Open Access Article
Open Access ArticleIsolation methods for particle protein corona complexes from protein-rich matrices
Linda
Böhmert
,
Linn
Voß
,
Valerie
Stock
,
Albert
Braeuning
,
Alfonso
Lampen
and
Holger
Sieg
 *
*
German Federal Institute for Risk Assessment, Dept. Food Safety, Max-Dohrn-Str. 8-10, 10589 Berlin, Germany. E-mail: holger.sieg@bfr.bund.de; Tel: +49 (30) 18412-25800
First published on 9th January 2020
Abstract
Background: Nanoparticles become rapidly encased by a protein layer when they are in contact with biological fluids. This protein shell is called a corona. The composition of the corona has a strong influence on the surface properties of the nanoparticles. It can affect their cellular interactions, uptake and signaling properties. For this reason, protein coronae are investigated frequently as an important part of particle characterization. Main body of the abstract: The protein corona can be analyzed by different methods, which have their individual advantages and challenges. The separation techniques to isolate corona-bound particles from the surrounding matrices include centrifugation, magnetism and chromatographic methods. Different organic matrices, such as blood, blood serum, plasma or different complex protein mixtures, are used and the approaches vary in parameters such as time, concentration and temperature. Depending on the investigated particle type, the choice of separation method can be crucial for the subsequent results. In addition, it is important to include suitable controls to avoid misinterpretation and false-positive or false-negative results, thus allowing the achievement of a valuable protein corona analysis result. Conclusion: Protein corona studies are an important part of particle characterization in biological matrices. This review gives a comparative overview about separation techniques, experimental parameters and challenges which occur during the investigation of the protein coronae of different particle types.
Introduction
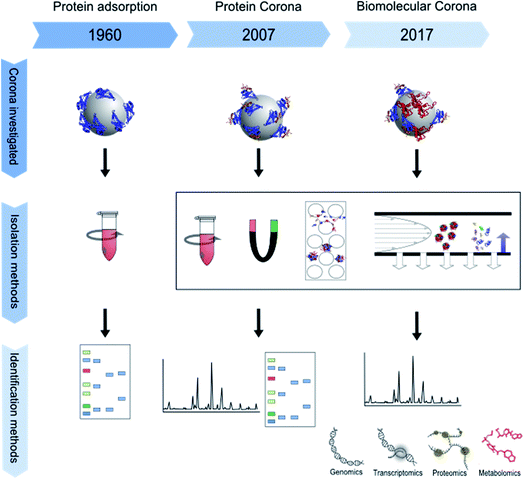
The impact of micro- and nano-particles on the human body has been the focus of research for almost two decades. In particular, nanoparticles, which are mostly defined by a size between 1 and 100 nanometers in at least one dimension, are of interest to the scientific community.1,2 Their higher surface-to-volume ratio, as compared to bigger particles, changes their physicochemical properties. Nanoparticles therefore may show higher biological reactivity.3 Furthermore, their small size may allow them to escape from the clearance mechanisms of the body, and thus they circulate longer in the blood and pass through cell membranes more easily.4 In combination with their greater reactivity, this increases their biological impact and implies a potential risk for undesired effects such as oxidative stress or cellular import of chemical substances adsorbed to the particle surface.5 Furthermore, the shape of particles is of special interest since it influences their bioavailability. For example, cylindrical nanoparticles have been shown to interact more strongly with cells than spherical ones.6 Surface modifications also affect nanoparticle reactivity by changing their zeta potential and influencing their binding kinetics and thus their biological interactions.5,7 Nowadays, most scientists agree that it is insufficient to interpret results from toxicological or biochemical studies without proper characterization of the respective nanoparticles and analysis of their interactions with the test environment.8 Particle composition, shape, size and size distribution, and ion release, as well as stability of particle dispersions are of major relevance, as are their surface chemistry and coating which are directly involved in building an interface between the particle and its biological environment.5,7 This biological interface involves physicochemical, thermodynamic and kinetic interactions between the nanomaterial surface and the surfaces of biological components such as proteins, phospholipids, or DNA contained in biological fluids or present in living cells. The interface represents the contact point between a particle and the biological system and is called the “nano-bio interface” or “protein corona”, even though the particle corona may also contain components other than proteins.9 The composition of the protein corona depends on the surrounding matrix, for example blood serum, plasma or the extra- and intra-cellular protein matrix and can change over time and after uptake or transportation into another cellular compartment.3 The corona can have a major influence on different processes, for example the attachment of particles to cell surfaces, the triggering of cellular signals, the modulation of particle uptake, distribution and excretion, the release of ions or other substances from the particles, or even immunological reactions.10–12 For example, some proteins make the nanoparticle surface more hydrophilic.13 The term “nanoparticle protein corona” was introduced by Cedervall et al. in 2007 to describe the formation of protein layers on a nanoparticle's surface.14 The concept of a biological interface for particles, however, had already been studied earlier using the terminology “interface”, or being described as protein adsorption to particles and its role in overall biological interactions.15–19 For a more detailed description of the history of protein corona research please see below. Over time, experimental set-ups and especially read-outs became more and more sophisticated, ranging from simple gel electrophoresis in the 1960s to “omics” approaches in recent years as schematically shown in Fig. 1.The number of publications on nanoparticle corona analysis has massively increased during the last decade (Fig. 2). Moreover, already about 100 reviews have been published that deal with the nanoparticle protein corona and the relationships between nanoparticles, their attached proteins and different downstream effects. Some discuss the evolution, composition and kinetics of the protein corona,20–23 some deal with implications for cellular uptake24–26 and some are focused on the relationship between the nanoparticle corona and the immune system.27,28 The authors of the aforementioned articles agree that information about the corona is essential for assessing and understanding the impact of particles on the human body, as through its (protein) composition the corona determines the biological identity of a particle which is recognized by the cell. All in all, it became clear that the corona composition of different particles is not only dependent on the particle material, its surface chemistry, and the composition of the surrounding media, but is also subject to time-dependent changes. Therefore, corona analytics does not only include the determination of a complex protein composition, but should also consider the complex kinetics of corona formation.
It has to be kept in mind that choosing a method for isolation of particles with their corona proteins from a protein-rich matrix, such as a cell culture medium containing bovine serum as a supplement or blood samples, is a critical step in the workflow. Isolation methods for the protein coronae of nanoparticles are not standardized. From many publications in the field it remains unclear whether the most appropriate isolation method for the particular scenario investigated has been chosen based on thorough pre-testing and method optimization. The separation step has a major influence on the results and their interpretation, as it decides whether proteins can be identified as part of the corona or not. False positive or false negative results may be obtained due to association or dissociation of proteins from the corona resulting from physical or chemical forces during the isolation and purification process. As an additional aspect in corona analytics it should be noted that most model matrices, e.g. cell culture medium, reflect the in vivo situation only to a limited degree.14,29–32 Model media can be used to simulate the behavior of nanoparticles, e.g. after direct injection into the bloodstream or when focusing on the cellular environment. They do not reproduce the fact that nanoparticles in vivo may be transferred through several body compartments with different compositions of body fluids, for example through the oral cavity, stomach and intestinal lumen, and then into enterocytes, and further via the blood stream into other organs.33 All these aspects determine how the protein corona is composed, and therefore how it influences cellular uptake and downstream effects.13,34
In the context described above, this review aims to give an overview of the different methods that have been developed to isolate nanoparticles with their protein corona from different environments, in order to enable the subsequent analysis of the protein corona composition. The main focus will be on the isolation methods for corona analytics in the context of specific research aims. To this end, particle sizes, surface modifications and materials are considered in relation to the respective isolation method used, and the results are compared among these different characteristics. This work thus provides a broad overview on the aspects that need to be considered in nanoparticle protein corona analytics.
Terminology
Even though the interest in protein corona analytics has increased in the context of nanotoxicology and nanomedicine in recent years, the adsorption of proteins to surfaces is not a new discovery. Protein films on surfaces were already reported in the 1920s.35 Research on protein adsorption and its role in biological interactions with surfaces and materials began in the 1950s by Bangham and Vroman.16,18 Subsequent studies on protein adsorption were focused on prolonging the blood circulation half-life of particles by reducing protein adsorption, in order to prevent their opsonization and recognition by cells.15 Compared to the history of research on protein adsorption, the term protein corona has been introduced rather recently. It was proposed in 2007 to describe spontaneous self-assembly and layering of proteins on nanoparticle surfaces.14 Other researchers describe it as the biomolecular coating of a particle that is actually in contact with a biological system (e.g. a cell or an organ), which interacts with a nanoparticle dispersed in a biological medium.29 The interest in investigating the protein corona increased since it was proposed that the protein corona determines the interaction with the surrounding biological matter.36,37 Aggregation and formation of a protein corona in the extracellular environment will alter nanoparticle size, shape, and surface properties, providing the particle with a “biological identity” that is distinct from its initial “synthetic identity”.38 The term corona is an appropriate description for proteins adsorbed to a particle surface as it comes from the Latin word for circle or wreath. However, “corona” is also used in other contexts in the nano field, like in “core-corona nanoparticles” as an alternative term to describe core–shell nanoparticles that are composed of different materials in concentric layers.39–41 In addition, the term corona is used for stabilized polymers in some publications.42 It also has to be kept in mind that “nanoparticle” is a rather new word for a nanoscale structure. Other terms used for particles in the same or comparable size range are, for example, colloid, submicron particle, dust, fine particulate matter, and others. While reviewing the literature, the above terminology has to be taken into account to prevent exclusion of relevant literature.The corona has become an important parameter within the characterization of nanoparticles and their interaction with biological systems. When a particle surface comes into contact with a protein-rich environment for the first time, it gets rapidly covered with proteins. At this moment, the protein corona composition is mainly driven by the abundance of the proteins in the surrounding medium. Over time, however, the specific affinity of individual proteins to the particle surface gains importance. Vroman and coworkers have postulated in the 1980s that during the adsorption of blood proteins to surfaces, small proteins cover the surface faster, due to their higher mobility, whereas bigger proteins with higher affinity are adsorbed later but are exchanged more slowly, and therefore get enriched over time.43,44 This was later termed the Vroman effect. The adsorption of proteins to surfaces is in a dynamic state and therefore proteins with long residence times and slow exchange rates become enriched on the particle surface and gradually replace more abundant proteins with lower affinities.
The protein corona is loosely subdivided by some researchers into the so-called “hard corona” and the “soft corona”. Sometimes, the soft and hard coronae are defined by their spatial arrangement, with the inner coating of particles (i.e. the proteins in direct contact with the particle surface) called the hard corona and the outer layer called the soft corona.45,46 In contrast, hard and soft coronae may also be defined based on protein affinities, with the hard corona consisting of proteins with a high affinity to the particle and having evolved over a longer period, while the soft corona is accordingly described as the (outer) layer of the protein corona with lower protein affinity to the particle surface, but high abundance in the biological fluid and a high rate of ad- and de-sorption.5,23,29,47–49
Another nano-specific term appeared in the literature about 10 years ago: the term “nanoparticle/biological interface” or for short the “nano-bio interface” was used by Nel et al. in their review discussing the interactions of nanoparticles with not only proteins but also other biological structures like membranes, cells, DNA or organelles.9 An issue of JACS Select was entitled ‘Chemistry at the Nano-Bio Interface’ and presented many studies about modification and manipulation of the interaction of nanoparticles with biological systems for medical and bio-sensing purposes.50 However, the composition of the protein corona was not the focus of most of these studies. Approaching the issue from the reverse point of view, the term cell “vision” has been established.51–53 It describes a complementary concept to the protein corona and emphasizes the role of the cell when it comes to a cellular reaction to nanoparticles. During the first contact of nanoparticles with cells, the structure of the cell membrane with its phospholipid composition, surface proteins and glycocalyx defines how the cell comes into contact with a nanoparticle and its protein corona. Depending on the cell type, the “vision” of cells is different and so is the cellular reaction to a specific nanoparticle.53,54
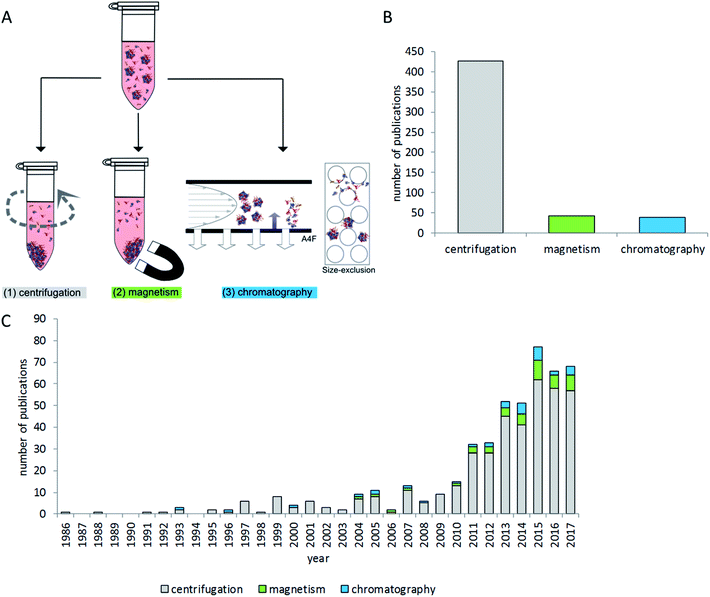
Due to the diversity of terminology, literature mining aiming to provide a broad overview of the critical steps of separating particles with their protein corona from the surrounding protein-rich matrix is a difficult task. Therefore the NCBI PubMed database was searched using multiple search terms (“nanoparticle corona”, “particle corona”, “protein corona”, “nano corona” and “particle protein adsorption”). The resulting 4427 hits were further screened manually for contents related to centrifugation and chromatographic and magnetic separation methods related to the separation of particles with proteins on their surface from protein-rich matrices. Studies dealing with fibers and larger surfaces were excluded. Studies which were only investigating binding affinities of single proteins to particles or beads were excluded as well. The remaining 412 studies were then evaluated to give an overview on the state of the art of protein corona isolation methods, as presented in the following sections.
