Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Extraction of chemical structures from literature and patent documents using open access chemistry toolkits: a case study with PFAS†
Shadrack J.
Barnabas
 a,
Timo
Böhme
a,
Stephen K.
Boyer
a,
Timo
Böhme
a,
Stephen K.
Boyer
 b,
Matthias
Irmer
b,
Matthias
Irmer
 a,
Christoph
Ruttkies
a,
Christoph
Ruttkies
 a,
Ian
Wetherbee
c,
Todor
Kondić
a,
Ian
Wetherbee
c,
Todor
Kondić
 d,
Emma L.
Schymanski
d,
Emma L.
Schymanski
 *d and
Lutz
Weber
*d and
Lutz
Weber
 *a
*a
aOntoChem GmbH, Blücherstrasse 24, 06120 Halle (Saale), Germany. E-mail: lutz.weber@ontochem.com
bCollabra Inc., San Jose, CA 95120, USA
cGoogle LLC, 1600 Amphitheatre Parkway, Mountain View, CA 94043, USA
dLuxembourg Centre for Systems Biomedicine (LCSB), University of Luxembourg, 6 avenue du Swing, L-4367 Belvaux, Luxembourg. E-mail: emma.schymanski@uni.lu
First published on 31st May 2022
Abstract
The extraction of chemical information from documents is a demanding task in cheminformatics due to the variety of text and image-based representations of chemistry. The present work describes the extraction of chemical compounds with unique chemical structures from the open access CORE (COnnecting REpositories) and Google Patents full text document repositories. The importance of structure normalization is demonstrated using three open access cheminformatics toolkits: the Chemistry Development Kit (CDK), RDKit and OpenChemLib (OCL). Each toolkit was used for structure parsing, normalization and subsequent substructure searching, using SMILES as structure representations of chemical molecules and International Chemical Identifiers (InChIs) for comparison. Per- and polyfluoroalkyl substances (PFAS) were chosen as a case study to perform the substructure search, due to their high environmental relevance, their presence in both literature and patent corpuses, and the current lack of community consensus on their definition. Three different structural definitions of PFAS were chosen to highlight the implications of various definitions from a cheminformatics perspective. Since CDK, RDKit and OCL implement different criteria and methods for SMILES parsing and normalization, different numbers of parsed compounds were extracted, which were then evaluated using the three PFAS definitions. A comparison of these toolkits and definitions is provided, along with a discussion of the implications for PFAS screening and text mining efforts in cheminformatics. Finally, the extracted PFAS (∼1.7 M PFAS from patents and ∼27 K from CORE) were compared against various existing PFAS lists and are provided in various formats for further community research efforts.
Introduction
Per- and polyfluoroalkyl substances (PFAS) are compounds of high public interest as there is increasing evidence that exposure to PFAS can lead to adverse human and environmental health effects.1,2 These concerns are accompanied by their documented accumulation in the environment (as so-called “forever chemicals”) due to their widespread use and stability.3 Well-known PFAS include older PFAS such as PFOA (perfluorooctanoic acid) and PFOS (perfluorooctane sulfonic acid), as well as newer PFAS such as GenX (a replacement product for the older PFAS). There is strong regulatory debate about PFAS, including calls to regulate them as a class4 and for better approaches to detect PFAS in humans and in the environment. Since PFAS and replacement PFAS products are a fast-moving business, cheminformatics tools are gaining importance in identifying candidate PFAS compounds from within scientific and other text sources such as patent repositories, including in-house confidential business documentation.Past efforts to identify and collect chemical structures of existing PFAS have resulted in several so-called “suspect” lists. The Organisation for Economic Co-operation and Development (OECD) released a PFAS list containing 4729 PFAS entities in 2017 (ref. 5 and 6) (hereafter “OECDPFAS”). The United States Environmental Protection Agency (EPA) “PFASMASTER” list currently (December 2021) contains 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 048 PFAS entries,7 merged from several PFAS lists on the EPA CompTox Chemicals Dashboard.8 Of these two lists, PFASMASTER contains 10
048 PFAS entries,7 merged from several PFAS lists on the EPA CompTox Chemicals Dashboard.8 Of these two lists, PFASMASTER contains 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 785 entries that can be represented by an International Chemical Identifier (InChI), while the OECDPFAS list contains 3741 entries with an InChI, using versions downloaded from the EPA website on 2021-12-11 (ref. 7 and 9) and provided in ref. 10 The other entities in the lists are substances without a clear composition, or with known composition that cannot be represented fully with an InChI. Of the 3741 OECD compounds with an InChI, 3731 are also contained in the PFASMASTER list (by matching InChI).
785 entries that can be represented by an International Chemical Identifier (InChI), while the OECDPFAS list contains 3741 entries with an InChI, using versions downloaded from the EPA website on 2021-12-11 (ref. 7 and 9) and provided in ref. 10 The other entities in the lists are substances without a clear composition, or with known composition that cannot be represented fully with an InChI. Of the 3741 OECD compounds with an InChI, 3731 are also contained in the PFASMASTER list (by matching InChI).
These lists and more are used in environmental assessments to gauge the extent of the “PFAS knowledge gap”. Such lists serve additional purposes, e.g., to search for the respective compounds in analytical data of environmental samples.11 The majority of PFAS suspect lists are hand curated, painstakingly compiled by experts and thus limited both by access to relevant information and by the manual nature of the efforts. Since the current definition of PFAS is strongly debated by the community, three different structural definitions of PFAS in use have been considered in this case study, clarified below and shown in Fig. 1:
 | ||
| Fig. 1 Schematic representation of the PFAS definitions A, B and C considered in this work. “AH” = hydrogen or any other atom; R1, R2, R3 represent any atom other than hydrogen. | ||
Definition A
Each compound that contains a CF2 group is considered a PFAS. This definition has been proposed recently by the OECD.12,13 This definition will lead to a large amount of chemicals that are considered to be PFAS.Definition B
Each compound that contains a (AH)(AH)(F)C–C(AH)F2 group is considered a PFAS, where the AH groups could be hydrogen or any other atom and the bond between both aliphatic carbon atoms is a single bond. This definition is used in this present work as a straightforward structural definition as a compromise between definitions A and C.Definition C
Each compound that contains a (R1)(R2)(F)C–C(R3)F2 group is considered a PFAS, where the R groups are any atom except hydrogen and the bond between both aliphatic carbon atoms is a single bond. This is a new, very recent EPA definition.14,15 This definition will lead to the least amount of PFAS molecules.Extracting chemical information from text documents is a challenging task. Unlike other natural language terms, chemistry-related terms pose additional challenges, as the number of known chemical compounds with unique structures is not only very high (e.g. PubChem16 currently contains 111 M unique compounds, which is only a tiny fraction of the estimated chemical space) but they may appear in text documents with a multiplicity of trivial names. Examples include perfluorooctanesulfonic acid (PFOS), International Union of Pure and Applied Chemistry (IUPAC) names (e.g. 1,1,2,2,3,3,4,4,5,5,6,6,7,7,8,8,8-heptadecafluorooctane-1-sulfonic acid), mixtures of trivial and IUPAC naming, enumerations of Markush17 structures, trade names and half formulas (e.g. Krytox oils, F–(CF(CF3)–CF2–O)n–CF2CF3 where n = 10–60), database identifiers such as Chemical Abstract Service (CAS) registry numbers (e.g. 1763-23-1), PubChem Compound Identifiers (CIDs, e.g. 74483), and even images that are referenced in the text with simple numeric labels. Advanced and flexible methods are required to capture all types of chemical information, with subsequent cheminformatic manipulation to ensure correct mapping to detailed structural information.
