachment. It is thought that the positively charged electrode material forms a strong adsorption interface to enhance the attachment of negatively charged bacteria due to electrostatic attraction. For example, an electrochemical modification of a glassy carbon electrode showed that the surface equipped with –N+(CH3)3 was the fastest to colonise and produce the highest current density compared to negatively charged and more hydrophobic surfaces.145 Electrostatic self-assembly of charged polymers was used to systematically modify the surface of indium-tin-coated electrodes, wherein the thickest biofilms, highest current density and shortest start-up time were achieved for negatively charged electrode surfaces or polystyrol sulfonate-coated electrodes, while positively charged chitosan, negatively charged alginate and positively charged polyethylene imine produced thinner biofilms with less current and a longer start-up time.146 Surface functional groups also affect the attachment of microorganisms, the formation of biofilm and electron transfer. C-, N-, O- and S-containing functional groups were reported to enhance microbial attachment at the interface.147S. loihica PV-4 expressed enhanced EET activity on the hydrophilically-functionalized surface. It proposed that the redox state of OMCs varies significantly at electrodes with different wettability, resulting in different EET activities.148
In brief, an efficient material–microorganism interface can afford a large surface area that offers sufficient locations for microorganism adhesion, contact with outer membrane components, stimulate the release of mediators, and thus establish an efficient electron transfer conduit through the microelectrodes to promote the chemical processes.
3.2. The bacterial engineering strategies
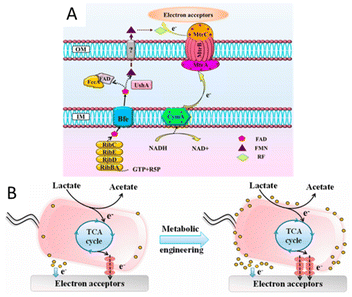
In addition to the approaches involving optimizing the materials to increase the specific surface area, biocompatibility, conductivity, etc., or enhancing the interaction between bacteria and electrodes through the special structure of the materials to promote electron transfer, the engineering of microorganisms is also a emerging method. In recent years, the studies on the modification of bacteria to promote electron transfer through metabolic engineering methods and synthetic biology strategies have attracted widespread attention.The electrons transferred extracellularly come from the cytoplasmic electrons generated by the intracellular metabolism. Promoting the use of carbon sources will also promote the generation of electrons. By expressing a proteorhodopsin in S. oneidensis MR-1, a significant increase in current generation was achieved during illumination. Because the light-driven proton pump proteorhodopsin activity enhanced the lactate uptake by increasing the proton motive force, the engineered strain was able to consume lactate at an increased rate to accelerate the EET rate.149 Nicotinamide adenine dinucleotide (NAD+) and its reduced form NADH are essential cofactors for the metabolism of microorganisms,150–153 and they have also been shown to be intracellular electron pools for EET. The increase of the electron pool can improve intracellular electron flux and the EET rate.154 The heterogeneous expression of formate dehydrogenase enabled enhanced NADH regeneration in Clostridium ljungdahlii to manipulate the [NADH]/[NAD+] ratio, thereby increasing the release of intracellular electrons and power generation.155 In order to further improve the EET rate, a modular synthetic biology approach was used to increase the de novo biosynthesis of the total endogenous NAD(H/+) pool in S. oneidensis MR-1, leading to a 2.1-fold increase in the total intracellular NAD(H/+) level in S. oneidensis (Fig. 4). The maximum power density of S. oneidensis was increased from 30.2 ± 3.4 mW m−2 to 162.8 ± 5.6 mW m−2, and the coulombic efficiency increased from 8.6% to 21.7%.154
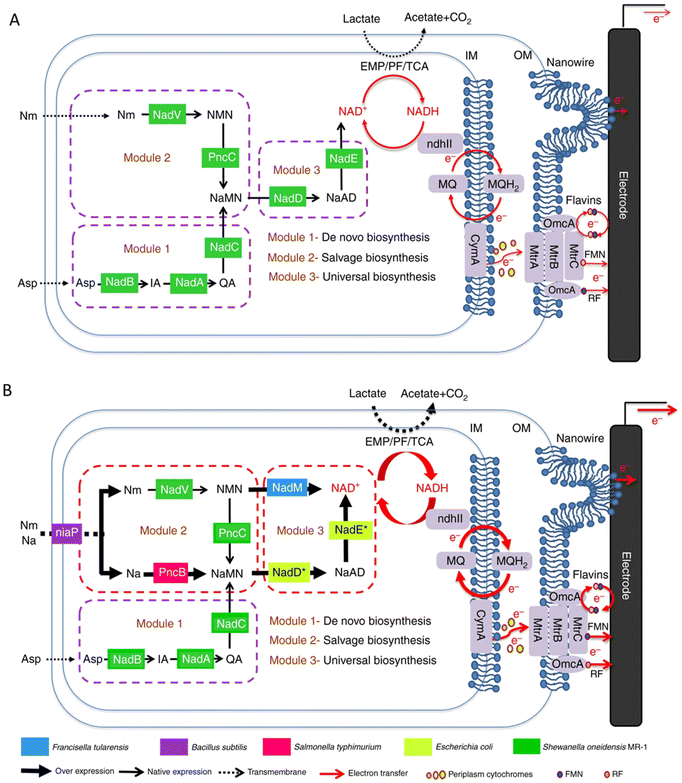 | ||
| Fig. 4 Schematic of modular design to enhance NAD+ biosynthesis and EET rate in S. oneidensis MR-1. (A) The NAD+ biosynthesis pathway of the wild-type S. oneidensis MR-1 (as revealed by genomic studies) is categorized into three modules. (B) Recombinant S. oneidensis SN5 harboring five homogeneously and heterogeneously introduced genes (ycel, pncB, nadM, nadD*, and nadE*) to enhance NAD+ biosynthesis. Reproduced from ref. 154 with permission from Nature Publishing Group, Copyright 2018. | ||
There have also been many studies focusing on the optimization of extracellular electron transfer, such as the overexpression of c-type cytochromes and shuttles and the promotion of the formation of biofilm. The overexpression of CymA of S. oneidensis improved the extracellular electron transfer rate and the resulting electrochemical performance.156 By the co-expression of the metal-reducing conduit biosynthesis gene cluster mtrC–mtrA–mtrB in S. oneidensis (Fig. 5), a 1.9-fold higher biodegradation rate than that of the wild-type strain was exhibited.157 The promotion of shuttles biosynthesis and secretion would also be an efficient approach to enhancing EET efficiency. Isoprenoid quinones are bioactive molecules that include an isoprenoid chain and a quinone head, which also act as electron transporters. Saccharomyces cerevisiae EPYFA3 was engineered to overproduce the endogenous isoprenoid quinone coenzyme Q6, resulting in a nearly three-fold production increase.158 A synthetic riboflavin biosynthesis pathway from Bacillus subtilis was integrated into S. oneidensis, resulting in a significant increase in secreted riboflavin and a subsequently increased EET rate.159,160 Similarly, increasing the biosynthesis of phenazines has been shown to improve the EET efficiency of Pseudomonas aeruginosa. The quorum sensing system regulates the current generation of P. aeruginosa by controlling the production of phenazines, and the overexpression of the quorum sensing system can increase the production of phenazines, thus increasing the EET efficiency.161 Studies have demonstrated that thicker biofilms generally exhibit better electrical conductivity than thinner ones. For example, the heterologous overexpression of the c-di-GMP biosynthesis gene ydeH from Escherichia coli in S. oneidensis MR-1 increased the production of cyclic-di-GMP, a key intracellular regulator for controlling biofilm formation, and generated ∼2.8 times more current in MFC than that of the wild-type S. oneidensis.162 In addition, cell-surface polysaccharides and other biofilm-related components were also reported to influence the EET efficiency of S. oneidensis. Destruction of the putative cell surface polysaccharide biosynthesis gene SO3177 in S. oneidensis MR-1 could enhance its hydrophobicity and ability to attach to graphite electrodes, resulting in larger colonies than the wild-type S. oneidensis MR-1, generating ∼1.5 times the current in the MFC.163
 | ||
| Fig. 5 (A) Schematic illustration of the flavin and metal-reducing conduit-mediated EET pathway in S. oneidensis MR-1. (B) Schematic of the EET rate in S. oneidensis MR-1 before and after metabolic engineering. Reproduced from ref. 157 with permission from American Chemical Society, Copyright 2017. | ||
Synthetic biology has played a good role in modifying microorganisms to improve EET at the material–microorganism interface, which is usually a rate-limiting step of bioelectrochemical systems. However, because of the lack of key knowledge and a systematic understanding of the molecular mechanism of electron transfer and the development of new gene editing tools, there is still much room for synthetic biology strategies to improve EET. More researches and attention should be paid to the optimization of EEU using synthetic biology, which is a more complex but important issue.