Application of separation methods in corona analytics
Research on nanoparticle-matrix interactions is very diverse and reflects the variety of nanoparticle applications. A brief overview of investigated particle types and experimental conditions is given in Fig. 2. Literature analysis revealed that the most widely used method of choice for nanoparticle–protein corona isolation is centrifugation. This technique is rather easy to implement with standard laboratory equipment, and it is also considered time- and cost-effective. In addition, magnetic separation is commonly used for particles with magnetic properties, for example iron oxides. Size exclusion chromatography is used rarely, mostly with polymers and silver nanoparticles.Most of the available studies focus on proteins which are generally assumed to play the most decisive roles in nanoparticle corona formation.5,23,49,55 A few studies also analyze other biomolecules interacting with nanoparticles, e.g. lipids or nucleic acids.3,56–58 Even though most studies have the analysis of particle–protein interactions in common, very different aspects related to protein adsorption and corona formation are investigated. For example, some studies focus on the relationship between particle properties like surface chemistry and protein corona formation14,23,59–65 and others analyze the protein corona as particle properties (like size or shape) needed to interpret cellular or biological effects.26,38,66–72 Further aspects include improving particle properties to change protein corona composition and biological response;73–75 targeting or characterizing of particles for usage as drug carriers or for other medical purposes;76–81 or the comparison of different particles.60,68,82 Some studies report on the topic of biological function by single-protein measurement or functionality analysis. Consequently, several studies investigated the binding properties of, for example, fibrinogen,83,84 lysozyme85,86 and others86–88 using colorimetric protein quantification or ELISA. Kondo et al. combined protein binding studies with enzymatic activity assays of catalase, trypsin and peroxidase.64 Jayaram et al. investigated cellular oxidative stress by the measurement of oxidized corona proteins.89 Li et al. focused on corona-dependent particle uptake and enzymatic activity in inflammatory processes.90 Taken together, drawing conclusions on biological effects in relation to protein corona composition remains challenging and a major focus of many research projects.91
Method development and validation for corona or protein adsorption analysis constitutes another important aspect.92–96 In addition, the investigation of conformational and structural changes of particle-adsorbed proteins;97–99 analyses of differences between model particles and the real-life environment;100,101 the search for in vitro methods to predict the in vivo haemocompatibility of nanoparticles;83 investigation of the relationships between the particle surface, protein adsorption and immune response;102 interaction analysis of proteins with particles for analytical purposes;103 surface functionalization of particles with proteins;104 kinetic analysis;105 enzymatic degradation of the protein corona;106 and the characterization of gold particles from immunocolloidal methods play a role.88
Particles used for corona studies
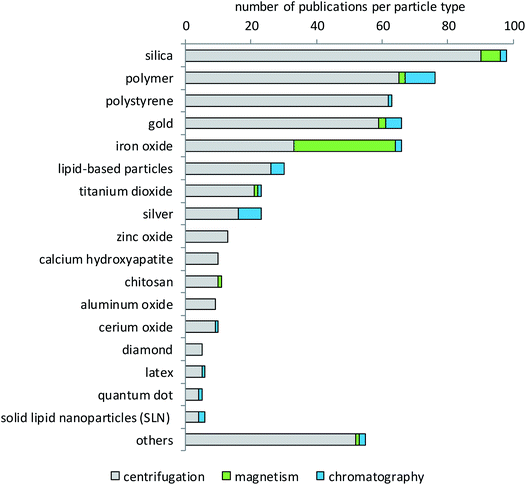
Literature evaluation was used to determine which type of particles has been studied more or less extensively for protein corona formation and composition. Fig. 3 shows that silica nanoparticles constitute the largest fraction of particles analyzed for their protein corona, which is not surprising since they are considered promising materials for diverse biomedical or industrial applications107–110 and are also frequently analyzed in toxicological studies.111 The big group of polymers contains very different materials. Most of them are of interest for research because they are being developed for a variety of diagnostic and therapeutic applications.14,60,112–119 These can be polystyrene (PS) with and without surface modifications, polyethylene glycol (PEG), polylactide-derivatives (PLGA), resins, crosslinked cellulose, carbon beads or polylactic acid (PLA). The same holds true for the separately listed group of chitosan particles.120,121 By contrast, the main reason for the high interest in polystyrene particles is their use as model particles that are available in many different sizes and narrow size distributions, labeled with different fluorophores and with different surface modifications.122–124 In addition, their potential use in pharmaceutical industries98,125,126 and also the growing concern about the environmental fate of plastic particles appear to underlie the interest in their protein corona.127,128 Gold nanoparticles are, in most cases, of interest due to their potential for drug delivery applications.129–133 In addition, gold is very applicable in analytical investigations due to its low reactivity, low solubility and high electron density. The same holds true for iron oxide nanoparticles that are used in high-sensitivity biomolecular magnetic resonance imaging (MRI) and tumor targeting.31,134–138 Additionally, iron oxide particles are also used for analytical purposes due to their magnetic properties.139,140 As shown in Fig. 3, mainly iron oxide nanoparticles are separated by magnetic methods. However, some particles from other materials have also been separated by magnetism-based techniques. This applies to cases where iron oxide nanoparticle cores are embedded in other materials to combine the magnetic properties of the iron oxide with the desired properties of the other materials. Such magnetic nanoparticles can be functionalized and used in different fields like biology, medicine, physics and materials science.104 Most particles that are subsumed in the group of lipid-based particles are of interest as drug targeting agents.141–145 These can be in the form of liposomes, lipoplexes or solid-lipid-nanoparticles, which show different properties in surface reactivity, protein binding and drug delivery. Titanium dioxide has a lot of applications, for example as a pigment or photocatalyst, in sunscreens, in food colorants, for electrical purposes or in the biomedical field, e.g. in dental and orthopedic reconstructive surgery.146–151 Titanium dioxide is also used as a comparator in studies dealing with airborne particulate matter.152 Zinc oxide is used in cosmetics, especially sunscreen products as a UV-protective agent, in the agricultural and food industries, and in packaging. Moreover, its use in food fortification is discussed, due to the essential role of zinc in many metabolic and enzymatic processes.153–155 Other particles used are, for example, aluminum,156 calcium compounds,157 copper oxide and rare elements such as cerium, bismuth, cobalt, gadolinium or tin oxide.82 Very recent studies investigated, for example, the corona formation on quantum dots,158 colloidal silica nanoparticles159 and BSA-stabilized gold nanoparticles.160Biological matrices used for corona studies
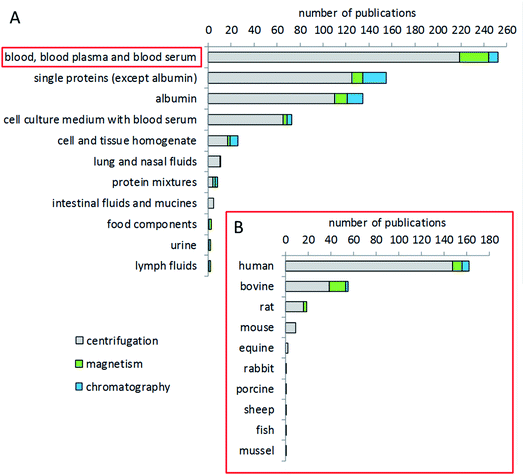
The most studied biological fluid in corona analytics is blood, either whole blood or, in the majority of cases, plasma or serum (Fig. 4). In particular, human blood has been of interest, congruent with the intended use of many particles for medical applications. In addition, animal blood from common test animal species such as rat or mouse is also frequently used for corona analysis in studies dealing with animal trials, or as a comparator to human blood.161,162 Bovine blood components, such as serum or plasma, are also of interest for corona studies, due to the fact that bovine serum is often used as a supplement in cell culture studies. Other, not blood-derived matrices include buffers with individual or mixed proteins, e.g. aimed at investigating the specific binding behavior of a certain protein.163,164 Few studies have been conducted dealing with fluids of the gastrointestinal tract, food components, urine, or the lymphatic system.165–167 Several studies provide a comparison of the coronae formed after incubation with different matrices. Some groups compared blood, blood plasma and serum as matrices.168,169 Other groups compared bovine and human serum.165,170 Jiang et al. compared bovine serum and a cell culture medium.171 Lundqvist et al. made a comparison between extracellular fluids (blood plasma) and the intracellular matrix (cytosolic fluid) to simulate corona changes after cellular uptake of nanoparticles.21 Some groups investigated the corona from blood components from humans and different animal species such as mice,172 rats, sheep and rabbits.173Choice of temperature and time for corona formation
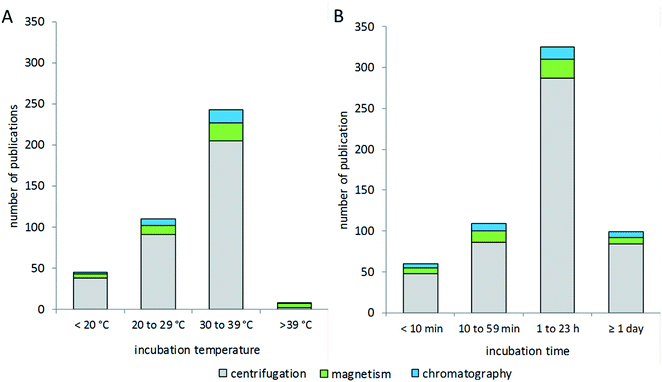
Research in the field of nanoparticle-bio interfaces focuses mainly on humans or other mammalian systems, and therefore the incubation temperature is often set to 37 °C,82,174 as shown in Fig. 5A. Nonetheless, incubation at room temperature has also been performed,140,148 and sometimes lower temperatures down to 4 °C have also been used.175–177 The latter ensure structural stability of proteins, while at the same time physiological relevance might be questioned. The incubation time for corona formation at the particle surface is often set to 60 minutes, assuming this to be an appropriate period to establish equilibrium.30,74,153,178,179 However, short-term and long-term studies have also been performed, with incubation times ranging from 1 minute to several days.110,180–183 Only a few studies investigated protein corona formation in vivo in mice,184,185 for which, in our view, the biggest challenge is the particle recovery and purification after in vivo administration.The process of corona formation does not only depend on biophysical parameters, but is also rather dependent on biological processes such as cellular uptake or intracellular distribution into different compartments. So, the extracellular corona differs strongly from the corona after uptake or, for example, vesicular delivery. This topic has been profoundly reviewed by Monopoli et al.3 Integrative approaches of bioinformatic simulations and corona formation experiments are performed to study protein corona kinetics.19 In particular, Tenzer and colleagues have demonstrated the early development of the corona, revealing that corona composition is established within the first few minutes of protein-particle contact, while the protein amount changes over time.183 Mortensen and co-workers monitored changes of the protein corona from 1 to 48 hours of incubation.110 In this context, corona formation is described as a very dynamic process based on the “Vroman effect”. In particular, very mobile, small and highly abundant proteins tend to form the corona in early kinetics, while bigger, less mobile and higher affinity proteins can replace them in later stages and form a thermodynamically stable protein corona. Mortensen et al. further stated that the presence and absence of specific proteins at a certain time point can influence interactions between nanoparticles and cells. This knowledge reinforces the importance of dynamic investigations on the kinetic process of corona formation. Nevertheless, based on our literature evaluation, the choice of isolation method appears to not be correlated with the incubation time or temperature used for corona formation.