The automated analysis of the increasing number of accessible scientific documents may provide input to fuel scientific studies to identify novel molecules with potentially desired or undesired properties. OC|processor18 is a modular semantic annotation toolkit, based on Apache UIMA.19 It is designed to annotate different document types such as PDF, images, HTML, XML, MS Office and plain text documents. It uses a range of established dictionaries and ontologies as well as rule-based algorithms to annotate and index scientific named entities such as diseases, genes, species and chemistry. The properties of concept synonyms as well as the hierarchy of ontological concepts are taken into account to provide more accurate context sensitive annotation. For example, the term “sting” could be annotated as a known musician, a species, a disease or a protein. OC|processor disambiguates based on the term environment and the presence of related concepts, assigning the annotation/knowledge domain with the highest confidence value. The precision and recall of OC|processor has been detailed elsewhere.20 For this study, the growing bodies of open access document repository CORE21,22 (COnnecting REpositories) and patent full text documents in Google Patents23 were selected to demonstrate the automated capability of identifying and analyzing scientific entities, applied to the case study of potential PFAS in documents. OC|processor18 was used to automatically identify and extract mentions of chemical compounds from patents and other open access scientific documents such as scientific articles and university documents in CORE. The resulting collection of diverse chemical compounds was subsequently filtered for small molecule compounds for which a unique InChI24 could be generated, thus removing incompletely-defined structures such as substances, polymers as well as mentions of chemical class terms and Markush-like17 structures. Of the three definitions presented above, definition B was used for most of the detailed investigations in this study. The final PFAS lists are available for all 3 definition versions described above and have been made public, together with additional results, in various formats10,25 (see also data availability) for general assessment and as input for future studies.
Experimental
Semantic annotation and extraction of chemical compounds
OC|processor18 comprises various modules that take the different modalities of chemistry into account, aiming at a comprehensive annotation of chemistry terms in documents. This allows the identification of novel concepts and compounds that were not yet known at the time before annotating a given document. If new compounds are identified, these are registered in Google BigQuery26 tables in the open access SciWalker-Open-Data project, giving access to >150 million small molecules with a unique standard InChI (version 1.03).24 These unique InChIs were generated from connection tables generated from the SMILES27–29 representations of chemical structures. SMILES containing a wildcard entry (i.e. “*”) were considered as representing a scaffold containing an undefined substituent and were not registered. Thus, the current approach is limited by the expressivity of SMILES as well as the InChI rules. For example, standard InChI will represent different tautomers of a molecule as one unique structure, while neither SMILES nor InChI consider coordinate (dative) or hydrogen bonds. Since valence isomerism is not handled by either system, this would result in different structures for molecules exhibiting valence isomerism.30 Hereafter, the use of “unique InChI” or InChI in this manuscript refers to a unique standard InChI (version 1.03).Document sets
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 963
963![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 421 de-duplicated documents were selected and downloaded from the CORE document set of open access documents.22 These documents, when annotated with OC|processor, resulted in the annotation of 818
421 de-duplicated documents were selected and downloaded from the CORE document set of open access documents.22 These documents, when annotated with OC|processor, resulted in the annotation of 818![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 compounds with an unique InChI.31 The SMILES extracted from CORE are from the text only, images were not extracted.
280 compounds with an unique InChI.31 The SMILES extracted from CORE are from the text only, images were not extracted.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 730
730![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 728 Google Patent documents semantically annotated with OC|processor in May 2021 using both the text and images found in these patents was used. The resulting annotations are available in a BigQuery table32 dated May 13, 2021 (see Big Query32patents-public-data in the google_patents_research dataset and table annotations_202105). In total, 51
728 Google Patent documents semantically annotated with OC|processor in May 2021 using both the text and images found in these patents was used. The resulting annotations are available in a BigQuery table32 dated May 13, 2021 (see Big Query32patents-public-data in the google_patents_research dataset and table annotations_202105). In total, 51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 928
928![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 230
230![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 588 annotations were found. Of those, 4
588 annotations were found. Of those, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 533
533![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 988
988![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 229 were compound annotations with associated SMILES and InChI. Of these 4.5 billion annotations, 18
229 were compound annotations with associated SMILES and InChI. Of these 4.5 billion annotations, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 032
032![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 261 had an unique InChI33 and respective Ontology Concept IDentifier (OCID)34 in the SciWalker-Open-Data project.35 As a next (pre-filtering) step, the 18
261 had an unique InChI33 and respective Ontology Concept IDentifier (OCID)34 in the SciWalker-Open-Data project.35 As a next (pre-filtering) step, the 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 032
032![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 261 unique compounds from the chemistry annotations of patents were reduced to a dataset of 4
261 unique compounds from the chemistry annotations of patents were reduced to a dataset of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 182
182![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 712 SMILES that contained an “F” character, resulting from a fluorine, iron or francium atom.
712 SMILES that contained an “F” character, resulting from a fluorine, iron or francium atom.
The quality of the chemistry-related annotations from the combined text and image patent data is lower than from the CORE set. Optical structure recognition and extraction from images often leads to erroneous structures such as compounds containing hypervalent atoms or wrong isotopes that arise from poor image quality.
Compound structure normalization
Normalization (or standardization) of compound structure representations is an important step in preparing compounds for further analysis, including reliable substructure searching. Thus, the various effects of parsing the SMILES strings from the steps above to create a molecule object, plus subsequent normalization, were investigated using three different open access chemistry toolkits: RDKit (version 2020.03.2),36 the Chemistry Development Kit (CDK, version 2.5)37,38 and OpenChemLib (OCL, version 2021.11.3).39 The approaches used were:• RDKit: with the two available standardizers – molVS40,41 and rdMol.
• CDK: via SMILES parsing, normalizing the SMILES with the kekulize option.
• OCL: via SMILES parsing and MoleculeStandardizer, writing the SMILES in a kekulized form.
After parsing the input SMILES, the resulting molecule object was again represented as SMILES as an intermediate step before parsing it again and performing the substructure search to classify it as a PFAS or non-PFAS. This procedure has an effect on the parsing results as described below; in a production environment this additional SMILES generation step would probably not be performed.
PFAS substructure search with graph-based atom-by-atom-search (ABAS)
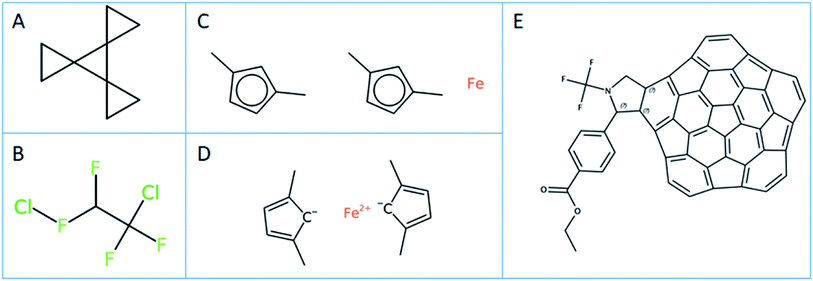
In-house Java code calling the respective CDK and OCL libraries and python scripts based on RDKit were used for the substructure calculations.42 To ensure that the substructure atom-by-atom-search (ABAS) graph based subroutines were implemented correctly, the code was tested using the query and SMILES set mentioned in the RDKit manual. The SMILES structure used to test the implementation was “C1CC12C3(C24CC4)CC3” (PubChem CID 141640; see Fig. 2A). A correct implementation of the SMILES substructure search should return 4 for the SMARTS query “*1**1”.The SMILES query definitions C(F)(F), C(F)(F)C(F) and C(*)(F)(F)C(F)(*)(*) were used to perform the substructure search to define the number of unique PFAS compounds.