3.3. The bio-inorganic hybrid system to construct efficient material–microorganism interface
Very recently, various nanomaterials and microorganisms have been integrated to construct bio-inorganic hybrid electrocatalysis systems. In bio-inorganic hybrid systems, nanomaterials are located on the outer membrane or inside the cell of the microorganism, establishing an effective material–microorganism interface at the electrode to facilitate electron transfer.Bio-inorganic hybrid systems have been used to improve conductivity to promote EET. A highly conductive polymer, polypyrrole (PPy), was coated on the surface of mixed culture acetogens in situ and was inoculated on the cathode of BES. The charge transfer resistance of the PPy-coated biocathode was found to be lower than that of the uncoated. The acetate production rate and faradaic efficiency in PPy-coated biocathodes were increased by 3 to 6 times.164 Similarly, a polymer/inorganic materials hybrid system was designed. A bacterial surface anchored electron collector (S collector), and a surface and periplasmic single cell in situ electron collector (SP collector) were proposed (Fig. 6). The former modification was realized by coating polydopamine on the cell surface for promoting electron transfer, while FeS was selected for SP collector assembly due to their ease of biosynthesis, high electroactivity and biocompatibility. The SP collector design was made to further wire up more “idle” conduits and thus improve interfacial electron transfer; the maximum power output of 3.21 W m−2 was achieved in the MFC inoculated with S. oneidensis MR-1@SP cells, which is the highest reported record with this model strain.165 As a kind of modification to the electrode, CDs can be used to improve the interfacial electron transfer in the bio-nano system. The highly conductive N-doped Fe3O4 with a carbon dot shell was combined with G. sulfurreducens to form a bio-inorganic hybrid system. Fe3O4@CDs formed an interaction network with both internal and external conductive proteins of G. sulfurreducens. In the microbial electrolytic cell, the maximum current was 6.37 times higher than that without nanomaterials. The bacterial surface was modified through electrostatic attraction between the positively charged IL–NH2-modified N,S-doped carbon dots (m-NSCDs) and negatively charged bacteria. The m-NSCDs located on the surface of bacteria were reported to enhance the EET between the anode and S. oneidensis MR-1, with a power output 2.6 times higher than that of the raw bacteria.166 An emerging field of hybridizing microorganisms and nanomaterials involves utilizing the self-generation properties of the microbes to produce nanomaterials by themselves. For example, Au NPs could be fabricated in situ into a G. sulfurreducens biofilm. This strategy greatly lowered the charge transfer resistance and enhanced the anodic limiting current, and the removal percentage of the organic substrate was enhanced 2.2 times.167
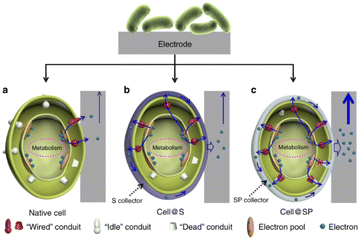 | ||
| Fig. 6 Schematic of the biointerfacial electron transfer between an electrode and cells. (a) Native cell, (b) S collector encapsulated cell (cell@S) and (c) SP collector encapsulated cell (cell@SP). Reproduced from ref. 165 with permission from Nature Publishing Group, Copyright 2020. | ||
Since the electron pool driving the EET pathway is located in the cytoplasm, nanomaterials entering the cytoplasm through the cell membrane may cause additional interactions that affect the physiological function of exoelectrogens and EET efficiency. For example, the cellular internalization of CDs into Shewanella xiamenensis was reported to afford enhanced conductivity and also trigger the cellular physiological response to secrete abundant electron shuttles to aid the boosting of the EET efficiency, with a maximum current density that was 2.4 times that of the S. xiamenensis electrode.168 CDs were found to be efficiently taken up by S. oneidensis MR-1 (Fig. 7). The CDs-fed cells showed accelerated metabolic rates, with increased intracellular charge, higher adenosine triphosphate levels, quicker substrate consumption, more abundant extracellular secretion, and maximum current and power output increases of 7.34 and 6.46 times, respectively.169 Palladium (Pd) nanoparticles were introduced into the periplasm of the sulfate-reducing bacterium Desulfovibrio desulfuricans. A small amount of Pd nanoparticles facilitated electron transfer in the periplasm and a large amount of Pd particles could directly act as a wire connecting the cytochromes in the inner membrane and outer membrane, leading to an increase in current.170
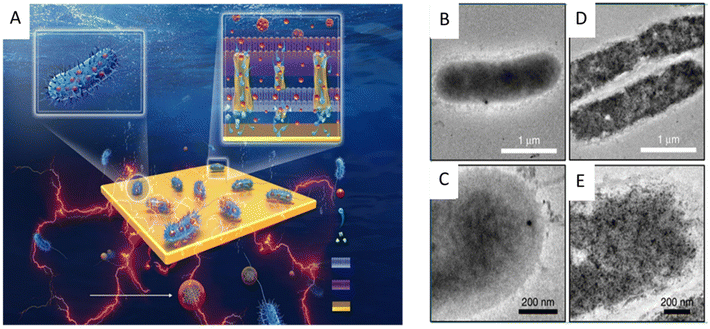 | ||
| Fig. 7 (A) Illustration of the synthesis of the carbon dots (CDs) and the CDs-fed S. oneidensis MR-1 for enhanced bioelectricity generation. Cellular uptake of the CDs by S. oneidensis MR-1. TEM images of S. oneidensis MR-1 cells (B and C) before and (D and E) after 6 h incubation with the CDs. Reproduced from ref. 169 with permission from Nature Publishing Group, Copyright 2020. | ||
Photo-responsive nanomaterials have also been used to facilitate electron transfer in microbial hybrid electrocatalytic systems. By utilizing the electrostatic layer-by-layer assembly strategy, the conductive Au NPs and photo-responsive CdS NPs were alternatively modified onto the surface of E. coli at the bioanodes in MFCs. The CdS layers can generate photocurrent without any loss of biocurrent because the outer Au layer could serve as a conductive channel for the photoelectron and bioelectron transfer between each bacterium. The output power of its MFC increased 1.9 times in the dark and 2.5 times in light.171 Similarly, Au nanoparticles were biosynthesized in situ inside cyanobacteria to promote light harvesting as a light absorber and facilitate extracellular electron transfer as a conductive conduit through the cell membrane (Fig. 8). The intracellular nanoparticles efficiently harvest sunlight and generate “hot” electrons via interband transition, and electronically connect the cells with the external electrode, increasing the maximum power density by 33.6 times.172 Introducing transmembrane and outer-membrane silver (Ag) nanoparticles into Shewanella was found to boost the charge-extraction efficiency in MFCs. The resulting Shewanella-silver MFCs delivered a maximum current density of 3.85 mA cm−2, a power density of 0.66 mW cm−2, and a single-cell turnover frequency of 8.6 × 105 s−1, which are all considerably higher than those of the best MFCs reported until then.173
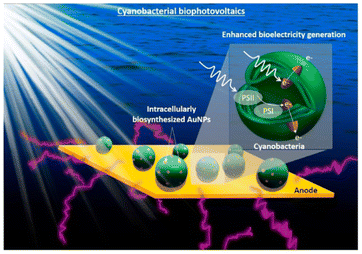 | ||
| Fig. 8 Conceptual image of the proposed work on the cyanobacterial biophotovoltaics for enhanced biophotoelectricity generation. Intracellularly biosynthesized Au NPs will improve cyanobacterial electron harvesting and extracellular electron transfer. Reproduced from ref. 172 with permission from Elsevier, Copyright 2021. | ||
In the bio-inorganic electrocatalysis system, the electron transfer efficiency is enhanced due to the efficient interfaces formed by the synergistic interaction of microorganisms and nanomaterials. Nanomaterials can not only provide highly conductive interfaces but also introduce additional functions, such as photogenerated electrons through photo-responsive materials, to facilitate electron transfer.