Separation techniques – centrifugation
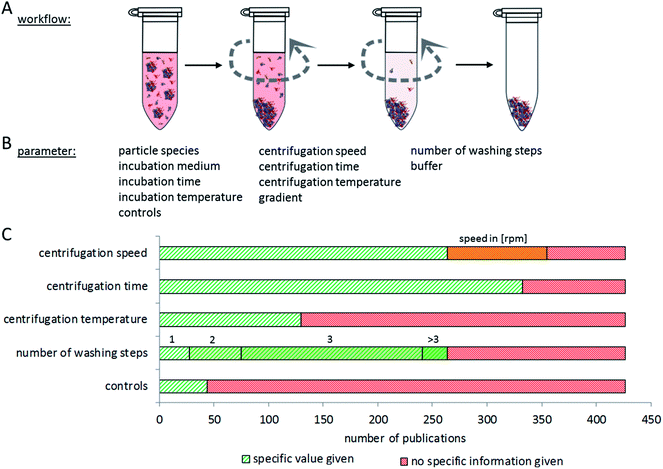
Proper interpretation and comparison of corona data from different particles and studies require the use of reproducible methods to isolate particles with their corona out of complex, protein-rich matrices. Particles should be isolated without losing the attached proteins, while at the same time false positives should also be omitted, making separation methodology a complex analytical challenge. Currently, a plethora of different experimental approaches is used, hampering direct comparisons between many studies. In addition, many papers lack information about the controls included in the experiment to exclude false positive or negative results. In the following, we summarize and compare current methods for nanoparticle–protein corona isolation, divided into three main categories: centrifugation, magnetism and chromatography.The most common method for separating particles and their protein corona from a matrix is centrifugation or ultracentrifugation. Here, the different densities of particles and proteins in the matrix are used for separation. Using centrifugation-based approaches, various aspects of particle–protein interactions have been studied.5,87,183 The major issue with this method is the risk of false positives: proteins, protein complexes which originally did not bind to the particle, or proteins that bind to particle-attached proteins but not to the particle itself, may sediment during the centrifugation process together with the particles and their corona. On the other hand, false negatives may occur due to dissociation of proteins from the nanoparticle–corona complex due to centrifugation forces. It is therefore very important to determine the number of washing steps and the centrifugation time which is appropriate for separating a specific type of nanoparticle–corona complex from a certain protein-rich medium. It appears, however, that this optimization is often not performed, or at least not described in the respective publications.
The speed and duration of centrifugation must be optimized to the respective particle species, to assure complete separation of nanoparticles with their corona from the surrounding media, while at the same time preventing protein aggregates ending up in the pellet. Konduru et al. noted that the density of the particles (which has a significant influence on the sedimentation properties) cannot always be directly estimated from the density of the material the particles are made of, due to agglomeration and media inclusion effects.186 Therefore, suitable controls are necessary to check for remaining nanoparticles in the supernatant and for unintentionally precipitated protein aggregates in the pellet.
While considering appropriate centrifugation conditions, different properties of the nanoparticles and the experimental set-up have to be taken into account. In general, one harsh centrifugation step is often not sufficient for complete isolation of particle-corona complexes without false positive results. These may, for example, result from protein complexes that have a higher density than the investigated particles or from proteins trapped in cavities of particle agglomerates.30 On the other hand, insufficient centrifugation speed may leave particles in the supernatant.187 Similar considerations are to be made for the washing procedure. Here, washing time, centrifugation speed and the number of repetitions need to be balanced. Furthermore, the choice of the washing buffer may influence the binding of proteins to the nanoparticles due to changing pH values, salt concentrations, and temperature.188,189Fig. 6 summarizes the aspects to consider when setting up a centrifugation-based separation method. Unfortunately, only a comparably low number of publications contain experimental details about the centrifugation procedure and its optimization. Here, it would be desirable to establish standards of reporting in order to allow the reader to judge the applied method.
Different variations and advancements of classic centrifugation methods have been introduced in nanoparticle corona research: sucrose cushions are used to reduce the interaction time of nanoparticles and media. Unbound proteins are separated from the nanoparticles by the sucrose cushion and agglomerates also tend to float above the cushion. This way, snapshots of nanoparticle–protein interactions can be analyzed with high resolution30,183 The sucrose cushion may be used either as a homogeneous sucrose solution with a defined density or as a gradient. The latter enables the harvesting of different fractions of nanoparticle-corona complexes to investigate the fate of nanoparticles, their protein corona and unbound proteins.30,190,191 In particular, for low-density particles, ultracentrifugation is the method of choice, sometimes in combination with a sucrose cushion.192 Ultracentrifugation is mostly referred to as centrifugation at >100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g, and can be subdivided into analytical and preparative ultracentrifugation. Analytical ultracentrifugation allows monitoring of the concentration of an analyte in the sample in real time, for example by analyzing fluorescent nanoparticles. This way, information about sedimentation properties and size distribution can be obtained.193 However, a high centrifugation speed may lead to aggregation of proteins, thereby increasing the risk of obtaining false positive results by agglomeration. Sequential centrifugation has been used to achieve neat separation of particle-corona complexes.194,195 This technique can be very useful, while at the same time multiple purification steps bear the risk of disruption of the corona equilibrium.
000×g, and can be subdivided into analytical and preparative ultracentrifugation. Analytical ultracentrifugation allows monitoring of the concentration of an analyte in the sample in real time, for example by analyzing fluorescent nanoparticles. This way, information about sedimentation properties and size distribution can be obtained.193 However, a high centrifugation speed may lead to aggregation of proteins, thereby increasing the risk of obtaining false positive results by agglomeration. Sequential centrifugation has been used to achieve neat separation of particle-corona complexes.194,195 This technique can be very useful, while at the same time multiple purification steps bear the risk of disruption of the corona equilibrium.
Separation techniques – magnetism
The second way to isolate particles with their corona is the use of magnetic force. Provided that the particle species is suitable for use with magnetic separation methods, magnetism is used, for example, as an easier and faster approach for separation.196 In general, the magnetic properties of the nanoparticle are provided by iron oxide (Fig. 3). Iron oxide nanoparticles are promising tools in targeted cancer diagnostics, and they are also available in versions coated with other materials such as silica to form hybrid particles that combine the different properties of the two materials.197 Nevertheless, centrifugation methods are also used for iron oxide nanoparticles, sometimes even for comparison. Bonvin et al. used both magnetic separation and centrifugation methods on the same nanoparticles.196 Different experimental setups are available: Luborsky and Drummond described an experimental setup with magnetic columns and magnetic gradients which has been adapted to nanoparticle-related experiments.198 Using magnetic forces for separation is thought to have less of an impact on the structure of the nanoparticle–protein corona complex than centrifugation.196 Nevertheless, it has to be noted that the risk of agglomeration increases with particle size. Because of this it is not recommended to use magnetic separation for nanoparticles with diameters greater than 10 nm. Thus, for larger particles, multi-step purification with increasing centrifugation intensities could be more appropriate.196 A big advantage of magnetic separation is the reduction of false-positive proteins due to aggregation under centrifugal forces. It also reduces the loss of protein after multiple washing steps. Nevertheless, washing steps are needed after magnetic separation too, but the loss of particles at each step might be lower. The limited number of applicable particle species as well as the possibility of interactions between magnetic particles and other required analytical methods are disadvantages of this separation technique.Separation techniques – chromatography
Chromatographic approaches are used less frequently than the above methods, probably because they are in general more time-consuming and cost-intensive and allow only a relatively low throughput of samples. In addition, they are not suitable for a variety of particle types with bigger sizes, for polydisperse particles, and for particles which adhere to the column material. However, they provide a possibility for investigating association/dissociation rates and affinity of individual proteins bound to nanoparticles, and they also allow collection of different fractions of a sample with less perturbation to particle–protein complexes.14 In general, two main methods are used, size exclusion chromatography and flow-field-flow-fractionation methods, such as asymmetric flow-field-flow-fractionation (A4F).199Described for the first time in 1950, the principle of size exclusion chromatography is separation by the hydrodynamic volume of the analyte.200 Smaller particles interact more with the stationary phase (mostly composed of porous particles) and therefore need more time to pass through the column while bigger particles elute faster. This principle helps in determining association and dissociation rates: complexes with a stable corona have an increased hydrodynamic diameter compared to blank particles, since proteins remain attached to the particle. In contrast, if particle–protein affinity is weak, proteins tend to dissociate quickly, causing no measurable increase in the hydrodynamic diameter. Therefore, the latter will elute at the same time as their non-incubated counterparts.14,201 Also, particle–protein complexes can be collected in fractions relative to their size and used for further analysis.201 However, interactions with the stationary phase may occur and lead to a change in the protein corona. Not only due to competing interactions between the particle, corona and stationary phase, but also due to shearing forces, this may result in a loss of protein–particle interaction.
A4F is a method to separate analytes in a wide size range (1 nm to 100 μm).202 It also helps to reduce potential non-specific interactions.87 A4F uses a liquid cross flow that is established in a channel with a nonporous and a porous wall. Particles are thus exposed to a laminar flow that pushes the particles along the tube, and a cross flow that forces them to the bottom of the channel. Due to their Brownian motion (which is more for smaller particles), the analytes can also float back into the channel, thereby avoiding passage through the porous membrane.203,204 A4F is especially used for analysis of complex samples and stable protein coronae.7,87,199 Fractions can be collected without much perturbation of particle–protein complexes. However, method establishment is usually very time-consuming due to the variety of parameters to be optimized.205
Ashby and colleagues combined the chromatographic method of flow field-flow fractionation with ultracentrifugation to obtain information about the composition of hard and soft coronae.87 However, ultracentrifugation tends to yield false positives or negatives due to dissociation of proteins owing to centrifugal forces or due to down-centrifugation of non-corona proteins. These often unconsidered issues aggravate the identification especially of the soft corona proteins, as already mentioned at an early stage of corona analytics by Lundqvist in 2008.5
Comparability of different methods
A few studies are available comparing the results obtained with different corona isolation methods. Bonvin et al. compared magnetic separation and a new multi-step centrifugation method for their iron oxide nanoparticles in human blood and lymph serum.166 The results showed that the hard corona obtained by magnetic separation differed from that obtained by the new centrifugation method, as only about half of the identified corona proteins were commonly detected using both methods. The parallel use of the two different methods was thus helpful to verify the presumably true positive results, but it was not possible to firmly distinguish whether the other proteins identified by only one single method represent false or true positives. Similarly, Pisani et al. compared magnetic separation with centrifugation and also concluded that the results differ substantially.93 By contrast, other analyses have revealed greater similarity of coronae obtained by using different separation methods: Monopoli et al. isolated nanoparticles by centrifugation, size exclusion chromatography, and magnetic separation. The protein composition of isolated hard coronae in the latter analyses was very similar between the different methods.206 Bekdemir et al. compared their centrifugation results using mercapto-undecanoic acid (MUA) gold nanoparticles with the results of Rocker et al., 2009 which were obtained using iron–platinum (FePt) nanoparticles of a similar size and with the same surface modification (carboxylated).193,207 The authors concluded that protein dissociation behavior mainly depends on the nanoparticle surface modification and size, and not on its core material. However, their centrifugation based method requires the adsorption of proteins to be reversible. In contrast to their finding that diluting the sample led to a lower number of proteins adsorbed to the particle, Milani et al. showed no reversibility of the protein corona after transfer into more diluted media and thus no Hill coefficient but material dependency was observed. The different results may also be due to the nature of the surface modifications since different materials may lead to different ligand packing densities with little change in the hydrodynamic diameter.193,208Approaches using two independent methods allow for a better assessment of the protein corona, but difficulties remain, as highlighted above. Often, false positives or false negatives result from the isolation/separation method. The variation between different isolation methods might, to a large extent, arise from different forces acting on NPs during the isolation process. In addition, the obtained results might be transferred to only a small selection of closely related nanoparticles, thus hampering more general conclusions and making it difficult to compare results from different corona studies.93,166,196