PFAS substructure search with fingerprint selection and ABAS
As a first step, molecular fingerprints were calculated for the extracted molecular structures to create a Lucene search index using Apache Lucene in the following manner. Fingerprints (FP) were calculated by the respective toolkit libraries as shown in Table 1. These fingerprints were then stored for each molecule as a “document” in a Lucene index, providing the necessary fingerprint index of the molecules. The fingerprint of the substructure query was then calculated in the same way, followed by searching the Lucene index for candidates. In a second step, the resulting candidate compounds were filtered by ABAS graph-based substructure search from above. Molecules passing both steps were considered as hits. This approach has recently been implemented in Sachem43 storing fingerprint data in an experimental Lucene implementation ported to C. In this study, a standard Lucene implementation in Java 1.8 was used with fingerprint libraries pattern fingerprinter (RDKit), DescriptorHandlerLongFFP512 (OCL) and CDKFingerprinter (CDK). The pattern fingerprint of RDKit uses SMARTS pattern to generate topological fingerprints of molecules.44 The DescriptorHandlerLongFFP512 of OCL is a binary fingerprint that depends on a dictionary of 512 predefined structure fragments.45 The CDKFingerprinter generates one-dimensional bit arrays, where bits are assigned based on the presence of a certain structural feature in a compound.46 The molecules were normalized using the options available in OCL and CDK, and the molVS standardizer for RDKit.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 compound CORE dataset
280 compound CORE dataset
| Toolkit | Normalizer | PFAS definition B: no normalization | PFAS definition B: with normalization | ||||
|---|---|---|---|---|---|---|---|
| True | False | Invalid | True | False | Invalid | ||
| CDK | Built-in | 4163 | 801![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 624 624 |
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 493 493 |
4192 | 814![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 081 081 |
7 |
| OCL | Standardizer | 4192 | 813![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 829 829 |
259 | 4192 | 813![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 834 834 |
254 |
| RDKit | molVS | 4191 | 813![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 463 463 |
626 | 4191 | 813![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 462 462 |
627 |
| RDKit | rdMol | 4191 | 813![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 463 463 |
626 | 4191 | 813![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 090 090 |
999 |
Results and discussion
Compound structure normalization
Several instances of different cheminformatics toolkits producing different normalized SMILES expressions were found. These inconsistencies influence later results and are described below with specific examples.The number of molecules rejected by parsing the SMILES with the different toolkits is quite different. A rejected SMILES cannot be used for subsequent substructure search, potentially reducing the number of identified PFAS molecules. Thus, the quality of the different SMILES parsers was checked by first parsing the input SMILES, then generating the corresponding InChI from the molecule object. In a second step, a normalized SMILES was written from the molecule object, parsed again and the InChI of these “reparsed” SMILES was calculated. Discrepancies between the InChIs from step one and step two in this procedure reveal issues in the quality of the parsing.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) COH or HNC(
COH or HNC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) are incorrect representations of [NH]C(
O) are incorrect representations of [NH]C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)). Since each chemistry toolkit uses somewhat different rules to normalize SMILES, this has an effect on the outcomes on the PFAS substructure search described below. Some normalization tasks may also be performed by specific “standardizer” modules of the toolkits that use rules (with varying degrees of available documentation) to transform SMILES into a normalized form.
O)). Since each chemistry toolkit uses somewhat different rules to normalize SMILES, this has an effect on the outcomes on the PFAS substructure search described below. Some normalization tasks may also be performed by specific “standardizer” modules of the toolkits that use rules (with varying degrees of available documentation) to transform SMILES into a normalized form.
PFAS substructure search (definition B) and effect of prior normalization
The effect of normalization on the PFAS substructure search using definition B (Fig. 1B) on the CORE dataset is given in Table 1. The maximum number of unique PFAS compounds found by CDK and OCL using normalization is the same, i.e. 4192 PFAS (according to definition B). RDKit finds one structure less, which has a SMILES ClFC(F)C(F)(F)Cl (OCID190000011511). This compound structure is actually an incorrect representation of 1,2-dichlorotetrafluoroethane, containing a hypervalent fluorine (see Fig. 2B). This structure was integrated into the OntoChem database of registered compounds when it was found in an early version of the Wikipedia Chemical infobox.50 Meanwhile, this entry has been corrected in Wikipedia Chemistry but still remains as a legacy in the OntoChem compound registry system, waiting for relinking to the correct structure and respective OCID190005899464.In general, the number of SMILES that are not accepted by the different toolkits as valid SMILES are quite different (see “Invalid” entries in Table 1) and also depend on whether or not normalization is used. CDK seems to be more “forgiving” than RDKit and OCL, but only if normalization is used.
Of the 7 SMILES in CDK that are characterized as invalid SMILES representations with normalization, 6 are ferrocenes with coordinative bonds, such as [Fe].Cc1ccc(C)c1.Cc1ccc(C)c1 (OCID190071023137, see Fig. 2C). A meaningful ferrocene SMILES should have an iron with 2 positive charges and two cyclopentadienes with a negative charge like for example [Fe++].CC1=CC=C(C)[C–]1.CC2=CC=C(C)[C–]2 (see Fig. 2D), however this “correct” SMILES does not truly reflect the aromatic structure with a distributed negative charge and its coordinative bonding nature. This problem will be seen for all coordinative compounds, as the current SMILES syntax does not allow for coordinative or hydrogen bonds like they are available in the MDL MOL file version V3000 definitions.51 This is a serious deficiency of the current SMILES notation, excluding most metal complexes from the universe of SMILES and InChI descriptions, and is a topic under discussion within the InChI committee and IUPAC. The 7th invalid SMILES was generated by OSRA, with the hypervalent carbon atoms as shown and discussed in Fig. 3A above (OCID190014261931).
For the 254 SMILES that were found to be invalid SMILES representations by OCL with normalization, all 254 contained an aromatic selenium atom “[se]” in a kekulized, non-aromatic SMILES string. In our opinion, this behaviour is correct, as there is no such thing as a single aromatic atom in a non-aromatic environment. However, this [se] is corrected to [Se] by the other toolkits at the normalization stage. In addition, the non-normalized OCL version finds 259 invalid SMILES – the 254 are as for the normalized OCL, while these 5 additional SMILES include atoms with excessive charges such as [As+8], [As+9], [O+8], [O+9], [I+9], which are corrected to their uncharged forms by the normalizer – a behaviour which likely undesirable. The invalid SMILES for CDK (7) and OCL (254) with normalization are the result of the initial SMILES parsing. The invalid SMILES from RDKit were not investigated further, however, these are provided in ref. 10. for further inspection. It is interesting to note that the number of PFAS compounds does not change when using OCL or RDKit, irrespective of whether normalization is applied or not. However, CDK clearly needs a structure normalization before performing substructure searching.
Mixed toolkit normalization and substructure searching on the CORE dataset
Table 2 presents the results of using different combinations of toolkits for the normalization and subsequent substructure search engines. The first line per toolkit (two lines in the case of RDKit) repeats the results from Table 1, where the normalization and substructure search is performed by the same toolkit. As for Table 1, definition B was used for parsing the PFAS query against the 818![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 CORE compound dataset.