3.4. Discussion of different strategies
The above three strategies promote electron transfer at the material–microbial interface in microbial hybrid electrocatalysis systems by optimizing the design of electrode materials, modifying bacteria through metabolic engineering methods and synthetic biology strategies, and constructing bio-inorganic hybrid systems.So far, various materials have been used as electrode materials for microbial electrocatalysis, including carbon-based materials, metal and metal oxide materials, conductive polymer materials, and so on. These materials have different dimensions, ranging from 0D quantum dots, 1D nanowires, and 2D films to 3D materials. Different kinds of materials are often made into composite materials or modified to improve performances, which has been reported to significantly reduce the charge transfer resistance and facilitate the adsorption of bacteria to promote the formation of biofilm, thus greatly improving the performances of MFC. So far, through the design and modification of materials, the electron transfer at the interface has been optimized but is still the limiting factor. There are some problems such as complex interface conditions and limited technical means. Most importantly, due to the limitation of the electron transfer ability of electroactive microorganisms, the restriction of electron transfer ability in the system cannot be fundamentally breaked through only the optimization of electrode materials. The cost should also be taken into account in the selection and design of electrode materials. Carbon-based materials are commonly used as electrodes also because of their low cost. Efficient electrode materials that can be synthesized on a large scale with a wide range of sources and low production costs are beneficial to the practical applications of microbial hybrid electrocatalysis systems.
Synthetic biology has become the focus for enhancing the electron transfer of electroactive microorganisms in recent years, especially the extracellular electron transfer of exoelectrogens. Using metabolic engineering methods and synthetic biology strategies to modify bacteria can promote intracellular electron production and extracellular electron transfer, thus significantly improving EET. Synthetic biology can impact economic feasibility by increasing power generation and product range.174 The engineering of exoelectrogens not only focuses on the extracellular electron transfer pathways but also starts from the intracellular metabolism. However, the internal metabolic reactions of microorganisms are numerous and complex, so the comprehension of molecular mechanisms of EET is still incomplete. Another concern is that the current researches on synthetic biology mainly focuse on EET, while less attention is paid to EEU at the biocathode. The mechanism of EEU is unclear, and how the electrons are transferred to the target reaction inside the cell is still unknown. A full understanding of the metabolisms and physiology of electroactive microorganisms is necessary for establishing synthetic biology approaches. It seems that there are bright prospects for using synthetic biology to improve the electron transfer of the material–microorganism interface in microbial hybrid electrocatalysis systems.
In bio-inorganic hybrid electrocatalysis systems, the conversion of energy and matter depends on the metabolism of microorganisms. Combined with the respective advantages of electrocatalysis and microbial conversion processes, bio-inorganic hybrid systems have been proven to improve energy conversion, electron transfer and product selectivity. In particular, the application of photocatalyst materials in hybrid systems, injecting the energy of photons into the microorganisms, not only promotes electron transfer but also promotes or even changes the intracellular metabolic process. To date, the research on bio-inorganic hybrid systems in microbial electrocatalysis is still limited, especially cathodic microbial electrocatalysis. In the future, more inorganic materials are expected to be used in hybrid systems. With the development of electroactive microorganisms, microbial electrocatalysis can involve more abundant microorganisms, including engineering bacteria through synthetic biology. However, now there are still problems to be solved. For example, the coating of a single bacterial cell is usually unstable, affecting the long-term viability and stability of the cell, which requires very close contact and a high interface area, etc. Moreover, elucidating the precise effects of physiological stress responses and internalized nanomaterials on metabolism remains a challenge. In bio-inorganic hybrid systems, microbial self-assembly is a low-cost and low-energy consumption method for synthesizing nanomaterials. However, the materials currently synthesized are mainly metals and semiconductors, and the precursors are often some expensive metal salts, such as HAuCl4. Alternatively, if nanomaterials can be synthesized by microbial synthesis from metal wastewater, it may also serve as an economic way to recycle waste metal.
Each of the above three strategies has its advantages and disadvantages. At present, the selection, design and modification of electrode materials to build an efficient interface have been developed, with the inputs of a variety of electrode materials and preparation methods. Reducing the cost of raw materials and synthesis processes is one of the concerns. Synthetic biology has great prospects for the construction of efficient interfaces, but its development still needs some time. The bio-inorganic hybrid electrocatalysis system has just emerged, and there are many problems to be solved. Of course, these three methods are not antagonistic and should be considered together to solve the problems of electron transfer at the interface.
4. Conclusions
This review presents a comprehensive summary of the material–microorganism interfaces in electrocatalysis hybrid systems. By coupling efficient living cell catalysis and material catalysts, such a hybrid system is expected to be an innovative platform for energy harvesting and chemical production, wherein the electron transfer efficiency is the key influencing factor for practical applications. The transfer pathways of electrons in the system are complicated and varied, depending on the microorganism species, the electrode construction, and particularly, the interfaces between the living cells and additional organic/inorganic materials. We have discussed the electron transfer mechanism at the material–microorganism interface, the strategies for constructing efficient material–microorganism interfaces including material designs and bacterial engineering and the bio-inorganic hybrid electrocatalysis system.The electron transfer at the interface between materials and microorganisms is one of the key issues to improve the efficiency of microbial hybrid electrocatalysis. Therefore, it is necessary to understand the mechanism of electron transfer at the interface. To date, there are many studies on the mechanism of EET of exoelectrogens, and some mechanisms of EET are well studied such as the porin–cytochrome pathway in Gram-negative exoelectrogens, S. oneidensis MR-1 and G. sulfurreducens. However, the exoelectrogens found are phylogenetically diverse, and the EET mechanisms of many other exoelectrogens are still unclear. Besides, there are still many exoelectrogens that have not been found. In addition, the process of EET is not only related to the pathways that transfer the electrons out of the cell but is also closely related to the intracellular metabolism, which requires a lot of in-depth researches. The studies on electron transfer have been mostly focused on EET, and the research on how electrons are transferred from the cathode to the microbes in the EEU remains at the initial stage. The EEU process is very important for the biological cathodic reduction of carbon dioxide and hydrogen production, which needs and deserves further studies. After the electrons are transferred inside the cell, how to interact with the intracellular metabolisms is also an issue that needs to be addressed. The underlying mechanisms of the relationship between the electron-transferring components and the intracellular metabolism remain unsettled.