Methods for identification of nanoparticle corona proteins
In general, the isolation of nanoparticle–protein corona complexes from biological media is considered to be the most critical point in corona research. However, interpretation of the data also depends on the method of “stripping” the particle for analyzing the composition of the corona by protein identification methods. Different approaches have been used for this purpose, depending on the experimental question addressed. Some methods quantify the total protein content, such as the Bradford and BCA assays209–211 and UV/vis absorption measurements.212 The protein amount can be measured by the determination of bound proteins or as a difference in the supernatant. Other methods separate proteins, for example electrophoresis or chromatographic techniques, followed by visualization methods such as Coomassie brilliant blue or silver staining.60,213 More detailed information about the corona, and thus possibly also insights into the biochemistry of particle–cell interactions via the corona, can be obtained using methods for protein identification, for example mass spectrometry. A standard protein identification technique is 2-dimensional gel electrophoresis followed by tryptic digestion and mass spectrometry. Therefore, protein identification by MALDI-TOF is usually used.91,156 Unfortunately, traditional mass spectrometric methods allow just the identification of a rather small number of highly abundant proteins and is very time-consuming. Advanced mass spectrometry methods, such as LC-MS, orbitrap- or triple-quad-MS can replace the gel electrophoresis steps and lead to a significantly increased sensitivity with lower detection limits. Up to now, using high-resolution mass spectrometry to identify proteins has been the recommended state-of-the-art method, even though this requires bioinformatics data evaluation of big data sets. For a more in-depth review of this topic, we recommend the recent review articles by Carrillo-Carrion and Pederzoli (Table 1).214,215| Method | Advantages | Disadvantages |
|---|---|---|
| Centrifugation | - Separation according to density and size | - Long centrifugation times can lead to false particle–protein interactions |
| - Most frequently used technique | - Several purification steps needed; modifications in the protein–corona system can occur | |
| - Widely used and easy to use216 | - Magnetic nanoparticles can agglomerate196 false-positive highly abundant proteins due to insufficient washing | |
| - High throughput216 | - Changing of centrifugation tubes is necessary to exclude carryover of proteins adsorbed to the tube walls | |
| - Centrifugal speeds and times can be optimized according to the nanoparticle material and media | - Outcome affected by centrifugation force, washing duration, washing solution and solution volumes; must be adjusted for each particle type | |
| - Tuning experimental conditions makes the method available for a wide range of nanoparticles | - The smaller and less dense the particles, the higher the centrifugation speeds chosen; thus aggregation30 occurs | |
| - High resolution results30 | - Not suited for very small (5–20 nm) or low density nanoparticles (1 g cm−3), because unbound proteins and protein corona complexes cannot be separated effectively30,217 | |
| - Possibility of separating different populations co-existing in situ30 | - Not preparative, so no populations can be recovered for further studies23,29,206 | |
| Size exclusion chromatography (SEC) | - Flexible technique, many stationary/mobile phases | - Interaction between analytes and the stationary phase can occur218 |
| - can be used with standard lab equipment | - SEC selectivity decreases when applied to analytes with a very high molar mass such as nanoparticles218 | |
| - Analyte resolution and recovery in SEC is generally superior to A4F218 | - Low throughput | |
| - Has been developed into a systematic methodology14 | - No full recovery of hard corona complexes for further studies30 | |
| - Less perturbing than centrifugation14 | ||
| Asymmetric flow-field-flow fractionation (A4F) | - Complex, heterogeneous and polydisperse dispersions can be investigated without extensive sample preparation205 | - Long establishment process |
| - Reduced to no destruction or alteration of the protein corona | - Must be adjusted for every particle type205 | |
| - Prior fractionation by AF4 allows size investigation of complex heterogeneous and polydisperse mixtures205 | - Low throughput205 | |
| - Several detection techniques can be coupled to AF4 (online and offline)205 | - Expensive | |
| - Possible automation205 | - Not routinely available in many analytical laboratories | |
| - Short measurement time205 | - Separation of particles from a very polydisperse sample leads to peak broadening and loss of resolution; must thus be divided into several experiments | |
| - Easy collection of fractions205 | - Sample loss due to adsorption on the membrane can occur, affecting retention and disturbing quantification of single fractions205 | |
| - Absence of a packaging material or a stationary phase205 | - No full recovery of fraction for further experiments30 | |
| - The potential of AF4 increases with increasing molar mass205 | ||
| - Once established, AF4 is a multifunctional technique for separation and characterization of nearly all nano-sized205particles | ||
| Magnetism | - Low impact on the structure | - Only practicable for small (∼10 nm), magnetic nanoparticles196 |
| - High throughput | - Degree of separation decreases with decreasing magnetism196 |
Conclusion
Individual nanomaterials exhibit very specific characteristics. Therefore, the choice of separation method for the isolation of protein–corona complexes depends strongly on the particle and its physicochemical parameters and also on the surrounding matrix, possible unintended interactions and the desired fate of the corona. Centrifugation as the most widely used technique may be the first choice for many particles but may, for example, not be feasible for low-density analytes. Centrifugation is easy to use, but care should be taken for proper controls to exclude false positive and negative results in protein identification. In addition, loss of particle fractions during repeated washing and centrifugation steps is an issue. However, this might be managed by analyzing all fractions for their particle contents to exclude the loss of particle fractions. Additionally, control samples with the particle-free matrix of interest, including stabilizing agents and ions that might be released from the particles, can help to prevent false positives. Nanoparticle corona analysis is a very diverse field of current research, investigating a dynamic process that depends on a plethora of different parameters. To enable comparative and reproducible results across different studies and laboratories, it would be desirable to have harmonized experimental setups for the scientific community, and detailed descriptions of experimental procedures including the use of controls.Conflicts of interest
There are no conflicts of interest to declare.Funding
This project was funded by the German Federal Institute for Risk Assessment as part of the Junior Research Group of Nanotoxicology (OAWO-NANO 1323-102).Author's contributions
The authors of this manuscript are Linda Böhmert (LB), Linn Voß (LV), Valerie Stock (VS), Albert Braeuning (AB), Alfonso Lampen (AL), and Holger Sieg (HS). The authors contributed to the conception and design of the work (LB, AB, AL, and HS) and data collection and interpretation (LB, LV, VS, and HS), wrote the manuscript (LB, LV, VS, and HS) and revised the manuscript (LB, AB, AL, and HS).References
- S. Foss Hansen, B. H. Larsen, S. I. Olsen and A. Baun, Categorization framework to aid hazard identification of nanomaterials, Nanotoxicology, 2007, 1(3), 243–250 CrossRef.
- RA/RAEng, Nanoscience and nanotechnologies: opportunities and uncertainties, The Royal Society and Royal Academy of Engineering, 2004, Contract No.: ISBN 0 85403 604 0 Search PubMed.
- M. P. Monopoli, C. Aberg, A. Salvati and K. A. Dawson, Biomolecular coronas provide the biological identity of nanosized materials, Nat. Nanotechnol., 2012, 7(12), 779–786 CrossRef CAS PubMed.
- M. Avila-Olias, C. Pegoraro, G. Battaglia and I. Canton, Inspired by nature: fundamentals in nanotechnology design to overcome biological barriers, Ther. Delivery, 2013, 4(1), 27–43 CrossRef CAS PubMed.
- M. Lundqvist, J. Stigler, G. Elia, I. Lynch, T. Cedervall and K. A. Dawson, Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts, Proc. Natl. Acad. Sci. U. S. A., 2008, 105(38), 14265–14270 CrossRef CAS.
- J. W. Yoo and S. Mitragotri, Polymer particles that switch shape in response to a stimulus, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(25), 11205–11210 CrossRef CAS PubMed.
- T. K. Mudalige, H. Qu and S. W. Linder, Asymmetric Flow-Field Flow Fractionation Hyphenated ICP-MS as an Alternative to Cloud Point Extraction for Quantification of Silver Nanoparticles and Silver Speciation: Application for Nanoparticles with a Protein Corona, Anal. Chem., 2015, 87(14), 7395–7401 CrossRef CAS.
- H. F. Krug and P. Wick, Nanotoxicology: an interdisciplinary challenge, Angew. Chem., Int. Ed. Engl., 2011, 50(6), 1260–1278 CrossRef CAS PubMed.
- A. E. Nel, L. Madler, D. Velegol, T. Xia, E. M. V. Hoek and P. Somasundaran, et al., Understanding biophysicochemical interactions at the nano-bio interface, Nat. Mater., 2009, 8(7), 543–557 CrossRef CAS PubMed.
- P. P. Karmali and D. Simberg, Interactions of nanoparticles with plasma proteins: implication on clearance and toxicity of drug delivery systems, Expert Opin. Drug Delivery, 2011, 8(3), 343–357 CrossRef CAS.
- I. Lynch, A. Salvati and K. A. Dawson, Protein-nanoparticle interactions: What does the cell see?, Nat. Nanotechnol., 2009, 4(9), 546–547 CrossRef CAS PubMed.
- K. H. Muller, M. Motskin, A. J. Philpott, A. F. Routh, C. M. Shanahan and M. J. Duer, et al., The effect of particle agglomeration on the formation of a surface-connected compartment induced by hydroxyapatite nanoparticles in human monocyte-derived macrophages, Biomaterials, 2014, 35(3), 1074–1088 CrossRef CAS.
- G. Hartmann, T. Baumgartner and M. Schuster, Influence of Particle Coating and Matrix Constituents on the Cloud Point Extraction Efficiency of Silver Nanoparticles (Ag-NPs) and Application for Monitoring the Formation of Ag-NPs from Ag+, Anal. Chem., 2014, 86(1), 790–796 CrossRef CAS PubMed.
- T. Cedervall, I. Lynch, S. Lindman, T. Berggard, E. Thulin and H. Nilsson, et al., Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles, Proc. Natl. Acad. Sci. U. S. A., 2007, 104(7), 2050–2055 CrossRef CAS PubMed.
- T. M. Allen, C. Hansen, F. Martin, C. Redemann and A. Yau-Young, Liposomes containing synthetic lipid derivatives of poly(ethylene glycol) show prolonged circulation half-lives in vivo, Biochim. Biophys. Acta, 1991, 1066(1), 29–36 CrossRef CAS.
- A. D. Bangham, B. A. Pethica and G. V. Seaman, The charged groups at the interface of some blood cells, Biochem. J., 1958, 69(1), 12–19 CrossRef CAS PubMed.
- R. Gref, M. Lück, P. Quellec, M. Marchand, E. Dellacherie and S. Harnisch, et al., ‘Stealth’ corona-core nanoparticles surface modified by polyethylene glycol (PEG): influences of the corona (PEG chain length and surface density) and of the core composition on phagocytic uptake and plasma protein adsorption, Colloids Surf., B, 2000, 18(3), 301–313 CrossRef CAS PubMed.
- L. Vroman, Effect of Adsorbed Proteins on the Wettability of Hydrophilic and Hydrophobic Solids, Nature, 1962, 196(4853), 476–477 CrossRef CAS PubMed.
- O. Vilanova, J. J. Mittag, P. M. Kelly, S. Milani, K. A. Dawson and J. O. Radler, et al., Understanding the Kinetics of Protein-Nanoparticle Corona Formation, ACS Nano, 2016, 10(12), 10842–10850 CrossRef CAS PubMed.
- D. Dell'Orco, M. Lundqvist, C. Oslakovic, T. Cedervall and S. Linse, Modeling the time evolution of the nanoparticle-protein corona in a body fluid, PLoS One, 2010, 5(6), e10949 CrossRef PubMed.
- M. Lundqvist, J. Stigler, T. Cedervall, T. Berggard, M. B. Flanagan and I. Lynch, et al., The evolution of the protein corona around nanoparticles: a test study, ACS Nano, 2011, 5(9), 7503–7509 CrossRef CAS PubMed.
- G. Maiorano, S. Sabella, B. Sorce, V. Brunetti, M. A. Malvindi and R. Cingolani, et al., Effects of cell culture media on the dynamic formation of protein-nanoparticle complexes and influence on the cellular response, ACS Nano, 2010, 4(12), 7481–7491 CrossRef CAS PubMed.
- M. P. Monopoli, D. Walczyk, A. Campbell, G. Elia, I. Lynch and F. B. Bombelli, et al., Physical-chemical aspects of protein corona: relevance to in vitro and in vivo biological impacts of nanoparticles, J. Am. Chem. Soc., 2011, 133(8), 2525–2534 CrossRef CAS PubMed.
- X. J. Cheng, X. Tian, A. Q. Wu, J. X. Li, J. Tian and Y. Chong, et al., Protein Corona Influences Cellular Uptake of Gold Nanoparticles by Phagocytic and Nonphagocytic Cells in a Size-Dependent Manner, ACS Appl. Mater. Interfaces, 2015, 7(37), 20568–20575 CrossRef CAS PubMed.
- V. Mirshafiee, R. Kim, S. Park, M. Mahmoudi and M. L. Kraft, Impact of protein pre-coating on the protein corona composition and nanoparticle cellular uptake, Biomaterials, 2016, 75, 295–304 CrossRef CAS PubMed.
- S. Ritz, S. Schottler, N. Kotman, G. Baier, A. Musyanovych and J. Kuharev, et al., Protein corona of nanoparticles: distinct proteins regulate the cellular uptake, Biomacromolecules, 2015, 16(4), 1311–1321 CrossRef CAS PubMed.
- F. Barbero, L. Russo, M. Vitali, J. Piella, I. Salvo and M. L. Borrajo, et al., Formation of the Protein Corona: The Interface between Nanoparticles and the Immune System, Semin. Immunol., 2017, 52–60 CrossRef CAS PubMed.
- D. Boraschi, P. Italiani, R. Palomba, P. Decuzzi, A. Duschl and B. Fadeel, et al., Nanoparticles and innate immunity: new perspectives on host defence, Semin. Immunol., 2017, 33–51 CrossRef CAS.
- D. Walczyk, F. B. Bombelli, M. P. Monopoli, I. Lynch and K. A. Dawson, What the cell “sees” in bionanoscience, J. Am. Chem. Soc., 2010, 132(16), 5761–5768 CrossRef CAS PubMed.
- D. Di Silvio, N. Rigby, B. Bajka, A. Mayes and A. Mackie, Baldelli Bombelli F. Technical tip: high-resolution isolation of nanoparticle-protein corona complexes from physiological fluids, Nanoscale, 2015, 7(28), 11980–11990 RSC.
- D. Bargheer, J. Nielsen, G. Gebel, M. Heine, S. C. Salmen and R. Stauber, et al., The fate of a designed protein corona on nanoparticles in vitro and in vivo, Beilstein J. Nanotechnol., 2015, 6, 36–46 CrossRef.
- P. Satzer, M. Wellhoefer and A. Jungbauer, Continuous separation of protein loaded nanoparticles by simulated moving bed chromatography, J. Chromatogr. A, 2014, 1349, 44–49 CrossRef CAS.
- Committee ES, Guidance on the risk assessment of the application of nanoscience and nanotechnologies in the food and feed chain, EFSA J., 2011, 9(5), 2140 CrossRef.