280 CORE compound dataset.
| SSS | Standardizer | True | False | Invalid |
|---|---|---|---|---|
| CDK | CDK normalizer | 4192 |
814![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 081 081
|
7 |
| CDK | OCL standardizer | 4192 | 813![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 834 834 |
256 |
| CDK | RDKit standardizer molVS | 3018 | 266![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 657 657 |
548![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 605 605 |
| CDK | RDKit standardizer rdMol | 3018 | 266![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 862 862 |
548![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 400 |
| OCL | OCL standardizer | 4192 |
813![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 834 834
|
254 |
| OCL | CDK normalizer | 4192 | 814![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 072 072 |
16 |
| OCL | RDKit standardizer molVS | 4191 | 813![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 220 220 |
869 |
| OCL | RDKit standardizer rdMol | 4191 | 813![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 220 220 |
869 |
| RDKit | RDKit standardizer molVS | 4191 |
813![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 462 462
|
627 |
| RDKit | RDKit standardizer rdMol | 4191 |
813![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 090 090
|
999 |
| RDKit | OCL standardizer | 4191 | 813![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 051 051 |
1038 |
| RDKit | CDK normalizer | 4191 | 813![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 453 453 |
636 |
For the CDK, while the combination of RDKit normalization and CDK substructure search does not appear to work well together, the CDK substructure search works well with its own CDK as well as with OCL normalization. For the OCL results, it is interesting to note that the syntactically wrong SMILES with aromatic selenium mentioned above are corrected to non-aromatic by CDK, therefore reducing the number of invalid SMILES for the CDK + OCL combination. For the RDKit results, while the number of identified PFAS molecules was not influenced by the normalization used, the least invalid SMILES were found when using RDKit for both normalization and substructure search. Since the molVS model from RDKit returned fewer invalid entries but the same number of PFAS, this was used subsequently. Not surprisingly, Table 2 shows that it seems to be meaningful to take normalization and substructure search from the same toolkit.
PFAS substructure search (definition B) on the patent dataset
Using the insights gained from Table 2, the larger, more heterogeneous SMILES data set of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 182
182![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 712 SMILES from the patent extraction was investigated. The results of normalization and PFAS substructure search using the CDK, OCL and RDKit toolkits are shown in Table 3.
712 SMILES from the patent extraction was investigated. The results of normalization and PFAS substructure search using the CDK, OCL and RDKit toolkits are shown in Table 3.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 182
182![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 712 patent compound dataset using CDK, OCK and RDKit with PFAS definition B
712 patent compound dataset using CDK, OCK and RDKit with PFAS definition B
| SSS | Standardizer | True | False | Invalid |
|---|---|---|---|---|
| CDK | CDK normalizer | 78![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 412 412 |
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 104 104![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 264 264 |
36 |
| OCL | OCL standardizer | 78![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 411 411 |
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 104 104![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 038 038 |
263 |
| RDKit | molVS | 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 762 762 |
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 988 988![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 584 584 |
118![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 366 366 |
Inspecting the invalid 36 SMILES obtained for the CDK results revealed that all structures are ferrocene type compounds as already observed with the CORE dataset. Of the 263 invalid OCL SMILES, 237 were the already known problematic aromatic selenium compounds within a non-aromatic SMILES, 25 had problems with the assignment of aromatic bonds, while one SMILES contained an incorrect nitrogen notation “[N-13]”. Again, it is interesting to note that the results from OCL and CDK are very close to each other. The invalid RDKit SMILES were too numerous for (detailed) further inspection, but are provided in ref. 10.
PFAS substructure search and effect of prior fingerprint selection
Tools that implement substructure searching for large chemical databases perform this task typically in two steps – first, fingerprints are generated and searched for a list of candidate molecules for step two, a full graph-based search also known as atom-by-atom search (ABAS). The reason for this is that ABAS is a NP complete problem and such searches can take quite some time, depending on the query structure. Thus, to achieve reasonable search results in a short time, the number of ABAS searches needs to be reduced to a minimum, which is achieved by a fast fingerprint compound pre-selection step. Thus, fingerprints should deliver a superset of compound candidates, which are then narrowed down by ABAS to the set of compounds that truly contain that substructure. The smaller the difference between this initial fingerprint list and the number of final compounds, the better and thus the more efficient the applied fingerprint algorithm. As a consequence, many fingerprint algorithms have been developed and optimized for pre-selection.It is not the goal of this work to qualify and compare different fingerprint algorithms, since the described substructure search results were obtained with an ABAS on all compounds of interest (not only on a subset), as accurate results were the prime interest and search time was not an issue. However, a combined compound normalization + fingerprinting + substructure search process was also used to identify PFAS compounds from the extracted structures, as this method would probably be used in the future by typical chemistry database users to identify PFAS compounds. Table 4 shows the effect of fingerprint screening in substructure search for PFAS definitions A, B and C across the two compound datasets (CORE and Patents). It is interesting to note that the combined use of fingerprint selection and subsequent substructure search on the selected list resulted in quite comparable results for all the chemistry toolkits when using the higher quality CORE dataset. The number of identified PFAS is the same for CDK and OCL, slightly lower for RDKit. The CDK fingerprint selection appears to be more efficient than using the OCL or RDKit fingerprints for PFAS definition A and B. For the more strict definition C, OCL fingerprints are most selective. Not surprising is the lower number of identified PFAS for the more heterogeneous patent SMILES dataset, since more molecules are sorted out by the RDKit parser as shown in Table 4.
PFAS hits from the 818![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 compound (CORE) dataset 280 compound (CORE) dataset |
PFAS hits from the 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 182 182![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 712 compound (patent) dataset 712 compound (patent) dataset |
|||
|---|---|---|---|---|
| FP | FP + ABAS | FP | FP + ABAS | |
| Definition A | ||||
| OCL | 58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 132 132 |
27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 287 287 |
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 044 044![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 452 452 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 844 844![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 193 193 |
| CDK | 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 632 632 |
27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 287 287 |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 658 658![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 045 045 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 844 844![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 254 254 |
| RDKit | 300![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 848 848 |
27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 282 282 |
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 047 047![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 047 047 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 792 792![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 598 598 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Definition B | ||||
| OCL | 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 830 830 |
4192 | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 225 225![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 142 142 |
78![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 411 411 |
| CDK | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 922 922 |
4192 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 335 335![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 409 409 |
78![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 412 412 |
| RDKit | 299![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 969 969 |
4191 | 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 041 041![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 432 432 |
75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 762 762 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Definition C | ||||
| OCL | 9043 | 3507 | 472![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 731 731 |
62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 553 553 |
| CDK | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 922 922 |
3507 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 335 335![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 409 409 |
62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 561 561 |
| RDKit | 215![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 514 514 |
3502 | 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 502 502![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 138 138 |
60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 426 426 |
The results of PFAS selection with the combined use of fingerprints and subsequent ABAS selection correspond exactly to the results when using ABAS on all input molecules – with one exception of CDK for definition A where the direct ABAS search finds one structure in addition to the fingerprint + ABAS process, which is OCID190080191030 (PubChem CID 117959248) with a very extensive polycyclic aromatic structure, shown in Fig. 2E.