Synthetic biology has great potential to improve electron transfer at the material–microorganism interface because it can break the limit of electron transfer by modifying microorganisms. We think synthetic biology will be the emphasis of the studies of microbial electrocatalysis. The bio-inorganic hybrid system has been proven to improve electron transfer and energy conversion in microbial electrocatalysis. The development of the above two methods requires a deep understanding of electron transfer, which still needs further experimental researches as mentioned. In addition to promoting electron transfer, the reductiones of carbon dioxide and hydrogen production need to be addressed through these two methods because it is difficult to control the transfer of electrons to the target reactions. In addition to further understanding electron transfer and metabolism in microorganisms, the cooperation of synthetic biology and the bio-inorganic hybrid system may be the direction taken for future development. Synthetic biology solves the directional problem of reactions, and the hybrid system increases the source of electrons, especially by injecting the energy of the photons into the system. The selection and design of electrode materials is a key part of microbial hybrid electrocatalysis, although it cannot fundamentally solve the problem of electron transfer limitation. However, it has a critical impact on the cost of the system, the long-term sustainability and the real applications.
The hybrid electrocatalysis systems combining inorganic catalysis and microbial catalysis have a broad development space for the improvement of electrocatalysis efficiency and the production of chemicals and energy. Extensive and in-depth researches are needed to contribute to the practical applications and underlying mechanisms. It is hoped that this review will deepen the understanding of material–microorganism hybrid electrocatalytic systems and attract more researchers to pay attention to this field.
Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
We acknowledge the financial support from the National Key R&D Program of China (2021YFA1202802, 2022YFF0712200), the China Postdoctoral Science Foundation Funded Project (E2891IR1) and the CAS Pioneer Hundred Talents Program.References
- S. Xu, Y. Gao, T. Liang, L. Zhang and B. Wang, Chin. Chem. Lett., 2022, 33, 5152–5157 CrossRef CAS.
- Z. W. Seh, J. Kibsgaard, C. F. Dickens, I. Chorkendorff, J. K. Norskov and T. F. Jaramillo, Science, 2017, 355, eaad4998 CrossRef PubMed.
- Z. Gao, J. Li, Z. Zhang and W. Hu, Chin. Chem. Lett., 2022, 33, 2270–2280 CrossRef CAS.
- J. Su, Y. Liu, Y. Song, L. Huang, W. Guo, X. Cao, Y. Dou, L. Cheng, G. Li, Q. Hu and R. Ye, SmartMat, 2022, 3, 35–53 CrossRef CAS.
- T. Xu, B. Ma, J. Liang, L. Yue, Q. Liu, T. Li, H. Zhao, Y. Luo, S. Lu and X. Sun, Acta Phys.-Chim. Sin., 2020, 2009043–2009040 Search PubMed.
- Y. Li, H. Wang, C. Priest, S. Li, P. Xu and G. Wu, Adv. Mater., 2021, 33, e2000381 CrossRef PubMed.
- S. K. Mazloomi and N. Sulaiman, Renewable Sustainable Energy Rev., 2012, 16, 4257–4263 CrossRef CAS.
- J.-F. Chang, Y. Xiao, Z.-Y. Luo, J.-J. Ge, C.-P. Liu and W. Xing, Acta Phys.-Chim. Sin., 2016, 32, 1556–1592 CAS.
- J. Wang, T. Liang, H. Li, J. Xiong, B. Liu, X. Xu, Y. Gao, Z. Yu, Q. Zheng, S. Zhang and B. Wang, Chin. Chem. Lett., 2022, 107826 CrossRef.
- S. Nitopi, E. Bertheussen, S. B. Scott, X. Liu, A. K. Engstfeld, S. Horch, B. Seger, I. E. L. Stephens, K. Chan, C. Hahn, J. K. Nørskov, T. F. Jaramillo and I. Chorkendorff, Chem. Rev., 2019, 119, 7610–7672 CrossRef CAS PubMed.
- A. Vasileff, C. Xu, Y. Jiao, Y. Zheng and S.-Z. Qiao, Chem, 2018, 4, 1809–1831 CAS.
- X. Yang, Q. X. Li, S. Y. Chi, H. F. Li, Y. B. Huang and R. Cao, SmartMat, 2022, 3, 163–172 CrossRef CAS.
- X. Fang, S. Kalathil and E. Reisner, Chem. Soc. Rev., 2020, 49, 4926–4952 RSC.
- K. Rabaey and R. A. Rozendal, Nat. Rev. Microbiol., 2010, 8, 706–716 CrossRef CAS PubMed.
- B. E. Logan, B. Hamelers, R. Rozendal, U. Schroder, J. Keller, S. Freguia, P. Aelterman, W. Verstraete and K. Rabaey, Environ. Sci. Technol., 2006, 40, 5181–5192 CrossRef CAS PubMed.
- H. Liu, S. Grot and B. E. Logan, Environ. Sci. Technol., 2005, 39, 4317–4320 CrossRef CAS PubMed.
- B. Bian, S. Bajracharya, J. Xu, D. Pant and P. E. Saikaly, Bioresour. Technol., 2020, 302, 122863 CrossRef CAS PubMed.
- P. Parkhey, in Scaling Up of Microbial Electrochemical Systems, 2022, pp. 173–193 Search PubMed.
- K. K. Sakimoto, A. B. Wong and P. Yang, Science, 2016, 351, 74–77 CrossRef CAS PubMed.
- H. Zhang, H. Liu, Z. Tian, D. Lu, Y. Yu, S. Cestellos-Blanco, K. K. Sakimoto and P. Yang, Nat. Nanotechnol., 2018, 13, 900–905 CrossRef CAS PubMed.
- B. Wang, C. Zeng, K. H. Chu, D. Wu, H. Y. Yip, L. Ye and P. K. Wong, Adv. Energy Mater., 2017, 7, 1700611 CrossRef.
- W. Wei, P. Sun, Z. Li, K. Song, W. Su, B. Wang, Y. Liu and J. Zhao, Sci. Adv., 2018, 4, eaap9253 CrossRef PubMed.
- S. Cestellos-Blanco, H. Zhang, J. M. Kim, Y.-X. Shen and P. Yang, Nat. Catal., 2020, 3, 245–255 CrossRef CAS.
- Z. Du, H. Li and T. Gu, Biotechnol. Adv., 2007, 25, 464–482 CrossRef CAS PubMed.
- F. Aulenta, E. Hao Yu, L. T. Angenent and M. Villano, Fuel Cells, 2017, 17, 582–583 CrossRef CAS.
- S. Srikanth, M. Kumar and S. K. Puri, Bioresour. Technol., 2018, 265, 506–518 CrossRef CAS PubMed.
- R. Kannaiah Goud, O. Sarkar and S. Venkata Mohan, Int. J. Hydrogen Energy, 2014, 39, 7572–7586 CrossRef CAS.
- A. S. Beliaev, D. A. Saffarini, J. L. McLaughlin and D. Hunnicutt, Mol. Microbiol., 2001, 39, 722–730 CrossRef CAS PubMed.
- Y. Liu, Z. Wang, J. Liu, C. Levar, M. J. Edwards, J. T. Babauta, D. W. Kennedy, Z. Shi, H. Beyenal, D. R. Bond, T. A. Clarke, J. N. Butt, D. J. Richardson, K. M. Rosso, J. M. Zachara, J. K. Fredrickson and L. Shi, Environ. Microbiol. Rep., 2014, 6, 776–785 CrossRef CAS PubMed.