- S.-j. Yu, J.-b. Chao, J. Sun, Y.-g. Yin, J.-f. Liu and G.-b. Jiang, Quantification of the uptake of silver nanoparticles and ions to HepG2 cells, Environ. Sci. Technol., 2013, 47(7), 3268–3274 CrossRef CAS.
- D. I. Hitchcock, Protein films on collodion membranes, J. Gen. Physiol., 1925, 8(2), 61–74 CrossRef CAS.
- M. Mahmoudi, J. Meng, X. Xue, X. J. Liang, M. Rahman and C. Pfeiffer, et al., Interaction of stable colloidal nanoparticles with cellular membranes, Biotechnol. Adv., 2014, 32(4), 679–692 CrossRef CAS PubMed.
- C. D. Walkey and W. C. Chan, Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment, Chem. Soc. Rev., 2012, 41(7), 2780–2799 RSC.
- A. Albanese, C. D. Walkey, J. B. Olsen, H. Guo, A. Emili and W. C. Chan, Secreted biomolecules alter the biological identity and cellular interactions of nanoparticles, ACS Nano, 2014, 8(6), 5515–5526 CrossRef CAS PubMed.
- H. Awada, L. Mezzasalma, S. Blanc, D. Flahaut, C. Dagron-Lartigau and J. Lyskawa, et al., Biomimetic Mussel Adhesive Inspired Anchor to Design ZnO@Poly(3-Hexylthiophene) Hybrid Core@Corona Nanoparticles, Macromol. Rapid Commun., 2015, 36(16), 1486–1491 CrossRef CAS PubMed.
- Z. Y. Guo, X. Hai, Y. T. Wang, Y. Shu, X. W. Chen and J. H. Wang, Core-Corona Magnetic Nanospheres Functionalized with Zwitterionic Polymer Ionic Liquid for Highly Selective Isolation of Glycoprotein, Biomacromolecules, 2018, 19(1), 53–61 CrossRef CAS PubMed.
- S. H. Jo, H. W. Kim, M. Song, N. J. Je, S. H. Oh and B. Y. Chang, et al., Core-Corona Functionalization of Diblock Copolymer Micelles by Heterogeneous Metal Nanoparticles for Dual Modality in Chemical Reactions, ACS Appl. Mater. Interfaces, 2015, 7(33), 18778–18785 CrossRef CAS PubMed.
- C. M. Harris, S. G. Miller, K. Andresen and L. B. Thompson, Quantitative measurement of sodium polystyrene sulfonate adsorption onto CTAB capped gold nanoparticles reveals hard and soft coronas, J. Colloid Interface Sci., 2018, 510, 39–44 CrossRef CAS PubMed.
- L. Vroman and A. L. Adams, Rapid identification of proteins on flat surfaces, using antibody-coated metal oxide suspensions, J. Immunol. Methods, 1986, 93(2), 213–216 CrossRef CAS PubMed.
- L. Vroman, A. L. Adams, G. C. Fischer and P. C. Munoz, Interaction of high molecular weight kininogen, factor XII, and fibrinogen in plasma at interfaces, Blood, 1980, 55(1), 156–159 CrossRef CAS PubMed.
- D. Docter, D. Westmeier, M. Markiewicz, S. Stolte, S. K. Knauer and R. H. Stauber, The nanoparticle biomolecule corona: lessons learned - challenge accepted?, Chem. Soc. Rev., 2015, 44(17), 6094–6121 RSC.
- J. Wolfram, Y. Yang, J. Shen, A. Moten, C. Chen and H. Shen, et al., The nano-plasma interface: Implications of the protein corona, Colloids Surf., B, 2014, 124, 17–24 CrossRef CAS PubMed.
- F. Darabi Sahneh, C. Scoglio and J. Riviere, Dynamics of nanoparticle-protein corona complex formation: analytical results from population balance equations, PLoS One, 2013, 8(5), e64690 CrossRef PubMed.
- N. Duran, C. P. Silveira, M. Duran and D. S. Martinez, Silver nanoparticle protein corona and toxicity: a mini-review, J. Nanobiotechnol., 2015, 13, 55 CrossRef PubMed.
- I. Lynch and K. A. Dawson, Protein-nanoparticle interactions, Nano Today, 2008, 3(1), 40–47 CrossRef CAS.
- T. E. Mallouk and P. Yang, Chemistry at the nano-bio interface, J. Am. Chem. Soc., 2009, 131(23), 7937–7939 CrossRef CAS PubMed.
- M. Mahmoudi, S. Laurent, M. A. Shokrgozar and M. Hosseinkhani, Toxicity evaluations of superparamagnetic iron oxide nanoparticles: cell “vision” versus physicochemical properties of nanoparticles, ACS Nano, 2011, 5(9), 7263–7276 CrossRef CAS PubMed.
- M. Mahmoudi, S. N. Saeedi-Eslami, M. A. Shokrgozar, K. Azadmanesh, M. Hassanlou and H. R. Kalhor, et al., Cell “vision”: complementary factor of protein corona in nanotoxicology, Nanoscale, 2012, 4(17), 5461–5468 RSC.
- S. Laurent, C. Burtea, C. Thirifays, U. O. Hafeli and M. Mahmoudi, Crucial ignored parameters on nanotoxicology: the importance of toxicity assay modifications and “cell vision”, PLoS One, 2012, 7(1), e29997 CrossRef CAS PubMed.
- B. B. Manshian, S. Munck, P. Agostinis, U. Himmelreich and S. J. Soenen, High content analysis at single cell level identifies different cellular responses dependent on nanomaterial concentrations, Sci. Rep., 2015, 5, 13890 CrossRef PubMed.
- I. Lynch, T. Cedervall, M. Lundqvist, C. Cabaleiro-Lago, S. Linse and K. A. Dawson, The nanoparticle-protein complex as a biological entity; a complex fluids and surface science challenge for the 21st century, Adv. Colloid Interface Sci., 2007, 134–135, 167–174 CrossRef CAS PubMed.
- G. Q. Hu, B. Jiao, X. H. Shi, R. P. Valle, Q. H. Fan and Y. Y. Zuo, Physicochemical Properties of Nanoparticles Regulate Translocation across Pulmonary Surfactant Monolayer and Formation of Lipoprotein Corona, ACS Nano, 2013, 7(12), 10525–10533 CrossRef CAS PubMed.
- M. E. Hughes, E. Brandin and J. A. Golovchenko, Optical absorption of DNA-carbon nanotube structures, Nano Lett., 2007, 7(5), 1191–1194 CrossRef CAS PubMed.
- A. A. Kapralov, W. H. Feng, A. A. Amoscato, N. Yanamala, K. Balasubramanian and D. E. Winnica, et al., Adsorption of Surfactant Lipids by Single-Walled Carbon Nanotubes in Mouse Lung upon Pharyngeal Aspiration, ACS Nano, 2012, 6(5), 4147–4156 CrossRef CAS PubMed.
- P. S. R. Naidu, M. Norret, N. M. Smith, S. A. Dunlop, N. L. Taylor and M. Fitzgerald, et al., The Protein Corona of PEGylated PGMA-Based Nanoparticles is Preferentially Enriched with Specific Serum Proteins of Varied Biological Function, Langmuir, 2017, 33(45), 12926–12933 CrossRef CAS PubMed.
- R. Ahmad Khanbeigi, T. F. Abelha, A. Woods, O. Rastoin, R. D. Harvey and M. C. Jones, et al., Surface chemistry of photoluminescent F8BT conjugated polymer nanoparticles determines protein corona formation and internalization by phagocytic cells, Biomacromolecules, 2015, 16(3), 733–742 CrossRef CAS PubMed.
- F. Meder, C. Brandes, L. Treccani and K. Rezwan, Controlling protein-particle adsorption by surface tailoring colloidal alumina particles with sulfonate groups, Acta Biomater., 2013, 9(3), 5780–5787 CrossRef CAS PubMed.
- K. Kandori, M. Oketani and M. Wakamura, Effects of Ti(IV) substitution on protein adsorption behaviors of calcium hydroxyapatite particles, Colloids Surf., B, 2013, 101, 68–73 CrossRef CAS PubMed.
- A. M. Clemments, P. Botella and C. C. Landry, Protein Adsorption From Biofluids on Silica Nanoparticles: Corona Analysis as a Function of Particle Diameter and Porosity, ACS Appl. Mater. Interfaces, 2015, 7(39), 21682–21689 CrossRef CAS.
- A. Kondo, F. Murakami, M. Kawagoe and K. Higashitani, Kinetic and circular dichroism studies of enzymes adsorbed on ultrafine silica particles, Appl. Microbiol. Biotechnol., 1993, 39(6), 726–731 CrossRef CAS PubMed.
- K. Kandori, M. Mukai, A. Fujiwara, A. Yasukawa and T. Ishikawa, Adsorption of Bovine Serum Albumin and Lysozyme on Hydrophobic Calcium Hydroxyapatites, J. Colloid Interface Sci., 1999, 212(2), 600–603 CrossRef CAS PubMed.
- A. Orlando, E. Cazzaniga, M. Tringali, F. Gullo, A. Becchetti and S. Minniti, et al., Mesoporous silica nanoparticles trigger mitophagy in endothelial cells and perturb neuronal network activity in a size- and time-dependent manner, Int. J. Nanomed., 2017, 12, 3547–3559 CrossRef CAS PubMed.
- Y. Hayashi, T. Miclaus, C. Scavenius, K. Kwiatkowska, A. Sobota and P. Engelmann, et al., Species differences take shape at nanoparticles: protein corona made of the native repertoire assists cellular interaction, Environ. Sci. Technol., 2013, 47(24), 14367–14375 CrossRef CAS PubMed.
- S. Sindhwani, A. M. Syed, S. Wilhelm, D. R. Glancy, Y. Y. Chen and M. Dobosz, et al., Three-Dimensional Optical Mapping of Nanoparticle Distribution in Intact Tissues, ACS Nano, 2016, 10(5), 5468–5478 CrossRef CAS PubMed.
- M. V. Zyuzin, Y. Yan, R. Hartmann, K. T. Gause, M. Nazarenus and J. Cui, et al., Role of the Protein Corona Derived from Human Plasma in Cellular Interactions between Nanoporous Human Serum Albumin Particles and Endothelial Cells, Bioconjugate Chem., 2017, 28(8), 2062–2068 CrossRef CAS PubMed.
- I. Kranz, J. B. Gonzalez, I. Dorfel, M. Gemeinert, M. Griepentrog and D. Klaffke, et al., Biological response to micron- and nanometer-sized particles known as potential wear products from artificial hip joints: Part II: Reaction of murine macrophages to corundum particles of different size distributions, J. Biomed. Mater. Res., 2009, 89(2), 390–401 CrossRef CAS.
- A. D. Abid, D. S. Anderson, G. K. Das, L. S. Van Winkle and I. M. Kennedy, Novel lanthanide-labeled metal oxide nanoparticles improve the measurement of in vivo clearance and translocation, Part. Fibre Toxicol., 2013, 10, 1 CrossRef CAS.
- B. Kang, P. Okwieka, S. Schottler, S. Winzen, J. Langhanki and K. Mohr, et al., Carbohydrate-Based Nanocarriers Exhibiting Specific Cell Targeting with Minimum Influence from the Protein Corona, Angew. Chem., Int. Ed. Engl., 2015, 54(25), 7436–7440 CrossRef CAS.
- M. Xu, J. Zhu, F. Wang, Y. Xiong, Y. Wu and Q. Wang, et al., Improved In Vitro and In Vivo Biocompatibility of Graphene Oxide through Surface Modification: Poly(Acrylic Acid)-Functionalization is Superior to PEGylation, ACS Nano, 2016, 10(3), 3267–3281 CrossRef CAS.
- O. Koshkina, D. Westmeier, T. Lang, C. Bantz, A. Hahlbrock and C. Wurth, et al., Tuning the Surface of Nanoparticles: Impact of Poly(2-ethyl-2-oxazoline) on Protein Adsorption in Serum and Cellular Uptake, Macromol. Biosci., 2016, 16(9), 1287–1300 CrossRef CAS.
- D. Estupinan, M. B. Bannwarth, S. E. Mylon, K. Landfester, R. Munoz-Espi and D. Crespy, Multifunctional clickable and protein-repellent magnetic silica nanoparticles, Nanoscale, 2016, 8(5), 3019–3030 RSC.
- S. Shahabi, S. Doscher, T. Bollhorst, L. Treccani, M. Maas and R. Dringen, et al., Enhancing Cellular Uptake and Doxorubicin Delivery of Mesoporous Silica Nanoparticles via Surface Functionalization: Effects of Serum, ACS Appl. Mater. Interfaces, 2015, 7(48), 26880–26891 CrossRef CAS PubMed.
- L. J. Peek, L. Roberts and C. Berkland, Poly(D,L-lactide-co-glycolide) nanoparticle agglomerates as carriers in dry powder aerosol formulation of proteins, Langmuir, 2008, 24(17), 9775–9783 CrossRef CAS.
- R. Shegokar, M. Jansch, K. K. Singh and R. H. Muller, In vitro protein adsorption studies on nevirapine nanosuspensions for HIV/AIDS chemotherapy, Nanomedicine, 2011, 7(3), 333–340 CrossRef CAS.