Finalized PFAS CORE and patent lists via OCL
Since compound structures may be described by syntactically correct SMILES strings but these may represent non-existing compounds, for example if they contain hypervalent atoms or non-existing isotopes (as discussed above), a final cleaning step was implemented based on the results above. Both input sets from CORE and Patents from above were used, along with the following procedure to derive a dataset of both valid normalized and standardized SMILES of PFAS classified molecules according to the three definitions using the OCL toolkit:• Parsing the input SMILES and eliminating erroneous wrong compound structures with hypervalent atoms or wrong isotopes
• Calculating the standard InChI of the input SMILES (“InChI-1”)
• Standardizing the parsed SMILES molecule object, writing a standardized SMILES and calculating the standard InChI of the standardized SMILES (“InChI-2”)
• De-duplicating structures based on “InChI-2”
• Running a ABAS substructure query on the standardized SMILES for PFAS definition A, B and C.
In the CORE set 974 structures were found with a wrong SMILES and 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 627 structures with a changed InChI after normalization using OCL – these were removed from the datasets. In the patent set, 108
627 structures with a changed InChI after normalization using OCL – these were removed from the datasets. In the patent set, 108![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 492 structures had incorrect SMILES and 81
492 structures had incorrect SMILES and 81![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 272 structures had a changed InChI after normalization with OCL.
272 structures had a changed InChI after normalization with OCL.
The results of the normalized structures classified as PFAS are shown in Table 5 and compared with the existing PFASMASTER and OECDPFAS lists (mentioned in the introduction) by InChIKey. The number of entries missing from PubChem was determined by matching InChIKeys in each PFAS dataset and the OCID-PubChem dataset in sciwalker: sciwalker-open-data.chemistry_compounds.ocid_pubchem_cid.
| Total | Not found in PFASMASTER (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 782 InChI) 782 InChI) |
Found in PFASMASTER (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 782 InChI) 782 InChI) |
Found in OECDPFAS (3741 InChI) | Not found in PubChema | |
|---|---|---|---|---|---|
| a Prior to deposition of the entire dataset to PubChem, to fill these gaps. | |||||
| CORE definition A | 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 058 058 |
25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 446 446 |
1612 (1686 IKFB) | 944 (988 IKFB) | 7119 |
| CORE definition B | 4139 | 2652 | 1487 | 939 | 1175 |
| CORE definition C | 3457 | 2095 | 1362 | 931 | 915 |
| Patents definition A | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 783 783![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 651 651 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 780 780![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 041 041 |
3610 | 1529 | 216![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 777 777 |
| Patents definition B | 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 108 108 |
71![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 818 818 |
3290 | 1520 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 809 809 |
| Patents definition C | 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 197 197 |
32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 564 564 |
1633 | 847 | 4882 |
The overlap of the PFAS in the CORE and patent datasets for the different definitions were (A) 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 876; (B) 1806; and (C) 866 PFAS entries, showing that the extraction of data from different sources reveals highly complementary results.
876; (B) 1806; and (C) 866 PFAS entries, showing that the extraction of data from different sources reveals highly complementary results.
The overlaps between the lists extracted here and the existing PFAS lists were much lower than expected. Likewise more entries were missing from PubChem than originally expected, especially for the CORE database. The results were reality checked – here documented with an example for the CORE set using the stringent definition C (915 compounds not in PubChem). One of these 915 compounds includes OCID190080091261 (InChIKey LZICQIXBOVBGMV-UHFFFAOYSA-N), shown in Fig. 4. This was published in a PhD thesis52 in Chemistry and extracted from the document section IV. Experimental part 240 16.8.2 via name to structure from “Trimethyl({4′-[(7,7,8,8,9,9,10,10,11,11,12,12,12-tridecafluorododecyl)oxy]-1,1′-biphenyl-4-yl}ethynyl)silane”, which has been interpreted correctly. This shows the potential for literature mining to capture structures that are real and worthy of further investigation, but not yet known to PFAS researchers or to large open databases such as PubChem.
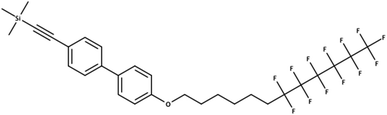 | ||
| Fig. 4 A PFAS classified compound (all definitions) that was indexed in a CORE publication but is not in PubChem (OCID190080091261). | ||
To enhance the discovery of these PFAS in environmental samples, both datasets have been made available as CSV files25 for use in mass spectrometry-based screening approaches, such as MetFrag53 and patRoon.54 Two separate files have been created, for the CORE and patent datasets respectively – with each entry tagged according to the PFAS definition that the given structure satisfies. The CORE dataset additionally includes the number of references in which the structure was found, which can be used for prioritization of candidate matches. The files were formatted as a MetFrag localCSV, where all entries that cause MetFrag to fail (formulas with digits preceding the carbon; certain unusual elements as removed in PubChemLite55) were removed. Where available, names and CIDs were filled in via PubChem, otherwise the OCID was assigned as a name. The resulting files contained 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 695 entries for CORE (of which 5903 entries are without CIDs and 363 entries were removed from the original CORE list) and 1
695 entries for CORE (of which 5903 entries are without CIDs and 363 entries were removed from the original CORE list) and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 778
778![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 470 entries for patents (of which 85
470 entries for patents (of which 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 277 are without CIDs and 5181 entries were removed). The number of PubChem CIDs is higher than above due to the different style of querying; here a combination of FTP files (InChIKey to CID mapping) and REST API (SMILES to CID mapping for remaining entries without CIDs) was used, as the REST API offers the SMILES standardization to match with the final version in PubChem. For the original lists, 5937 CIDs were missing in the CORE set of 27
277 are without CIDs and 5181 entries were removed). The number of PubChem CIDs is higher than above due to the different style of querying; here a combination of FTP files (InChIKey to CID mapping) and REST API (SMILES to CID mapping for remaining entries without CIDs) was used, as the REST API offers the SMILES standardization to match with the final version in PubChem. For the original lists, 5937 CIDs were missing in the CORE set of 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 058 SMILES (21.9%), while 85
058 SMILES (21.9%), while 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 472 CIDs were missing in the patents set of 1
472 CIDs were missing in the patents set of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 783
783![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 651 SMILES (4.8%). The ratio of missing CIDs was very similar in the final MetFrag files. Both datasets were deposited to PubChem (Feb. 12, 2022, submissions 112
651 SMILES (4.8%). The ratio of missing CIDs was very similar in the final MetFrag files. Both datasets were deposited to PubChem (Feb. 12, 2022, submissions 112![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 615 and 112
615 and 112![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 624) to fill these gaps and the CID mappings were updated on April 20, 2022 to include these new CIDs. The MetFrag CSV files are available on Zenodo25 for use in all mass spectrometry workflows, and are also available in the dropdown menu of the MetFrag Web interface (https://msbi.ipb-halle.de/MetFrag/).
624) to fill these gaps and the CID mappings were updated on April 20, 2022 to include these new CIDs. The MetFrag CSV files are available on Zenodo25 for use in all mass spectrometry workflows, and are also available in the dropdown menu of the MetFrag Web interface (https://msbi.ipb-halle.de/MetFrag/).