- A. Jain, X. Zhang, G. Pastorella, J. O. Connolly, N. Barry, R. Woolley, S. Krishnamurthy and E. Marsili, Bioelectrochemistry, 2012, 87, 28–32 CrossRef CAS PubMed.
- N. S. Malvankar and D. R. Lovley, ChemSusChem, 2012, 5, 1039–1046 CrossRef CAS PubMed.
- S. Pirbadian, S. E. Barchinger, K. M. Leung, H. S. Byun, Y. Jangir, R. A. Bouhenni, S. B. Reed, M. F. Romine, D. A. Saffarini, L. Shi, Y. A. Gorby, J. H. Golbeck and M. Y. El-Naggar, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 12883–12888 CrossRef CAS PubMed.
- R. M. Snider, S. M. Strycharz-Glaven, S. D. Tsoi, J. S. Erickson and L. M. Tender, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 15467–15472 CrossRef CAS PubMed.
- E. D. Brutinel and J. A. Gralnick, Appl. Microbiol. Biotechnol., 2012, 93, 41–48 CrossRef PubMed.
- D. K. Newman and R. Kolter, Nature, 2000, 405, 94–97 CrossRef CAS PubMed.
- A. Okamoto, S. Kalathil, X. Deng, K. Hashimoto, R. Nakamura and K. H. Nealson, Sci. Rep., 2014, 4, 5628 CrossRef CAS PubMed.
- C. M. Paquete, B. M. Fonseca, D. R. Cruz, T. M. Pereira, I. Pacheco, C. M. Soares and R. O. Louro, Front. Microbiol., 2014, 5, 318 Search PubMed.
- J. M. Dantas, L. Morgado, T. Catarino, O. Kokhan, P. Raj Pokkuluri and C. A. Salgueiro, Biochim. Biophys. Acta, Bioenerg., 2014, 1837, 750–760 CrossRef CAS PubMed.
- L. Shi, H. Dong, G. Reguera, H. Beyenal, A. Lu, J. Liu, H. Q. Yu and J. K. Fredrickson, Nat. Rev. Microbiol., 2016, 14, 651–662 CrossRef CAS PubMed.
- A. S. Beliaev and D. A. Saffarini, J. Bacteriol., 1998, 180, 6292–6297 CrossRef CAS PubMed.
- C. R. Myers and J. M. Myers, Appl. Environ. Microbiol., 2002, 68, 5585–5594 CrossRef CAS PubMed.
- D. Coursolle and J. A. Gralnick, Mol. Microbiol., 2010, 77, 995–1008 CAS.
- L. Shi, K. M. Rosso, T. A. Clarke, D. J. Richardson, J. M. Zachara and J. K. Fredrickson, Front. Microbiol., 2012, 3, 50 Search PubMed.
- D. G. G. McMillan, S. J. Marritt, J. N. Butt and L. J. C. Jeuken, J. Biol. Chem., 2012, 287, 14215–14225 CrossRef CAS PubMed.
- S. J. Marritt, T. G. Lowe, J. Bye, D. G. G. McMillan, L. Shi, J. Fredrickson, J. Zachara, D. J. Richardson, M. R. Cheesman, L. J. C. Jeuken and J. N. Butt, Biochem. J., 2012, 444, 465–474 CrossRef CAS PubMed.
- D. G. G. McMillan, S. J. Marritt, M. A. Firer-Sherwood, L. Shi, D. J. Richardson, S. D. Evans, S. J. Elliott, J. N. Butt and L. J. C. Jeuken, J. Am. Chem. Soc., 2013, 135, 10550–10556 CrossRef CAS PubMed.
- M. A. Firer-Sherwood, K. D. Bewley, J.-Y. Mock and S. J. Elliott, Metallomics, 2011, 3, 344–348 CrossRef CAS PubMed.
- G. Sturm, K. Richter, A. Doetsch, H. Heide, R. O. Louro and J. Gescher, ISME J., 2015, 9, 1802–1811 CrossRef PubMed.
- D. Leys, T. E. Meyer, A. S. Tsapin, K. H. Nealson, M. A. Cusanovich and J. J. Van Beeumen, J. Biol. Chem., 2002, 277, 35703–35711 CrossRef CAS PubMed.
- D. Leys, A. S. Tsapin, K. H. Nealson, T. E. Meyer, M. A. Cusanovich and J. J. V. Beeumen, Nat. Struct. Biol., 1999, 6, 1113–1117 CrossRef CAS PubMed.
- D. J. Richardson, J. N. Butt, J. K. Fredrickson, J. M. Zachara, L. Shi, M. J. Edwards, G. White, N. Baiden, A. J. Gates, S. J. Marritt and T. A. Clarke, Mol. Microbiol., 2012, 85, 201–212 CrossRef CAS PubMed.
- D. E. Ross, S. S. Ruebush, S. L. Brantley, R. S. Hartshorne, T. A. Clarke, D. J. Richardson and M. Tien, Appl. Environ. Microbiol., 2007, 73, 5797–5808 CrossRef CAS PubMed.
- R. S. Hartshorne, C. L. Reardon, D. Ross, J. Nuester, T. A. Clarke, A. J. Gates, P. C. Mills, J. K. Fredrickson, J. M. Zachara, L. Shi, A. S. Beliaev, M. J. Marshall, M. Tien, S. Brantley, J. N. Butt and D. J. Richardson, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 22169–22174 CrossRef CAS PubMed.
- G. F. White, Z. Shi, L. Shi, Z. Wang, A. C. Dohnalkova, M. J. Marshall, J. K. Fredrickson, J. M. Zachara, J. N. Butt, D. J. Richardson and T. A. Clarke, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 6346–6351 CrossRef CAS PubMed.
- L. Shi, B. Chen, Z. Wang, D. A. Elias, M. U. Mayer, Y. A. Gorby, S. Ni, B. H. Lower, D. W. Kennedy, D. S. Wunschel, H. M. Mottaz, M. J. Marshall, E. A. Hill, A. S. Beliaev, J. M. Zachara, J. K. Fredrickson and T. C. Squier, J. Bacteriol., 2006, 188, 4705–4714 CrossRef CAS PubMed.
- L. Shi, S. Deng, M. J. Marshall, Z. Wang, D. W. Kennedy, A. C. Dohnalkova, H. M. Mottaz, E. A. Hill, Y. A. Gorby, A. S. Beliaev, D. J. Richardson, J. M. Zachara and J. K. Fredrickson, J. Bacteriol., 2008, 190, 5512–5516 CrossRef CAS PubMed.
- B. H. Lower, L. Shi, R. Yongsunthon, T. C. Droubay, D. E. McCready and S. K. Lower, J. Bacteriol., 2007, 189, 4944–4952 CrossRef CAS PubMed.
- B. H. Lower, R. Yongsunthon, L. Shi, L. Wildling, H. J. Gruber, N. S. Wigginton, C. L. Reardon, G. E. Pinchuk, T. C. Droubay, J.-F. Boily and S. K. Lower, Appl. Environ. Microbiol., 2009, 75, 2931–2935 CrossRef CAS PubMed.
- Y. Xiong, L. Shi, B. Chen, M. U. Mayer, B. H. Lower, Y. Londer, S. Bose, M. F. Hochella, J. K. Fredrickson and T. C. Squier, J. Am. Chem. Soc., 2006, 128, 13978–13979 CrossRef CAS PubMed.
- H. Zhang, X. Tang, G. R. Munske, N. Zakharova, L. Yang, C. Zheng, M. A. Wolff, N. Tolic, G. A. Anderson, L. Shi, M. J. Marshall, J. K. Fredrickson and J. E. Bruce, J. Proteome Res., 2008, 7, 1712–1720 CrossRef CAS PubMed.