- T. M. Goppert and R. H. Muller, Polysorbate-stabilized solid lipid nanoparticles as colloidal carriers for intravenous targeting of drugs to the brain: comparison of plasma protein adsorption patterns, J. Drug Targeting, 2005, 13(3), 179–187 CrossRef.
- Q. Peng, S. Zhang, Q. Yang, T. Zhang, X. Q. Wei and L. Jiang, et al., Preformed albumin corona, a protective coating for nanoparticles based drug delivery system, Biomaterials, 2013, 34(33), 8521–8530 CrossRef CAS.
- J. Guan, Z. Zhang, X. Hu, Y. Yang, Z. Chai and X. Liu, et al., Cholera Toxin Subunit B Enabled Multifunctional Glioma-Targeted Drug Delivery, Adv. Healthcare Mater., 2017, 6, 1700709 CrossRef.
- M. Horie, K. Fujita, H. Kato, S. Endoh, K. Nishio and L. K. Komaba, et al., Association of the physical and chemical properties and the cytotoxicity of metal oxide nanoparticles: metal ion release, adsorption ability and specific surface area, Metallomics, 2012, 4(4), 350–360 RSC.
- N. Ajdari, C. Vyas, S. L. Bogan, B. A. Lwaleed and B. G. Cousins, Gold nanoparticle interactions in human blood: a model evaluation, Nanomedicine, 2017, 13(4), 1531–1542 CrossRef CAS.
- J. Deng, M. Sun, J. Zhu and C. Gao, Molecular interactions of different size AuNP-COOH nanoparticles with human fibrinogen, Nanoscale, 2013, 5(17), 8130–8137 RSC.
- T. S. Anirudhan and T. A. Rauf, Lysozyme immobilization via adsorption process using sulphonic acid functionalized silane grafted copolymer, Colloids Surf., B, 2013, 107, 1–10 CrossRef CAS.
- M. Huang and W. Wang, Factors affecting alum-protein interactions, Int. J. Pharm., 2014, 466(1–2), 139–146 CrossRef CAS PubMed.
- J. Ashby, S. Schachermeyer, S. Q. Pan and W. W. Zhong, Dissociation-Based Screening of Nanoparticle-Protein Interaction via Flow Field-Flow Fractionation, Anal. Chem., 2013, 85(15), 7494–7501 CrossRef CAS.
- C. De Roe, P. J. Courtoy and P. Baudhuin, A model of protein-colloidal gold interactions, J. Histochem. Cytochem., 1987, 35(11), 1191–1198 CrossRef CAS.
- D. T. Jayaram, S. Runa, M. L. Kemp and C. K. Payne, Nanoparticle-induced oxidation of corona proteins initiates an oxidative stress response in cells, Nanoscale, 2017, 9(22), 7595–7601 RSC.
- Y. Li and N. A. Monteiro-Riviere, Mechanisms of cell uptake, inflammatory potential and protein corona effects with gold nanoparticles, Nanomedicine, 2016, 11(24), 3185–3203 CrossRef CAS PubMed.
- S. Juling, A. Niedzwiecka, L. Bohmert, D. Lichtenstein, S. Selve and A. Braeuning, et al., Protein Corona Analysis of Silver Nanoparticles Links to Their Cellular Effects, J. Proteome Res., 2017, 16(11), 4020–4034 CrossRef CAS PubMed.
- K. Thode, M. Luck, W. Schroder, W. Semmler, T. Blunk and R. H. Muller, et al., The influence of the sample preparation on plasma protein adsorption patterns on polysaccharide-stabilized iron oxide particles and N-terminal microsequencing of unknown proteins, J. Drug Targeting, 1997, 5(1), 35–43 CrossRef CAS.
- C. Pisani, J. C. Gaillard, C. Dorandeu, C. Charnay, Y. Guari and J. Chopineau, et al., Experimental separation steps influence the protein content of corona around mesoporous silica nanoparticles, Nanoscale, 2017, 9(18), 5769–5772 RSC.
- T. M. Goppert and R. H. Muller, Alternative sample preparation prior to two-dimensional electrophoresis protein analysis on solid lipid nanoparticles, Electrophoresis, 2004, 25(1), 134–140 CrossRef.
- A. Gessner, B. R. Paulke and R. H. Muller, Analysis of plasma protein adsorption onto polystyrene particles by two-dimensional electrophoresis: comparison of sample application and isoelectric focusing techniques, Electrophoresis, 2000, 21(12), 2438–2442 CrossRef CAS.
- K. Lindl, M. Kresse and R. H. Muller, Evaluation of desorption of proteins adsorbed to hydrophilic surfaces by two-dimensional electrophoresis, Proteomics, 2001, 1(9), 1059–1066 CrossRef CAS.
- A. Kondo and J. Mihara, Comparison of Adsorption and Conformation of Hemoglobin and Myoglobin on Various Inorganic Ultrafine Particles, J. Colloid Interface Sci., 1996, 177(1), 214–221 CrossRef CAS.
- R. Cukalevski, M. Lundqvist, C. Oslakovic, B. Dahlback, S. Linse and T. Cedervall, Structural changes in apolipoproteins bound to nanoparticles, Langmuir, 2011, 27(23), 14360–14369 CrossRef CAS PubMed.
- E. Sanfins, C. Augustsson, B. Dahlback, S. Linse and T. Cedervall, Size-dependent effects of nanoparticles on enzymes in the blood coagulation cascade, Nano Lett., 2014, 14(8), 4736–4744 CrossRef CAS PubMed.
- N. Ali, K. Mattsson, J. Rissler, H. M. Karlsson, C. R. Svensson and A. Gudmundsson, et al., Analysis of nanoparticle-protein coronas formed in vitro between nanosized welding particles and nasal lavage proteins, Nanotoxicology, 2016, 10(2), 226–234 CrossRef CAS PubMed.
- M. Levak, P. Buric, M. Dutour Sikiric, D. Domazet Jurasin, N. Mikac and N. Bacic, et al., Effect of Protein Corona on Silver Nanoparticle Stabilization and Ion Release Kinetics in Artificial Seawater, Environ. Sci. Technol., 2017, 51(3), 1259–1266 CrossRef CAS.
- A. Almalik, H. Benabdelkamel, A. Masood, I. O. Alanazi, I. Alradwan and M. A. Majrashi, et al., Hyaluronic Acid Coated Chitosan Nanoparticles Reduced the Immunogenicity of the Formed Protein Corona, Sci. Rep., 2017, 7(1), 10542 CrossRef.
- S. Amiri, M. R. Mehrnia and F. P. Roudsari, Enhancing purification efficiency of affinity functionalized composite agarose micro beads using Fe3O4 nanoparticles, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2017, 1041–1042, 27–36 CrossRef CAS.
- Z. Wang, T. Yue, Y. Yuan, R. Cai, C. Niu and C. Guo, Kinetics of adsorption of bovine serum albumin on magnetic carboxymethyl chitosan nanoparticles, Int. J. Biol. Macromol., 2013, 58, 57–65 CrossRef CAS PubMed.
- M. Jansch, P. Stumpf, C. Graf, E. Ruhl and R. H. Muller, Adsorption kinetics of plasma proteins on ultrasmall superparamagnetic iron oxide (USPIO) nanoparticles, Int. J. Pharm., 2012, 428(1–2), 125–133 CrossRef CAS PubMed.
- Z. Ma, J. Bai and X. Jiang, Monitoring of the Enzymatic Degradation of Protein Corona and Evaluating the Accompanying Cytotoxicity of Nanoparticles, ACS Appl. Mater. Interfaces, 2015, 7(32), 17614–17622 CrossRef CAS PubMed.
- J. Shi, Y. Hedberg, M. Lundin, I. Odnevall Wallinder, H. L. Karlsson and L. Moller, Hemolytic properties of synthetic nano- and porous silica particles: the effect of surface properties and the protection by the plasma corona, Acta Biomater., 2012, 8(9), 3478–3490 CrossRef CAS PubMed.
- Z. Ma, J. Bai, Y. Wang and X. Jiang, Impact of shape and pore size of mesoporous silica nanoparticles on serum protein adsorption and RBCs hemolysis, ACS Appl. Mater. Interfaces, 2014, 6(4), 2431–2438 CrossRef CAS.
- S. Shahabi, L. Treccani, R. Dringen and K. Rezwan, Utilizing the protein corona around silica nanoparticles for dual drug loading and release, Nanoscale, 2015, 7(39), 16251–16265 RSC.
- N. P. Mortensen, G. B. Hurst, W. Wang, C. M. Foster, P. D. Nallathamby and S. T. Retterer, Dynamic development of the protein corona on silica nanoparticles: composition and role in toxicity, Nanoscale, 2013, 5(14), 6372–6380 RSC.
- E. Izak-Nau, M. Voetz, S. Eiden, A. Duschl and V. F. Puntes, Altered characteristics of silica nanoparticles in bovine and human serum: the importance of nanomaterial characterization prior to its toxicological evaluation, Part. Fibre Toxicol., 2013, 10(1), 56 CrossRef.
- T. I. Armstrong, M. C. Davies and L. Illum, Human serum albumin as a probe for protein adsorption to nanoparticles: relevance to biodistribution, J. Drug Targeting, 1997, 4(6), 389–398 CrossRef CAS PubMed.
- N. Bertrand, P. Grenier, M. Mahmoudi, E. M. Lima, E. A. Appel and F. Dormont, et al., Mechanistic understanding of in vivo protein corona formation on polymeric nanoparticles and impact on pharmacokinetics, Nat. Commun., 2017, 8(1), 777 CrossRef PubMed.
- S. Dimchevska, N. Geskovski, R. Koliqi, N. Matevska-Geskovska, V. Gomez Vallejo and B. Szczupak, et al., Efficacy assessment of self-assembled PLGA-PEG-PLGA nanoparticles: Correlation of nano-bio interface interactions, biodistribution, internalization and gene expression studies, Int. J. Pharm., 2017, 533(2), 389–401 CrossRef CAS PubMed.
- R. Gossmann, E. Fahrlander, M. Hummel, D. Mulac, J. Brockmeyer and K. Langer, Comparative examination of adsorption of serum proteins on HSA- and PLGA-based nanoparticles using SDS-PAGE and LC-MS, Eur. J. Pharm. Biopharm., 2015, 93, 80–87 CrossRef CAS PubMed.
- S. Kamei and J. Kopecek, Prolonged blood circulation in rats of nanospheres surface-modified with semitelechelic poly[N-(2-hydroxypropyl)methacrylamide], Pharm. Res., 1995, 12(5), 663–668 CrossRef CAS PubMed.
- P. Zhang, X. Y. Xu, Y. P. Chen, M. Q. Xiao, B. Feng and K. X. Tian, et al., Protein corona between nanoparticles and bacterial proteins in activated sludge: Characterization and effect on nanoparticle aggregation, Bioresour. Technol., 2017, 250, 10–16 CrossRef.
- M. F. Zambaux, F. Bonneaux, R. Gref, E. Dellacherie and C. Vigneron, Preparation and characterization of protein C-loaded PLA nanoparticles, J. Controlled Release, 1999, 60(2–3), 179–188 CrossRef CAS.
- C. Tripisciano, T. Eichhorn, S. Harm and V. Weber, Adsorption of the inflammatory mediator high-mobility group box 1 by polymers with different charge and porosity, BioMed Res. Int., 2014, 2014, 238160 Search PubMed.
- R. Shelma and C. P. Sharma, Development of lauroyl sulfated chitosan for enhancing hemocompatibility of chitosan, Colloids Surf., B, 2011, 84(2), 561–570 CrossRef CAS PubMed.
- A. V. Il'ina, A. A. Gubaidullina, A. I. Melent'ev and V. P. Varlamov, [Preparation of chitosan microparticles and study of their interaction with interferon], Prikl. Biokhim. Mikrobiol., 2008, 44(2), 250–254 Search PubMed.
- M. Kendall, N. J. Hodges, H. Whitwell, J. Tyrrell and H. Cangul, Nanoparticle growth and surface chemistry changes in cell-conditioned culture medium, Philos. Trans. R. Soc. London, Ser. B, 2015, 370(1661), 20140100 CrossRef PubMed.
- S. K. Kloet, A. P. Walczak, J. Louisse, H. H. van den Berg, H. Bouwmeester and P. Tromp, et al., Translocation of positively and negatively charged polystyrene nanoparticles in an in vitro placental model, Toxicol. In Vitro, 2015, 29(7), 1701–1710 CrossRef CAS.
- M. Kokkinopoulou, J. Simon, K. Landfester, V. Mailander and I. Lieberwirth, Visualization of the protein corona: towards a biomolecular understanding of nanoparticle-cell-interactions, Nanoscale, 2017, 9(25), 8858–8870 RSC.
- F. Meng, G. H. Engbers and J. Feijen, Polyethylene glycol-grafted polystyrene particles, J. Biomed. Mater. Res., 2004, 70(1), 49–58 CrossRef.
- J. Müller, K. N. Bauer, D. Prozeller, J. Simon, V. Mailander and F. R. Wurm, et al., Coating nanoparticles with tunable surfactants facilitates control over the protein corona, Biomaterials, 2017, 115, 1–8 CrossRef PubMed.