Comparison of CORE with OECDPFAS classification
Finally, the PFAS structures extracted from the CORE database were investigated using the OECDPFAS classification system via the PubChem Classification Browser56 to determine whether particular PFAS classes were under or over-represented in the extracted data sets compared with the entire OECDPFAS list. The CORE set of 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 058 InChIKeys was uploaded to the PubChem ID Exchange,57 which returned 20
058 InChIKeys was uploaded to the PubChem ID Exchange,57 which returned 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 907 matches via Entrez History. This was then used to browse the NORMAN SLE Classification tree in PubChem.56 Since the influence of searching via InChIKey first block (structural skeleton) versus full InChIKey was not dramatic (only an additional 44 entries found, see row 1 of Table 5), this analysis was kept at the InChIKey level for consistency with the rest of this article. The OECDPFAS list is split into many categories; of primary interest for data extraction is the “Structure Category”, which covers 8 major PFAS categories (denoted 100 through 800), with several subcategories in each. The major categories and the number of matches in CORE are shown in Table 6.
907 matches via Entrez History. This was then used to browse the NORMAN SLE Classification tree in PubChem.56 Since the influence of searching via InChIKey first block (structural skeleton) versus full InChIKey was not dramatic (only an additional 44 entries found, see row 1 of Table 5), this analysis was kept at the InChIKey level for consistency with the rest of this article. The OECDPFAS list is split into many categories; of primary interest for data extraction is the “Structure Category”, which covers 8 major PFAS categories (denoted 100 through 800), with several subcategories in each. The major categories and the number of matches in CORE are shown in Table 6.
| OECD structure category | Total | In CORE | Ratio |
|---|---|---|---|
| a Neither mapping captures polymers, due to use of InChIKeys. PFAA = perfluoroalkyl acids. | |||
| S25|OECDPFAS|list of PFAS from the OECD | 3677 | 940 | 26% |
| 100 perfluoroalkyl carbonyl compounds | 490 | 126 | 26% |
| 200 perfluoroalkane sulfonyl compounds | 458 | 193 | 42% |
| 300 perfluoroalkyl phosphate compounds | 16 | 7 | 44% |
| 400 fluorotelomer-related compounds | 1392 | 350 | 25% |
| 500 per- and polyfluoroalkyl ether-based compounds | 322 | 52 | 16% |
| 600 other PFAA precursors or related – perfluoroalkyl | 282 | 129 | 46% |
| 700 other PFAA precursors or related – semifluorinated | 716 | 83 | 12% |
| 800 fluoropolymersa | 1 | 0 | 0% |
Table 6 shows that PFAS in the categories 200, 300 and 600 are found quite well in the CORE documents (approx. 40% coverage). In contrast, categories 500 (per- and polyfluoroalkyl ether-based compounds) and 700 (semifluorinated perfluoroalkyl acid (PFAA) precursors), are underrepresented (16 and 12%, respectively). Even within categories, different subcategories were underrepresented, for instance very few entries were found from subcategory 103 “other perfluoroalkyl carbonyl-based nonpolymers” (only 13 of 168 entries in OECDPFAS, i.e. 8%). Likewise, only 3 of 127 (2%) of subcategory 701.2 “Semi-fluorinated alkanes (SFAs) and derivatives (n ≥ 4)” were found, and only 26 of 405 (6%) of 705 “side-chain fluorinated aromatics”. It would be interesting future work to investigate whether the CORE and patent datasets could capture additional knowledge to add more PFAS to these categories, for instance by expanding the “splitPFAS” work at categorizing PFAS58 (prototyped so far on only 4 of the OECDPFAS categories) for this context.
Conclusions
This article details methods to extract mentions of potential PFAS compounds and their structures as SMILES strings from scientific documents and patents, along with the use of three open access chemistry toolkits to identify PFAS structures in these compound lists by parsing, removing wrong structures, normalizing, standardizing and substructure searching these SMILES. Of the extracted mentions, FCC(F)(F)F [1,1,1,2-tetrafluoroethane] was the most frequently detected compound – overall 6323 times in the CORE dataset. The resulting PFAS lists have been compiled, together with their references and chemical structures using three different structural definitions of PFAS (A, B and C), where A is a very broad definition, B is a narrower definition and a subset of A, and C is a subset of B. These definitions came from the PFAS community, with A being recently proposed by the OECD, and both B and C deriving from definitions used by the US EPA. These definitions did not always contain sufficient cheminformatic detail to clarify certain edge cases, such as unsaturation or hybridization. As such, the results here are intended to contribute to the current debate surrounding the definition of PFAS and help further refine these definitions.The resulting PFAS lists have been compared with two of the largest publicly available lists of PFAS molecules, PFASMASTER from the US EPA and the OECDPFAS list, released by the OECD. The overlap between the lists and the data extracted from scientific documents and patents is lower than expected, showing that many molecules on these lists are not found in the scientific documents and patents investigated, while also many molecules from the document extraction are not found in the published PFAS lists. Several thousand were also not in PubChem, but have since been deposited. The CORE and Patents datasets have been provided as CSV files on Zenodo25 for mass spectral screening. This information will add to the number of known potential PFAS substances and hopefully help contribute to alleviating the “PFAS knowledge gap”. The provision of public datasets will allow the integration of this information into various non-target mass spectrometry workflows, such as the open workflows MetFrag53 and patRoon,54 thus enabling other researchers to investigate the potential occurrence of the identified PFAS compounds in humans and the environment in future studies.
Data availability
Due to the size and nature of the supporting information files, URLs to access these are given in ref. 10, 25, 31, 32, 33 and 42. All input files and results are on FigShare (https://doi.org/10.6084/m9.figshare.17168960.v1), the final CSV lists are also available on Zeondo (https://doi.org/10.5281/zenodo.6034586) and available in MetFrag online (https://msbi.ipb-halle.de/MetFrag/). The code associated with this work is on GitHub (https://github.com/ontochem/PFAS). Finally, in addition to the deposit on FigShare, the patent annotations and the unique compounds from patents and CORE can be accessed via the embedded URLs (also given in the reference section, ref. 31–33). A (free) login is required for these URLs, which enables more powerful analysis than was possible via other repositories.Author contributions
SJB: investigation, software, data curation; TB: software; SB: conceptualization, data curation; MI: software; CR: software; IW: software, data curation; TK: software, data curation; ELS: conceptualization, data curation, writing – original draft, writing – review & editing; LW: conceptualization, data curation, writing – original draft, writing – review & editing.Conflicts of interest
SJB, TB, SB, MI, CR, IW, LW declare that they are involved in the creation of the commercial products OC|processor and Google Patents discussed in this paper.Acknowledgements
ELS and TK acknowledge funding support from the Luxembourg National Research Fund (FNR) for project A18/BM/12341006 and the European Union's Horizon 2020 Research and Innovation Programme under grant agreement No 101036756 for ZeroPM. ELS gratefully acknowledges deposition assistance from Ben Shoemaker and discussions with Paul Thiessen and Evan Bolton, PubChem (NCBI/NLM/NIH). The authors also thank Jane Frommer (Collabra) and the reviewers for their efforts and helpful comments.References
- S. E. Fenton, A. Ducatman, A. Boobis, J. C. DeWitt, C. Lau, C. Ng, J. S. Smith and S. M. Roberts, Per- and Polyfluoroalkyl Substance Toxicity and Human Health Review: Current State of Knowledge and Strategies for Informing Future Research, Environ. Toxicol. Chem., 2021, 40, 606–630 CrossRef CAS PubMed.
- E. M. Sunderland, X. C. Hu, C. Dassuncao, A. K. Tokranov, C. C. Wagner and J. G. Allen, A review of the pathways of human exposure to poly- and perfluoroalkyl substances (PFASs) and present understanding of health effects, J. Exposure Sci. Environ. Epidemiol., 2019, 29, 131–147 CrossRef CAS PubMed.