- L. A. Meitl, C. M. Eggleston, P. J. S. Colberg, N. Khare, C. L. Reardon and L. Shi, Geochim. Cosmochim. Acta, 2009, 73, 5292–5307 CrossRef CAS.
- S. Pirbadian and M. Y. El-Naggar, Phys. Chem. Chem. Phys., 2012, 14, 13802–13808 RSC.
- E. Marsili, D. B. Baron, I. D. Shikhare, D. Coursolle, J. A. Gralnick and D. R. Bond, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 3968–3973 CrossRef CAS PubMed.
- H. V. Canstein, J. Ogawa, S. Shimizu and J. R. Lloyd, Appl. Environ. Microbiol., 2008, 74, 615–623 CrossRef PubMed.
- D. Coursolle, D. B. Baron, D. R. Bond and J. A. Gralnick, J. Bacteriol., 2010, 192, 467–474 CrossRef CAS PubMed.
- C. E. Levar, C. H. Chan, M. G. Mehta-Kolte, D. R. Bond and D. K. Newman, mBio, 2014, 5, e02034–e02014 CrossRef CAS PubMed.
- L. Zacharoff, C. H. Chan and D. R. Bond, Bioelectrochemistry, 2016, 107, 7–13 CrossRef CAS PubMed.
- J. R. Lloyd, C. Leang, A. L. H. Myerson, M. V. Coppi, S. Cuifo, B. Methe, S. J. Sandler and D. R. Lovley, Biochem. J., 2003, 369, 153–161 CrossRef CAS PubMed.
- L. Morgado, M. Bruix, M. Pessanha, Y. Y. Londer and C. A. Salgueiro, Biophys. J., 2010, 99, 293–301 CrossRef CAS PubMed.
- C. Leang, M. V. Coppi and D. R. Lovley, J. Bacteriol., 2003, 185, 2096–2103 CrossRef CAS PubMed.
- X. Qian, G. Reguera, T. Mester and D. R. Lovley, FEMS Microbiol. Lett., 2007, 277, 21–27 CrossRef CAS PubMed.
- Y. Liu, J. K. Fredrickson, J. M. Zachara and L. Shi, Front. Microbiol., 2015, 6, 1075 Search PubMed.
- C. Leang, X. Qian, T. Mester and D. R. Lovley, Appl. Environ. Microbiol., 2010, 76, 4080–4084 CrossRef CAS PubMed.
- O. Choi, T. Kim, H. M. Woo and Y. Um, Sci. Rep., 2014, 4, 6961 CrossRef CAS PubMed.
- C. W. Marshall and H. D. May, Energy Environ. Sci., 2009, 2, 699–705 RSC.
- K. C. Wrighton, J. C. Thrash, R. A. Melnyk, J. P. Bigi, K. G. Byrne-Bailey, J. P. Remis, D. Schichnes, M. Auer, C. J. Chang and J. D. Coates, Appl. Environ. Microbiol., 2011, 77, 7633–7639 CrossRef CAS PubMed.
- K. G. Byrne-Bailey, K. C. Wrighton, R. A. Melnyk, P. Agbo, T. C. Hazen and J. D. Coates, J. Bacteriol., 2010, 192, 4078–4079 CrossRef CAS PubMed.
- H. K. Carlson, A. T. Iavarone, A. Gorur, B. S. Yeo, R. Tran, R. A. Melnyk, R. A. Mathies, M. Auer and J. D. Coates, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 1702–1707 CrossRef CAS PubMed.
- S. H. Light, L. Su, R. Rivera-Lugo, J. A. Cornejo, A. Louie, A. T. Iavarone, C. M. Ajo-Franklin and D. A. Portnoy, Nature, 2018, 562, 140–144 CrossRef CAS PubMed.
- D. R. Lovley and D. E. Holmes, Nat. Rev. Microbiol., 2022, 20, 5–19 CrossRef CAS PubMed.
- D. E. Ross, J. M. Flynn, D. B. Baron, J. A. Gralnick and D. R. Bond, PLoS One, 2011, 6, e16649 CrossRef CAS PubMed.
- O. Choi and B. I. Sang, Biotechnol. Biofuels, 2016, 9, 11 CrossRef PubMed.
- A. Bose, E. J. Gardel, C. Vidoudez, E. A. Parra and P. R. Girguis, Nat. Commun., 2014, 5, 3391 CrossRef CAS PubMed.
- D. Gupta, M. C. Sutherland, K. Rengasamy, J. M. Meacham, R. G. Kranz and A. Bose, mBio, 2019, 10, e02668–e02619 CrossRef CAS PubMed.
- M. S. Guzman, K. Rengasamy, M. M. Binkley, C. Jones, T. O. Ranaivoarisoa, R. Singh, D. A. Fike, J. M. Meacham and A. Bose, Nat. Commun., 2019, 10, 1355 CrossRef PubMed.
- J. Liu, Z. Wang, S. M. Belchik, M. J. Edwards, C. Liu, D. W. Kennedy, E. D. Merkley, M. S. Lipton, J. N. Butt, D. J. Richardson, J. M. Zachara, J. K. Fredrickson, K. M. Rosso and L. Shi, Front. Microbiol., 2012, 3, 37 Search PubMed.
- C. R. Beckwith, M. J. Edwards, M. Lawes, L. Shi, J. N. Butt, D. J. Richardson and T. A. Clarke, Front. Microbiol., 2015, 6, 332 CrossRef PubMed.
- A. L. McCully and A. M. Spormann, Environ. Microbiol., 2020, 22, 4794–4807 CrossRef CAS PubMed.
- A. A. Karbelkar, A. R. Rowe and M. Y. El-Naggar, Electrochim. Acta, 2019, 324, 134838 CrossRef CAS.
- S. M. Strycharz, R. H. Glaven, M. V. Coppi, S. M. Gannon, L. A. Perpetua, A. Liu, K. P. Nevin and D. R. Lovley, Bioelectrochemistry, 2011, 80, 142–150 CrossRef CAS PubMed.
- O. Choi and B.-I. Sang, Biotechnol. Biofuels, 2016, 9, 11 CrossRef PubMed.
- V. Agostino and M. A. Rosenbaum, Front. Energy Res., 2018, 6, 55 CrossRef.
- H. Li, P. H. Opgenorth, D. G. Wernick, S. Rogers, T. Y. Wu, W. Higashide, P. Malati, Y. X. Huo, K. M. Cho and J. C. Liao, Science, 2012, 335, 1596 CrossRef CAS PubMed.
- K. J. J. Steinbusch, H. V. M. Hamelers, J. D. Schaap, C. Kampman and C. J. N. Buisman, Environ. Sci. Technol., 2010, 44, 513–517 CrossRef CAS PubMed.
- H. Liu, S. Matsuda, K. Hashimoto and S. Nakanishi, ChemSusChem, 2012, 5, 1054–1058 CrossRef CAS PubMed.
- D. H. Kim and M. S. Kim, Bioresour. Technol., 2011, 102, 8423–8431 CrossRef CAS PubMed.
- F. Aulenta, L. Catapano, L. Snip, M. Villano and M. Majone, ChemSusChem, 2012, 5, 1080–1085 CrossRef CAS PubMed.
- E. Lojou, M. C. Durand, A. Dolla and P. Bianco, Electroanalysis, 2002, 14, 913–922 CrossRef CAS.
- M. Rosenbaum, F. Aulenta, M. Villano and L. T. Angenent, Bioresour. Technol., 2011, 102, 324–333 CrossRef CAS PubMed.