- L. Canesi, C. Ciacci, R. Fabbri, T. Balbi, A. Salis and G. Damonte, et al., Interactions of cationic polystyrene nanoparticles with marine bivalve hemocytes in a physiological environment: Role of soluble hemolymph proteins, Environ. Res., 2016, 150, 73–81 CrossRef CAS PubMed.
- F. Nasser and I. Lynch, Secreted protein eco-corona mediates uptake and impacts of polystyrene nanoparticles on Daphnia magna, J. Proteomics, 2016, 137, 45–51 CrossRef CAS PubMed.
- M. T. Ortega, J. E. Riviere, K. Choi and N. A. Monteiro-Riviere, Biocorona formation on gold nanoparticles modulates human proximal tubule kidney cell uptake, cytotoxicity and gene expression, Toxicol. In Vitro, 2017, 42, 150–160 CrossRef CAS PubMed.
- J. Piella, N. G. Bastus and V. Puntes, Size-Dependent Protein-Nanoparticle Interactions in Citrate-Stabilized Gold Nanoparticles: The Emergence of the Protein Corona, Bioconjugate Chem., 2017, 28(1), 88–97 CrossRef CAS PubMed.
- K. Saha, M. Rahimi, M. Yazdani, S. T. Kim, D. F. Moyano and S. Hou, et al., Regulation of Macrophage Recognition through the Interplay of Nanoparticle Surface Functionality and Protein Corona, ACS Nano, 2016, 10(4), 4421–4430 CrossRef CAS.
- U. Sakulkhu, M. Mahmoudi, L. Maurizi, G. Coullerez, M. Hofmann-Amtenbrink and M. Vries, et al., Significance of surface charge and shell material of superparamagnetic iron oxide nanoparticle (SPION) based core/shell nanoparticles on the composition of the protein corona, Biomater. Sci., 2015, 3(2), 265–278 RSC.
- R. R. Arvizo, K. Giri, D. Moyano, O. R. Miranda, B. Madden and D. J. McCormick, et al., Identifying new therapeutic targets via modulation of protein corona formation by engineered nanoparticles, PLoS One, 2012, 7(3), e33650 CrossRef CAS.
- V. Escamilla-Rivera, M. Uribe-Ramirez, S. Gonzalez-Pozos, O. Lozano, S. Lucas and A. De Vizcaya-Ruiz, Protein corona acts as a protective shield against Fe3O4-PEG inflammation and ROS-induced toxicity in human macrophages, Toxicol. Lett., 2016, 240(1), 172–184 CrossRef CAS.
- C. Grafe, A. Weidner, M. V. Luhe, C. Bergemann, F. H. Schacher and J. H. Clement, et al., Intentional formation of a protein corona on nanoparticles: Serum concentration affects protein corona mass, surface charge, and nanoparticle-cell interaction, Int. J. Biochem. Cell Biol., 2016, 75, 196–202 CrossRef PubMed.
- H. Groult, J. Ruiz-Cabello, A. V. Lechuga-Vieco, J. Mateo, M. Benito and I. Bilbao, et al., Phosphatidylcholine-coated iron oxide nanomicelles for in vivo prolonged circulation time with an antibiofouling protein corona, Chemistry, 2014, 20(50), 16662–16671 CrossRef CAS PubMed.
- A. Hill and C. K. Payne, Impact of Serum Proteins on MRI Contrast Agents: Cellular Binding and T2 relaxation, RSC Adv., 2014, 4, 31735–31744 RSC.
- C. Vogt, M. Pernemalm, P. Kohonen, S. Laurent, K. Hultenby and M. Vahter, et al., Proteomics Analysis Reveals Distinct Corona Composition on Magnetic Nanoparticles with Different Surface Coatings: Implications for Interactions with Primary Human Macrophages, PLoS One, 2015, 10(10), e0129008 CrossRef PubMed.
- X. Ma, C. Ding, X. Yao and L. Jia, Ethylene glycol assisted preparation of Ti(4+)-modified polydopamine coated magnetic particles with rough surface for capture of phosphorylated proteins, Anal. Chim. Acta, 2016, 929, 23–30 CrossRef CAS PubMed.
- Q. Gai, F. Qu, T. Zhang and Y. Zhang, Integration of carboxyl modified magnetic particles and aqueous two-phase extraction for selective separation of proteins, Talanta, 2011, 85(1), 304–309 CrossRef CAS PubMed.
- A. L. Barran-Berdon, D. Pozzi, G. Caracciolo, A. L. Capriotti, G. Caruso and C. Cavaliere, et al., Time evolution of nanoparticle-protein corona in human plasma: relevance for targeted drug delivery, Langmuir, 2013, 29(21), 6485–6494 CrossRef CAS PubMed.
- M. Jansch, A. B. Jindal, B. M. Sharmila, A. Samad, P. V. Devarajan and R. H. Muller, Influence of particle shape on plasma protein adsorption and macrophage uptake, Pharmazie, 2013, 68(1), 27–33 CAS.
- K. W. Kasongo, M. Jansch, R. H. Muller and R. B. Walker, Evaluation of the in vitro differential protein adsorption patterns of didanosine-loaded nanostructured lipid carriers (NLCs) for potential targeting to the brain, J. Liposome Res., 2011, 21(3), 245–254 CrossRef CAS PubMed.
- N. R. Kuznetsova and E. L. Vodovozova, Differential binding of plasma proteins by liposomes loaded with lipophilic prodrugs of methotrexate and melphalan in the bilayer, Biochemistry, 2014, 79(8), 797–804 CAS.
- M. Luck, W. Schroder, S. Harnisch, K. Thode, T. Blunk and B. R. Paulke, et al., Identification of plasma proteins facilitated by enrichment on particulate surfaces: analysis by two-dimensional electrophoresis and N-terminal microsequencing, Electrophoresis, 1997, 18(15), 2961–2967 CrossRef CAS PubMed.
- D. E. MacDonald, N. Deo, B. Markovic, M. Stranick and P. Somasundaran, Adsorption and dissolution behavior of human plasma fibronectin on thermally and chemically modified titanium dioxide particles, Biomaterials, 2002, 23(4), 1269–1279 CrossRef CAS PubMed.
- A. Marquez, T. Berger, A. Feinle, N. Husing, M. Himly and A. Duschl, et al., Bovine Serum Albumin Adsorption on TiO2 Colloids: The Effect of Particle Agglomeration and Surface Composition, Langmuir, 2017, 33(10), 2551–2558 CrossRef CAS PubMed.
- L. Song, K. Yang, W. Jiang, P. Du and B. Xing, Adsorption of bovine serum albumin on nano and bulk oxide particles in deionized water, Colloids Surf., B, 2012, 94, 341–346 CrossRef CAS PubMed.
- J. Sund, H. Alenius, M. Vippola, K. Savolainen and A. Puustinen, Proteomic characterization of engineered nanomaterial-protein interactions in relation to surface reactivity, ACS Nano, 2011, 5(6), 4300–4309 CrossRef CAS PubMed.
- S. Xiong, S. George, Z. Ji, S. Lin, H. Yu and R. Damoiseaux, et al., Size of TiO(2) nanoparticles influences their phototoxicity: an in vitro investigation, Arch. Toxicol., 2013, 87(1), 99–109 CrossRef CAS PubMed.
- D. Pasqui, L. Golini, C. D. Giovampaola, A. Atrei and R. Barbucci, Chemical and biological properties of polysaccharide-coated titania nanoparticles: the key role of proteins, Biomacromolecules, 2011, 12(4), 1243–1249 CrossRef CAS PubMed.
- H. Whitwell, R. M. Mackay, C. Elgy, C. Morgan, M. Griffiths and H. Clark, et al., Nanoparticles in the lung and their protein corona: the few proteins that count, Nanotoxicology, 2016, 10(9), 1385–1394 CrossRef CAS PubMed.
- J. Yu, H. J. Kim, M. R. Go, S. H. Bae and S. J. Choi, ZnO Interactions with Biomatrices: Effect of Particle Size on ZnO-Protein Corona, Nanomaterials, 2017, 7(11), 377 CrossRef PubMed.
- N. V. Konduru, K. M. Murdaugh, A. Swami, R. J. Jimenez, T. C. Donaghey and P. Demokritou, et al., Surface modification of zinc oxide nanoparticles with amorphous silica alters their fate in the circulation, Nanotoxicology, 2016, 10(6), 720–727 CrossRef CAS PubMed.
- M. Luo, C. Shen, B. N. Feltis, L. L. Martin, A. E. Hughes and P. F. Wright, et al., Reducing ZnO nanoparticle cytotoxicity by surface modification, Nanoscale, 2014, 6(11), 5791–5798 RSC.
- H. Sieg, C. Braeuning, B. M. Kunz, H. Daher, C. Kastner and B. C. Krause, et al., Uptake and molecular impact of aluminum-containing nanomaterials on human intestinal caco-2 cells, Nanotoxicology, 2018, 12(9), 992–1013 CrossRef CAS PubMed.
- K. Kandori, T. Kuroda, S. Togashi and E. Katayama, Preparation of calcium hydroxyapatite nanoparticles using microreactor and their characteristics of protein adsorption, J. Phys. Chem. B, 2011, 115(4), 653–659 CrossRef CAS PubMed.
- P. Foroozandeh, A. A. Aziz and M. Mahmoudi, Effect of Cell Age on Uptake and Toxicity of Nanoparticles: The Overlooked Factor at the Nanobio Interface, ACS Appl. Mater. Interfaces, 2019, 11(43), 39672–39687 CrossRef CAS PubMed.
- R. Petry, V. M. Saboia, L. S. Franqui, C. A. Holanda, T. R. R. Garcia and M. A. de Farias, et al., On the formation of protein corona on colloidal nanoparticles stabilized by depletant polymers, Mater. Sci. Eng., C, 2019, 105, 110080 CrossRef CAS PubMed.
- I. Matei, C. M. Buta, I. M. Turcu, D. Culita, C. Munteanu and G. Ionita, Formation and Stabilization of Gold Nanoparticles in Bovine Serum Albumin Solution, Molecules, 2019, 24(18), 3395 CrossRef PubMed.
- N. V. Konduru, R. J. Jimenez, A. Swami, S. Friend, V. Castranova and P. Demokritou, et al., Silica coating influences the corona and biokinetics of cerium oxide nanoparticles, Part. Fibre Toxicol., 2015, 12, 31 CrossRef PubMed.
- D. Pozzi, G. Caracciolo, L. Digiacomo, V. Colapicchioni, S. Palchetti and A. L. Capriotti, et al., The biomolecular corona of nanoparticles in circulating biological media, Nanoscale, 2015, 7(33), 13958–13966 RSC.
- X. D. Zhu, H. J. Zhang, H. S. Fan, W. Li and X. D. Zhang, Effect of phase composition and microstructure of calcium phosphate ceramic particles on protein adsorption, Acta Biomater., 2010, 6(4), 1536–1541 CrossRef CAS PubMed.
- F. Meder, T. Daberkow, L. Treccani, M. Wilhelm, M. Schowalter and A. Rosenauer, et al., Protein adsorption on colloidal alumina particles functionalized with amino, carboxyl, sulfonate and phosphate groups, Acta Biomater., 2012, 8(3), 1221–1229 CrossRef CAS PubMed.
- J. Martel, C. Y. Wu, C. Y. Hung, T. Y. Wong, A. J. Cheng and M. L. Cheng, et al., Fatty acids and small organic compounds bind to mineralo-organic nanoparticles derived from human body fluids as revealed by metabolomic analysis, Nanoscale, 2016, 8(10), 5537–5545 RSC.
- D. Bonvin, U. Aschauer, D. T. L. Alexander, D. Chiappe, M. Moniatte and H. Hofmann, et al., Protein Corona: Impact of Lymph Versus Blood in a Complex In Vitro Environment, Small, 2017, 13(29), 1700409 CrossRef PubMed.
- D. Di Silvio, N. Rigby, B. Bajka, A. Mackie and F. Baldelli Bombelli, Effect of protein corona magnetite nanoparticles derived from bread in vitro digestion on Caco-2 cells morphology and uptake, Int. J. Biochem. Cell Biol., 2016, 75, 212–222 CrossRef CAS PubMed.
- M. Aoyama, K. Hata, K. Higashisaka, K. Nagano, Y. Yoshioka and Y. Tsutsumi, Clusterin in the protein corona plays a key role in the stealth effect of nanoparticles against phagocytes, Biochem. Biophys. Res. Commun., 2016, 480(4), 690–695 CrossRef CAS PubMed.
- M. Lundqvist, C. Augustsson, M. Lilja, K. Lundkvist, B. Dahlback and S. Linse, et al., The nanoparticle protein corona formed in human blood or human blood fractions, PLoS One, 2017, 12(4), e0175871 CrossRef PubMed.
- M. S. Ehrenberg, A. E. Friedman, J. N. Finkelstein, G. Oberdorster and J. L. McGrath, The influence of protein adsorption on nanoparticle association with cultured endothelial cells, Biomaterials, 2009, 30(4), 603–610 CrossRef CAS PubMed.