- R. C. Buck, J. Franklin, U. Berger, J. M. Conder, I. T. Cousins, P. de Voogt, A. A. Jensen, K. Kannan, S. A. Mabury and S. P. van Leeuwen, Perfluoroalkyl and polyfluoroalkyl substances in the environment: Terminology, classification, and origins, Integr. Environ. Assess. Manage., 2011, 7, 513–541 CrossRef CAS PubMed.
- I. T. Cousins, J. C. DeWitt, J. Glüge, G. Goldenman, D. Herzke, R. Lohmann, C. A. Ng, M. Scheringer and Z. Wang, The high persistence of PFAS is sufficient for their management as a chemical class, Environ. Sci.: Processes Impacts, 2020, 22, 2307–2312 RSC.
- OECD, Toward a new comprehensive global database of per- and polyfluoroalkyl substances (PFASs): summary report on updating the OECD 2007 list of per- and polyfluorinated substances (PFASs), Report ENV/JM/MONO(2018)7, 2018, https://www.oecd.org/officialdocuments/publicdisplaydocumentpdf/?cote=ENV-JM-MONO(2018)7%26doclanguage=en, accessed 15 January 2022 Search PubMed.
- Z. Wang, S25|OECDPFAS|List of PFAS from the OECD, Version Number: NORMAN-SLE-S25.0.1.2, 2018, DOI:10.5281/zenodo.2648775.
- US EPA, CompTox Chemicals Dashboard|PFASMASTER Chemicals, https://comptox.epa.gov/dashboard/chemical_lists/PFASMASTER, accessed 14 November 2021 Search PubMed.
- A. J. Williams, C. M. Grulke, J. Edwards, A. D. McEachran, K. Mansouri, N. C. Baker, G. Patlewicz, I. Shah, J. F. Wambaugh, R. S. Judson and A. M. Richard, The CompTox Chemistry Dashboard: a community data resource for environmental chemistry, J. Cheminf., 2017, 9, 61 Search PubMed.
- US EPA and OECD, CompTox Chemicals Dashboard|PFASOECD Chemicals, https://comptox.epa.gov/dashboard/chemical-lists/PFASOECD, accessed 29 December 2021 Search PubMed.
- L. Weber and E. Schymanski, Supplementary Material: PFAS tables, 2021, DOI:10.6084/m9.figshare.17168960.v1.
- Y. Liu, L. A. D'Agostino, G. Qu, G. Jiang and J. W. Martin, High-Resolution Mass Spectrometry (HRMS) Methods for Nontarget Discovery and Characterization of Poly- and Per-fluoroalkyl Substances (PFASs) in Environmental and Human Samples, TrAC, Trends Anal. Chem., 2019, 121, 115420, DOI:10.1016/j.trac.2019.02.021.
- OECD, Reconciling Terminology of the Universe of Per- and Polyfluoroalkyl Substances: Recommendations and Practical Guidance, OECD Publishing, Paris, 2021, Report 61, https://www.oecd.org/chemicalsafety/portal-perfluorinated-chemicals/terminology-per-and-polyfluoroalkyl-substances.pdf, accessed 14 November 2021 Search PubMed.
- Z. Wang, A. M. Buser, I. T. Cousins, S. Demattio, W. Drost, O. Johansson, K. Ohno, G. Patlewicz, A. M. Richard, G. W. Walker, G. S. White and E. Leinala, A New OECD Definition for Per- and Polyfluoroalkyl Substances, Environ. Sci. Technol., 2021, 55, 23DOI, DOI:10.1021/acs.est.1c06896.
- US EPA, National PFAS Testing Strategy, https://www.epa.gov/assessing-and-managing-chemicals-under-tsca/national-pfas-testing-strategy, accessed 14 November 2021 Search PubMed.
- US EPA, National PFAS Testing Strategy: Identification of Candidate Per- and Poly- fluoroalkyl Substances (PFAS) for Testing, Washington, DC, 2021 Search PubMed.
- S. Kim, J. Chen, T. Cheng, A. Gindulyte, J. He, S. He, Q. Li, B. A. Shoemaker, P. A. Thiessen, B. Yu, L. Zaslavsky, J. Zhang and E. E. Bolton, PubChem in 2021: new data content and improved web interfaces, Nucleic Acids Res., 2021, 49, D1388–D1395 CrossRef CAS PubMed.
- J. M. Barnard, A comparison of different approaches to Markush structure handling, J. Chem. Inf. Model., 1991, 31, 64–68 CrossRef CAS PubMed.
- M. Irmer, C. Bobach, T. Böhme, U. Laube, A. Püschel and L. Weber, in BioCreative Challenge Evaluation Workshop, 2013, vol. 2, p. 92 Search PubMed.
- Apache UIMA – Apache UIMA, https://uima.apache.org/, accessed 14 November 2021 Search PubMed.
- S. A. Akhondi, H. Rey, M. Schwörer, M. Maier, J. Toomey, H. Nau, G. Ilchmann, M. Sheehan, M. Irmer, C. Bobach, M. Doornenbal, M. Gregory and J. A. Kors, Automatic identification of relevant chemical compounds from patents, Database, 2019, 2019, baz001, DOI:10.1093/database/baz001.
- P. Knoth and Z. Zdrahal, in CERN Workshop on Innovations in Scholarly Communication (OAI7), https://oro.open.ac.uk/32560/, 2011, accessed 14 November 2021 Search PubMed.
- The Open University and Jisc, CORE – Aggregating the world's open access research papers, https://core.ac.uk/, accessed 14 November 2021 Search PubMed.
- Google, Google Patents, https://patents.google.com/advanced, accessed 14 November 2021 Search PubMed.
- S. Heller, A. McNaught, S. Stein, D. Tchekhovskoi and I. Pletnev, InChI – the worldwide chemical structure identifier standard, J. Cheminf., 2013, 5, 7 CAS.
- S. J. Barnabas, T. Böhme, S. Boyer, M. Irmer, C. Ruttkies, I. Wetherbee, T. Kondic, E. L. Schymanski and L. Weber, OntoChem PFAS CORE and Patent Files for MetFrag, 2022, DOI:10.5281/zenodo.6034586.
- Google, BigQuery, https://cloud.google.com/bigquery, accessed 14 November 2021 Search PubMed.
- D. Weininger, SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules, J. Chem. Inf. Model., 1988, 28, 31–36 CrossRef CAS.
- Daylight Chemical Information Systems, Inc., SMILES – A Simplified Chemical Language, https://www.daylight.com/dayhtml/doc/theory/theory.smiles.html, accessed 13 April 2019 Search PubMed.
- Blue Obelisk, OpenSMILES Home Page, https://opensmiles.org/, accessed 14 November 2021 Search PubMed.
- L. Weber, R. Szargan, B. Schulze and M. Mühlstädt, Nitrogen-15 NMR, 2D NMR and ESCA characterization of a new stable 6a-thia(SIV)-1,6-diazapentalene, Magn. Reson. Chem., 1990, 28, 419–422 CrossRef CAS.
- OntoChem, OntoChem SciWalker-Open-Data: 818,280 compounds extracted from CORE documents, https://console.cloud.google.com/bigquery?project=sciwalker-open-data%26organizationId=359740966731%26d=chemistry_compounds%26p=sciwalker-open-data%26t=CORE_compounds%26page=table%26ws=!1m5!1m4!4m3!1ssciwalker-open-data!2schemistry_compounds!3sCORE_compounds, accessed 15 January 2022 Search PubMed.