- J. F. Heidelberg, R. Seshadri, S. A. Haveman, C. L. Hemme, I. T. Paulsen, J. F. Kolonay, J. A. Eisen, N. Ward, B. Methe, L. M. Brinkac, S. C. Daugherty, R. T. Deboy, R. J. Dodson, A. S. Durkin, R. Madupu, W. C. Nelson, S. A. Sullivan, D. Fouts, D. H. Haft, J. Selengut, J. D. Peterson, T. M. Davidsen, N. Zafar, L. Zhou, D. Radune, G. Dimitrov, M. Hance, K. Tran, H. Khouri, J. Gill, T. R. Utterback, T. V. Feldblyum, J. D. Wall, G. Voordouw and C. M. Fraser, Nat. Biotechnol., 2004, 22, 554–559 CrossRef CAS PubMed.
- M. V. Coppi, Microbiology, 2005, 151, 1239–1254 CrossRef CAS PubMed.
- J. S. Geelhoed and A. J. Stams, Environ. Sci. Technol., 2011, 45, 815–820 CrossRef CAS PubMed.
- H. W. Kreuzer, E. A. Hill, J. J. Moran, R. A. Bartholomew, H. Yang and E. L. Hegg, FEMS Microbiol. Lett., 2014, 352, 18–24 CrossRef CAS PubMed.
- Y. Fu, Y. Zhang, B. Li, D. Liang, S. Lu, Y. Xiang, B. Xie, H. Liu and K. H. Nealson, Electrochim. Acta, 2019, 305, 528–533 CrossRef CAS.
- S. A. Cheng, D. F. Xing, D. F. Call and B. E. Logan, Environ. Sci. Technol., 2009, 43, 3953–3958 CrossRef CAS PubMed.
- F. Mayer, F. Enzmann, A. M. Lopez and D. Holtmann, Bioresour. Technol., 2019, 289, 121706 CrossRef CAS PubMed.
- W. Liu, L. Wang, L. Gao and A.-J. Wang, in Microbial Electrochemical Technology, 2019, pp. 921–953 Search PubMed.
- M. Villano, F. Aulenta, C. Ciucci, T. Ferri, A. Giuliano and M. Majone, Bioresour. Technol., 2010, 101, 3085–3090 CrossRef CAS PubMed.
- J. Madjarov, R. Soares, C. M. Paquete and R. O. Louro, Front. Microbiol., 2022, 13, 913311 CrossRef PubMed.
- P. L. Tremblay, D. Hoglund, A. Koza, I. Bonde and T. Zhang, Sci. Rep., 2015, 5, 16168 CrossRef CAS PubMed.
- S. Y. Lim, W. Shen and Z. Gao, Chem. Soc. Rev., 2015, 44, 362–381 RSC.
- H. Wu, H. Tan, L. Chen, B. Yang, Y. Hou, L. Lei and Z. Li, Chin. Chem. Lett., 2021, 32, 2499–2502 CrossRef CAS.
- S. Li, C. Cheng and A. Thomas, Adv. Mater., 2017, 29, 1602547 CrossRef PubMed.
- S. Wen, B. Liu, W. Li, T. Liang, X. Li, D. Yi, B. Luo, L. Zhi, D. Liu and B. Wang, Adv. Funct. Mater., 2022, 32, 2203960 CrossRef CAS.
- K. Zhang, Z. Ma, H. Song, M. Zhang, H. Xu and N. Zhao, Int. J. Hydrogen Energy, 2020, 45, 12121–12129 CrossRef CAS.
- Y.-F. Guan, F. Zhang, B.-C. Huang and H.-Q. Yu, J. Cleaner Prod., 2019, 229, 412–419 CrossRef CAS.
- A. A. Yazdi, L. D'Angelo, N. Omer, G. Windiasti, X. Lu and J. Xu, Biosens. Bioelectron., 2016, 85, 536–552 CrossRef PubMed.
- S. Ci, Z. Wen, J. Chen and Z. He, Electrochem. Commun., 2012, 14, 71–74 CrossRef CAS.
- C. Soldano, A. Mahmood and E. Dujardin, Carbon, 2010, 48, 2127–2150 CrossRef CAS.
- H. Li, B. Liu, X. Yang, Y. Gao, X. Luo, X. Guan, Z. Zhang, Z. Yu and B. Wang, Prog. Nat. Sci.: Mater. Int., 2022, 32(6), 700–704 CrossRef CAS.
- L. Xiao, J. Damien, J. Luo, H. D. Jang, J. Huang and Z. He, J. Power Sources, 2012, 208, 187–192 CrossRef CAS.
- F. A. A. Alatraktchi, Y. Zhang and I. Angelidaki, Appl. Energy, 2014, 116, 216–222 CrossRef CAS.
- P. Zhang, X. Zhou, R. Qi, P. Gai, L. Liu, F. Lv and S. Wang, Adv. Electron. Mater., 2019, 5, 1900320 CrossRef.
- Q. Li, Q. Fu, H. Kobayashi, Y. He, Z. Li, J. Li, Q. Liao and X. Zhu, Sustainable Energy Fuels, 2020, 4, 2987–2997 RSC.
- L. Zou, Y. Qiao, X.-S. Wu and C. M. Li, J. Power Sources, 2016, 328, 143–150 CrossRef CAS.
- X. Fang, S. Kalathil, G. Divitini, Q. Wang and E. Reisner, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 5074–5080 CrossRef CAS PubMed.
- X. Xie, L. Hu, M. Pasta, G. F. Wells, D. Kong, C. S. Criddle and Y. Cui, Nano Lett., 2011, 11, 291–296 CrossRef CAS PubMed.
- H. Wang, G. Wang, Y. Ling, F. Qian, Y. Song, X. Lu, S. Chen, Y. Tong and Y. Li, Nanoscale, 2013, 5, 10283–10290 RSC.
- X. Wu, Y. Qiao, Z. Shi, W. Tang and C. M. Li, ACS Appl. Mater. Interfaces, 2018, 10, 11671–11677 CrossRef CAS PubMed.
- L. Jourdin, S. Freguia, B. C. Donose, J. Chen, G. G. Wallace, J. Keller and V. Flexer, J. Mater. Chem. A, 2014, 2, 13093–13102 RSC.
- Y. Y. Yu, D. D. Zhai, R. W. Si, J. Z. Sun, X. Liu and Y. C. Yong, Int. J. Mol. Sci., 2017, 18, 90 CrossRef PubMed.
- X. Zhang, H. Liu, J. Wang, G. Ren, B. Xie, H. Liu, Y. Zhu and L. Jiang, Nanoscale, 2015, 7, 18763–18769 RSC.
- R. Bian, Y. Jiang, Y. Wang, J. K. Sun, J. Hu, L. Jiang and H. Liu, Adv. Funct. Mater., 2018, 28, 1707408 CrossRef.
- R. Wang, H. Li, J. Sun, L. Zhang, J. Jiao, Q. Wang and S. Liu, Adv. Mater., 2021, 33, 2004051 CrossRef CAS PubMed.
- M. N. Norizan, M. H. Moklis, S. Z. Ngah Demon, N. A. Halim, A. Samsuri, I. S. Mohamad, V. F. Knight and N. Abdullah, RSC Adv., 2020, 10, 43704–43732 RSC.
- C. Erbay, X. Pu, W. Choi, M.-J. Choi, Y. Ryu, H. Hou, F. Lin, P. de Figueiredo, C. Yu and A. Han, J. Power Sources, 2015, 280, 347–354 CrossRef CAS.
- M. T. Amen, N. A. Barakat, M. A. H. M. Jamal, S.-T. Hong, I. M. Mohamed and A. Salama, Appl. Energy, 2018, 228, 167–175 CrossRef CAS.