- W. Jiang, K. Lai, Y. Wu and Z. Gu, Protein corona on magnetite nanoparticles and internalization of nanoparticle-protein complexes into healthy and cancer cells, Arch. Pharmacal Res., 2014, 37(1), 129–141 CrossRef CAS PubMed.
- C. L. Oliveira, F. Veiga, C. Varela, F. Roleira, E. Tavares and I. Silveira, et al., Characterization of polymeric nanoparticles for intravenous delivery: Focus on stability, Colloids Surf., B, 2017, 150, 326–333 CrossRef CAS PubMed.
- L. K. Muller, J. Simon, C. Rosenauer, V. Mailander, S. Morsbach and K. Landfester, The Transferability from Animal Models to Humans: Challenges Regarding Aggregation and Protein Corona Formation of Nanoparticles, Biomacromolecules, 2018, 19(2), 374–385 CrossRef PubMed.
- D. A. Mbeh, L. K. Mireles, D. Stanicki, L. Tabet, K. Maghni and S. Laurent, et al., Human alveolar epithelial cell responses to core-shell superparamagnetic iron oxide nanoparticles (SPIONs), Langmuir, 2015, 31(13), 3829–3839 CrossRef CAS PubMed.
- L. Landgraf, C. Christner, W. Storck, I. Schick, I. Krumbein and H. Dahring, et al., A plasma protein corona enhances the biocompatibility of Au@Fe3O4 Janus particles, Biomaterials, 2015, 68, 77–88 CrossRef CAS PubMed.
- J. Liu, F. Zeng and C. Allen, Influence of serum protein on polycarbonate-based copolymer micelles as a delivery system for a hydrophobic anti-cancer agent, J. Controlled Release, 2005, 103(2), 481–497 CrossRef CAS PubMed.
- G. Miotto, M. Magro, M. Terzo, M. Zaccarin, L. Da Dalt and E. Bonaiuto, et al., Protein corona as a proteome fingerprint: The example of hidden biomarkers for cow mastitis, Colloids Surf., B, 2016, 140, 40–49 CrossRef CAS PubMed.
- F. Lu, A. Mencia, L. Bi, A. Taylor, Y. Yao and H. HogenEsch, Dendrimer-like alpha-d-glucan nanoparticles activate dendritic cells and are effective vaccine adjuvants, J. Controlled Release, 2015, 204, 51–59 CrossRef CAS PubMed.
- M. Mahmoudi, N. Bertrand, H. Zope and O. C. Farokhzad, Emerging understanding of the protein corona at the nano-bio interfaces, Nano Today, 2016, 11(6), 817–832 CrossRef CAS.
- A. Weidner, C. Grafe, M. von der Luhe, H. Remmer, J. H. Clement and D. Eberbeck, et al., Preparation of Core-Shell Hybrid Materials by Producing a Protein Corona Around Magnetic Nanoparticles, Nanoscale Res. Lett., 2015, 10(1), 992 CrossRef CAS PubMed.
- S. Deville, B. Bare, J. Piella, K. Tirez, P. Hoet and M. P. Monopoli, et al., Interaction of gold nanoparticles and nickel(II) sulfate affects dendritic cell maturation, Nanotoxicology, 2016, 10(10), 1395–1403 CrossRef CAS PubMed.
- F. Branda, B. Silvestri, A. Costantini and G. Luciani, The fate of silica based Stober particles soaked into growth media (RPMI and M254): A DLS and zeta-potential study, Colloids Surf., B, 2015, 135, 840–845 CrossRef CAS PubMed.
- S. Tenzer, D. Docter, J. Kuharev, A. Musyanovych, V. Fetz and R. Hecht, et al., Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology, Nat. Nanotechnol., 2013, 8(10), 772–781 CrossRef CAS PubMed.
- Z. S. Al-Ahmady, M. Hadjidemetriou, J. Gubbins and K. Kostarelos, Formation of protein corona in vivo affects drug release from temperature-sensitive liposomes, J. Controlled Release, 2018, 276, 157–167 CrossRef CAS PubMed.
- R. Garcia-Alvarez, M. Hadjidemetriou, A. Sanchez-Iglesias, L. M. Liz-Marzan and K. Kostarelos, In vivo formation of protein corona on gold nanoparticles. The effect of their size and shape, Nanoscale, 2018, 10(3), 1256–1264 RSC.
- N. V. Konduru, R. M. Molina, A. Swami, F. Damiani, G. Pyrgiotakis and P. Lin, et al., Protein corona: implications for nanoparticle interactions with pulmonary cells, Part. Fibre Toxicol., 2017, 14(1), 42 CrossRef PubMed.
- G. Caracciolo, L. Callipo, S. C. De Sanctis, C. Cavaliere, D. Pozzi and A. Lagana, Surface adsorption of protein corona controls the cell internalization mechanism of DC-Chol-DOPE/DNA lipoplexes in serum, Biochim. Biophys. Acta, 2010, 1798(3), 536–543 CrossRef CAS PubMed.
- M. Mahmoudi, A. M. Abdelmonem, S. Behzadi, J. H. Clement, S. Dutz and M. R. Ejtehadi, et al., Temperature: the “ignored” factor at the NanoBio interface, ACS Nano, 2013, 7(8), 6555–6562 CrossRef CAS PubMed.
- M. Mahmoudi, M. A. Shokrgozar and S. Behzadi, Slight temperature changes affect protein affinity and cellular uptake/toxicity of nanoparticles, Nanoscale, 2013, 5(8), 3240–3244 RSC.
- D. Docter, U. Distler, W. Storck, J. Kuharev, D. Wunsch and A. Hahlbrock, et al., Quantitative profiling of the protein coronas that form around nanoparticles, Nat. Protoc., 2014, 9(9), 2030–2044 CrossRef CAS PubMed.
- K. E. Moore, M. Pfohl, F. Hennrich, V. S. Chakradhanula, C. Kuebel and M. M. Kappes, et al., Separation of double-walled carbon nanotubes by size exclusion column chromatography, ACS Nano, 2014, 8(7), 6756–6764 CrossRef CAS PubMed.
- M. Werwie, N. Fehr, X. Xu, T. Basche and H. Paulsen, Comparison of quantum dot-binding protein tags: affinity determination by ultracentrifugation and FRET, Biochim. Biophys. Acta, 2014, 1840(6), 1651–1656 CrossRef CAS PubMed.
- A. Bekdemir and F. Stellacci, A centrifugation-based physicochemical characterization method for the interaction between proteins and nanoparticles, Nat. Commun., 2016, 7, 13121 CrossRef CAS PubMed.
- V. Sherwood, D. Di Silvio and F. B. Bombelli, Nanoscopic Agents in a Physiological Environment: The Importance of Understanding Their Characteristics, Top. Med. Chem., 2016, 20, 29–54 CAS.
- S. R. Saptarshi, A. Duschl and A. L. Lopata, Interaction of nanoparticles with proteins: relation to bio-reactivity of the nanoparticle, J. Nanobiotechnol., 2013, 11 Search PubMed.
- D. Bonvin, D. Chiappe, M. Moniatte, H. Hofmann and M. Mionic Ebersold, Methods of protein corona isolation for magnetic nanoparticles, Analyst, 2017, 142(20), 3805–3815 RSC.
- C. Pisani, J. Gaillard, M. Odorico, J. Nyalosaso, C. Charnay and Y. Guari, et al., Corona interactome: A key for deciphering protein adsorption kinetics on silica nanocarriers surface, Toxicol. Lett., 2016, 258, S281 CrossRef.
- F. E. Luborsky and B. J. Drummond, Buildup of Particles on Fibers in a High Field High Gradient Separator, IEEE Trans. Magn., 1976, 12(5), 463–465 CrossRef.
- U. Hansen and A. F. Thunemann, Characterization of Silver Nanoparticles in Cell Culture Medium Containing Fetal Bovine Serum, Langmuir, 2015, 31(24), 6842–6852 CrossRef CAS PubMed.
- R. L. M. Synge and A. Tiselius, Fractionation of Hydrolysis Products of Amylose by Electrokinetic Ultrafiltration in an Agar-Agar Jelly, Biochem. J., 1950, 46(5), R41–R42 Search PubMed.
- A. L. Capriotti, G. Caracciolo, C. Cavaliere, V. Colapicchioni, S. Piovesana and D. Pozzi, et al., Analytical Methods for Characterizing the Nanoparticle-Protein Corona, Chromatographia, 2014, 77(11–12), 755–769 CrossRef CAS.
- G. Yohannes, M. Jussila, K. Hartonen and M. L. Riekkola, Asymmetrical flow field-flow fractionation technique for separation and characterization of biopolymers and bioparticles, J. Chromatogr. A, 2011, 1218(27), 4104–4116 CrossRef CAS PubMed.
- J. C. Giddings, X. R. Chen, K. G. Wahlund and M. N. Myers, Fast Particle Separation by Flow Steric Field-Flow Fractionation, Anal. Chem., 1987, 59(15), 1957–1962 CrossRef CAS.
- K. G. Wahlund and J. C. Giddings, Properties of an Asymmetrical Flow Field-Flow Fractionation Channel Having One Permeable Wall, Anal. Chem., 1987, 59(9), 1332–1339 CrossRef CAS PubMed.
- M. Wagner, S. Holzschuh, A. Traeger, A. Fahr and U. S. Schubert, Asymmetric Flow Field-Flow Fractionation in the Field of Nanomedicine, Anal. Chem., 2014, 86(11), 5201–5210 CrossRef CAS PubMed.
- M. P. Monopoli, A. S. Pitek, I. Lynch and K. A. Dawson, Formation and characterization of the nanoparticle-protein corona, Methods Mol. Biol., 2013, 1025, 137–155 CrossRef CAS PubMed.
- C. Rocker, M. Potzl, F. Zhang, W. J. Parak and G. U. Nienhaus, A quantitative fluorescence study of protein monolayer formation on colloidal nanoparticles, Nat. Nanotechnol., 2009, 4(9), 577–580 CrossRef PubMed.
- S. Milani, F. B. Bombelli, A. S. Pitek, K. A. Dawson and J. Radler, Reversible versus Irreversible Binding of Transferrin to Polystyrene Nanoparticles: Soft and Hard Corona, ACS Nano, 2012, 6(3), 2532–2541 CrossRef CAS PubMed.
- M. Horie, K. Nishio, K. Fujita, S. Endoh, A. Miyauchi and Y. Saito, et al., Protein adsorption of ultrafine metal oxide and its influence on cytotoxicity toward cultured cells, Chem. Res. Toxicol., 2009, 22(3), 543–553 Search PubMed.
- R. De Palma, J. Trekker, S. Peeters, M. J. Van Bael, K. Bonroy and R. Wirix-Speetjens, et al., Surface modification of gamma-Fe2O3@SiO2 magnetic nanoparticles for the controlled interaction with biomolecules, J. Nanosci. Nanotechnol., 2007, 7(12), 4626–4641 CAS.
- S. Staufenbiel, M. Merino, W. Li, M. D. Huang, S. Baudis and A. Lendlein, et al., Surface characterization and protein interaction of a series of model poly[acrylonitrile-co-(N-vinyl pyrrolidone)] nanocarriers for drug targeting, Int. J. Pharm., 2015, 485(1–2), 87–96 CrossRef CAS PubMed.
- A. Elaissari, L. Holt, F. Meunier, C. Voisset, C. Pichot and B. Mandrand, et al., Hydrophilic and cationic latex particles for the specific extraction of nucleic acids, J. Biomater. Sci., Polym. Ed., 1999, 10(4), 403–420 CrossRef CAS PubMed.
- T. X. Zhang, G. Y. Zhu, B. Y. Lu, C. L. Zhang and Q. Peng, Concentration-dependent protein adsorption at the nano-bio interfaces of polymeric nanoparticles and serum proteins, Nanomedicine, 2017, 12(22), 2757–2769 CrossRef CAS PubMed.
- F. Pederzoli, G. Tosi, M. A. Vandelli, D. Belletti, F. Forni and B. Ruozi, Protein corona and nanoparticles: how can we investigate on?, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2017, 9(6), e1467 Search PubMed.
- C. Carrillo-Carrion, M. Carril and W. J. Parak, Techniques for the experimental investigation of the protein corona, Curr. Opin. Biotechnol., 2017, 46, 106–113 CrossRef CAS PubMed.
- J. D. Robertson, L. Rizzello, M. Avila-Olias, J. Gaitzsch, C. Contini and M. S. Magon, et al., Purification of Nanoparticles by Size and Shape, Sci. Rep., 2016, 6, 27494 CrossRef CAS PubMed.
- O. Akbulut, C. R. Mace, R. V. Martinez, A. A. Kumar, Z. H. Nie and M. R. Patton, et al., Separation of Nanoparticles in Aqueous Multiphase Systems through Centrifugation, Nano Lett., 2012, 12(8), 4060–4064 CrossRef CAS.
- L. Pitkanen and A. M. Striegel, Size-exclusion chromatography of metal nanoparticles and quantum dots, TrAC, Trends Anal. Chem., 2016, 80, 311–320 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2020 |