- OntoChem, OntoChem SciWalker-Open-Data: Annotations in Patent Documents, https://console.cloud.google.com/bigquery?project=sciwalker-open-data&d=google_patents_research&p=patents-public-data&t=annotations_202101&page=table&ws=!1m30!1m4!4m3!1ssciwalker-open-data!2schemistry_compounds!3soc_registry_flagged!1m4!4m3!1ssciwalker-open-data!2schemistry_compounds!3sfda_unii!1m4!4m3!1spatents-public-data!2sgoogle_patents_research!3sannotations_202105!1m4!4m3!1ssciwalker-open-data!2schemistry_compounds!3sCORE_compounds!1m4!4m3!1ssciwalker-open-data!2schemistry_compounds!3sPatents_compounds_202101!1m4!4m3!1spatents-public-data!2sgoogle_patents_research!3sannotations_202101, accessed 15 January 2022.
- OntoChem, OntoChem SciWalker-Open-Data: 18,032,261 unique compounds (by InChI) extracted from Google Patents documents, https://console.cloud.google.com/bigquery?project=sciwalker-open-data&d=chemistry_compounds&p=sciwalker-open-data&t=Patents_compounds_202101&page=table&ws=!1m25!1m4!4m3!1ssciwalker-open-data!2schemistry_compounds!3soc_registry_flagged!1m4!4m3!1ssciwalker-open-data!2schemistry_compounds!3sfda_unii!1m4!4m3!1spatents-public-data!2sgoogle_patents_research!3sannotations_202105!1m4!4m3!1ssciwalker-open-data!2schemistry_compounds!3sCORE_compounds!1m4!4m3!1ssciwalker-open-data!2schemistry_compounds!3sPatents_compounds_202101, accessed 15 January 2022.
- EMBL-EBI, Ontology Concept Identifiers: Identifiers.org, https://registry.identifiers.org/registry/ocid, accessed 20 November 2021 Search PubMed.
- Google, SciWalker Open Data – SQL workspace – BigQuery – Google Cloud Platform, https://console.cloud.google.com/bigquery?project=sciwalker-open-data/chemistry_compounds/oc_registry, accessed 20 November 2021 Search PubMed.
- Greg Landrum, RDKit, https://www.rdkit.org/, accessed 29 December 2021 Search PubMed.
- C. Steinbeck, Y. Han, S. Kuhn, O. Horlacher, E. Luttmann and E. Willighagen, The Chemistry Development Kit (CDK): An Open-Source Java Library for Chemo- and Bioinformatics, J. Chem. Inf. Comput. Sci., 2003, 43, 493–500 CrossRef CAS PubMed.
- E. L. Willighagen, J. W. Mayfield, J. Alvarsson, A. Berg, L. Carlsson, N. Jeliazkova, S. Kuhn, T. Pluskal, M. Rojas-Chertó, O. Spjuth, G. Torrance, C. T. Evelo, R. Guha and C. Steinbeck, The Chemistry Development Kit (CDK) v2.0: atom typing, depiction, molecular formulas, and substructure searching, J. Cheminf., 2017, 9, 33 Search PubMed.
- Actelion Pharmaceuticals Ltd, GitHub: actelion/openchemlib, Actelion Pharmaceuticals Ltd, https://github.com/Actelion/openchemlib, 2021, accessed 29 December 2021 Search PubMed.
- M. Swain, MolVS: Molecule Validation and Standardization, https://github.com/mcs07/MolVS, 2021, accessed 29 December 2021 Search PubMed.
- M. Swain, Introduction — MolVS 0.1.1 documentation, https://molvs.readthedocs.io/en/latest/guide/intro.html, accessed 29 December 2021 Search PubMed.
- OntoChem, OntoChem PFAS Code, OntoChem, https://github.com/ontochem/PFAS, 2022, accessed 15 January 2022 Search PubMed.
- M. Kratochvíl, J. Vondrášek and J. Galgonek, Sachem: a chemical cartridge for high-performance substructure search, J. Cheminf., 2018, 10, 27 Search PubMed.
- G. Landrum, Fingerprinting and Molecular Similarity (RDKit), https://rdkit.readthedocs.io/en/latest/GettingStartedInPython.html#fingerprinting-and-molecular-similarity, accessed 13 May 2022 Search PubMed.
- T. Sander, DataWarrior User Manual: Molecule or Reaction Similarity and Descriptors (openmolecules.org), https://openmolecules.org/help/similarity.html, accessed 13 May 2022 Search PubMed.
- C. Steinbeck, Fingerprinter (CDK API - version 20070216), http://cdk.sourceforge.net/cdk-0.99.1/api/org/openscience/cdk/fingerprint/Fingerprinter.html, accessed 13 May 2022 Search PubMed.
- ChemAxon, ChemAxon – Software Solutions and Services for Chemistry & Biology, https://chemaxon.com/, accessed 29 December 2021 Search PubMed.
- I. Filippov, OSRA (Optical Structure Recognition Application), https://sourceforge.net/projects/osra/, accessed 29 December 2021 Search PubMed.
- I. V. Filippov and M. C. Nicklaus, Optical Structure Recognition Software To Recover Chemical Information: OSRA, An Open Source Solution, J. Chem. Inf. Model., 2009, 49, 740–743 CrossRef CAS PubMed.
- Wikipedia, Dichlorotetrafluoroethane, https://en.wikipedia.org/w/index.php?title=1,2-Dichlorotetrafluoroethane%26oldid=35140760, Wikipedia, 2006, accessed 29 December 2021 Search PubMed.
- Dassault Systèmes, BIOVIA CTfile formats, 2016, https://help.accelrysonline.com/ulm/onelab/1.0/content/ulm_pdfs/direct/reference/ctfileformats2016.pdf, accessed 29 December 2021 Search PubMed.
- B. Alameddine, PhD thesis, Université de Fribourg, 2007, oai:doc.rero.ch:20070803113704-NT.
- C. Ruttkies, E. L. Schymanski, S. Wolf, J. Hollender and S. Neumann, MetFrag relaunched: incorporating strategies beyond in silico fragmentation, J. Cheminf., 2016, 8, 3 Search PubMed.
- R. Helmus, T. L. ter Laak, A. P. van Wezel, P. de Voogt and E. L. Schymanski, patRoon: open source software platform for environmental mass spectrometry based non-target screening, J. Cheminf., 2021, 13, 1 CAS.
- E. L. Schymanski, T. Kondić, S. Neumann, P. A. Thiessen, J. Zhang and E. E. Bolton, Empowering large chemical knowledge bases for exposomics: PubChemLite meets MetFrag, J. Cheminf., 2021, 13, 19 CAS.
- NORMAN Network and NCBI/NLM/NIH, NORMAN SLE Classification Browser, https://pubchem.ncbi.nlm.nih.gov/classification/#hid=101, accessed 7 May 2020 Search PubMed.
- NCBI/NLM/NIH, PubChem Identifier Exchange, https://pubchem.ncbi.nlm.nih.gov/idexchange/idexchange.cgi, accessed 23 March 2021 Search PubMed.
- B. Sha, E. L. Schymanski, C. Ruttkies, I. T. Cousins and Z. Wang, Exploring open cheminformatics approaches for categorizing per- and polyfluoroalkyl substances (PFASs), Environ. Sci.: Processes Impacts, 2019, 21, 1835–1851 RSC.
Footnote |
| † Details on the supporting information are summarised in the Data availability section. |
| This journal is © The Royal Society of Chemistry 2022 |