- W. Wang, S. You, X. Gong, D. Qi, B. K. Chandran, L. Bi, F. Cui and X. Chen, Adv. Mater., 2016, 28, 270–275 CrossRef CAS PubMed.
- J. R. Kim, B. Min and B. E. Logan, Appl. Microbiol. Biotechnol., 2005, 68, 23–30 CrossRef CAS PubMed.
- S. Zhou, J. Tang and Y. Yuan, Bioelectrochemistry, 2015, 102, 29–34 CrossRef CAS PubMed.
- R. Nakamura, F. Kai, A. Okamoto, G. J. Newton and K. Hashimoto, Angew. Chem., Int. Ed., 2009, 48, 508–511 CrossRef CAS PubMed.
- J. Ji, Y. Jia, W. Wu, L. Bai, L. Ge and Z. Gu, Colloids Surf., A, 2011, 390, 56–61 CrossRef CAS.
- I. H. Park, M. Christy, P. Kim and K. S. Nahm, Biosens. Bioelectron., 2014, 58, 75–80 CrossRef CAS PubMed.
- B. Yu, Y. Li and L. Feng, J. Hazard. Mater., 2019, 377, 70–77 CrossRef CAS PubMed.
- K. Guo, S. Freguia, P. G. Dennis, X. Chen, B. C. Donose, J. Keller, J. J. Gooding and K. Rabaey, Environ. Sci. Technol., 2013, 47, 7563–7570 CrossRef CAS PubMed.
- H. M. Fruhauf, D. Holtmann and M. Stockl, Bioelectrochemistry, 2022, 147, 108213 CrossRef PubMed.
- E. Griškonis, A. Ilginis, I. Jonuškienė, L. Raslavičius, R. Jonynas and K. Kantminienė, Processes, 2020, 8, 939 CrossRef.
- C. M. Ding, M. L. Lv, Y. Zhu, L. Jiang and H. Liu, Angew. Chem., Int. Ed., 2015, 54, 1446–1451 CrossRef CAS.
- E. T. Johnson, D. B. Baron, B. Naranjo, D. R. Bond, C. Schmidt-Dannert and J. A. Gralnick, Appl. Environ. Microbiol., 2010, 76, 4123–4129 CrossRef CAS.
- S. J. Berrıos-Rivera, G. N. Bennett and K.-Y. San, Metab. Eng., 2002, 4, 217–229 CrossRef PubMed.
- S. J. Berríos-Rivera, K.-Y. San and G. Bennett, Metab. Eng., 2002, 4, 238–247 CrossRef PubMed.
- S. J. Berrıos-Rivera, G. N. Bennett and K.-Y. San, Metab. Eng., 2002, 4, 230–237 CrossRef PubMed.
- X. Chen, S. Li and L. Liu, Trends Biotechnol., 2014, 32, 337–343 CrossRef CAS PubMed.
- F. Li, Y.-X. Li, Y.-X. Cao, L. Wang, C.-G. Liu, L. Shi and H. Song, Nat. Commun., 2018, 9, 1–13 CrossRef PubMed.
- S. Han, X.-Y. Gao, H.-J. Ying and C. C. Zhou, Green Chem., 2016, 18, 2473–2478 RSC.
- A. Vellingiri, Y. E. Song, G. Munussami, C. Kim, C. Park, B.-H. Jeon, S.-G. Lee and J. R. Kim, J. Chem. Technol. Biotechnol., 2019, 94, 2115–2122 CrossRef CAS.
- D. Min, L. Cheng, F. Zhang, X. N. Huang, D. B. Li, D. F. Liu, T. C. Lau, Y. Mu and H. Q. Yu, Environ. Sci. Technol., 2017, 51, 5082–5089 CrossRef CAS PubMed.
- D. Kaur, D. Alkhder, C. Corre and F. Alberti, ACS Synth. Biol., 2020, 9, 2239–2245 CrossRef CAS PubMed.
- Y. Yang, Y. Ding, Y. Hu, B. Cao, S. A. Rice, S. Kjelleberg and H. Song, ACS Synth. Biol., 2015, 4, 815–823 CrossRef CAS PubMed.
- T. Lin, W. Ding, L. Sun, L. Wang, C.-G. Liu and H. Song, Nano Energy, 2018, 50, 639–648 CrossRef CAS.
- Y.-C. Yong, Y.-Y. Yu, C.-M. Li, J.-J. Zhong and H. Song, Biosens. Bioelectron., 2011, 30, 87–92 CrossRef CAS PubMed.
- T. Liu, Y. Y. Yu, X. P. Deng, C. K. Ng, B. Cao, J. Y. Wang, S. A. Rice, S. Kjelleberg and H. Song, Biotechnol. Bioeng., 2015, 112, 2051–2059 CrossRef CAS PubMed.
- A. Kouzuma, X.-Y. Meng, N. Kimura, K. Hashimoto and K. Watanabe, Appl. Environ. Microbiol., 2010, 76, 4151–4157 CrossRef CAS PubMed.
- H. Luo, J. Qi, M. Zhou, G. Liu, Y. Lu, R. Zhang and C. Zeng, Bioresour. Technol., 2020, 309, 123322 CrossRef CAS PubMed.
- Y. Y. Yu, Y. Z. Wang, Z. Fang, Y. T. Shi, Q. W. Cheng, Y. X. Chen, W. Shi and Y. C. Yong, Nat. Commun., 2020, 11, 4087 CrossRef CAS PubMed.
- D. Guo, H.-F. Wei, R.-B. Song, J. Fu, X. Lu, R. Jelinek, Q. Min, J.-R. Zhang, Q. Zhang and J.-J. Zhu, Nano Energy, 2019, 63, 103875 CrossRef CAS.
- M. Chen, X. Zhou, X. Liu, R. J. Zeng, F. Zhang, J. Ye and S. Zhou, Biosens. Bioelectron., 2018, 108, 20–26 CrossRef CAS PubMed.
- S. Liu, X. Yi, X. Wu, Q. Li and Y. Wang, Small, 2020, 16, e2004194 CrossRef PubMed.
- C. Yang, H. Aslan, P. Zhang, S. Zhu, Y. Xiao, L. Chen, N. Khan, T. Boesen, Y. Wang, Y. Liu, L. Wang, Y. Sun, Y. Feng, F. Besenbacher, F. Zhao and M. Yu, Nat. Commun., 2020, 11, 1379 CrossRef CAS PubMed.
- X. Wu, F. Zhao, N. Rahunen, J. R. Varcoe, C. Avignone-Rossa, A. E. Thumser and R. C. Slade, Angew. Chem., Int. Ed., 2011, 50, 427–430 CrossRef CAS PubMed.
- P. Li, Y. Jiang, R. B. Song, J. R. Zhang and J. J. Zhu, J. Mater. Chem. B, 2021, 9, 1638–1646 RSC.
- L. Liu and S. Choi, J. Power Sources, 2021, 506, 230251 CrossRef CAS.
- B. Cao, Z. Zhao, L. Peng, H. Y. Shiu, M. Ding, F. Song, X. Guan, C. K. Lee, J. Huang, D. Zhu, X. Fu, G. C. L. Wong, C. Liu, K. Nealson, P. S. Weiss, X. Duan and Y. Huang, Science, 2021, 373, 1336–1340 CrossRef CAS PubMed.
- S. M. Glaven, Microb. Biotechnol., 2019, 12, 819–823 CrossRef PubMed.
Footnote |
| † These authors made major equal contributions to this review. |
| This journal is © The Royal Society of Chemistry 2023 |

