Open Access Article
Open Access ArticleAmphipathic trans-acting phosphorothioate DNA elements mediate the delivery of uncharged nucleic acid sequences in mammalian cells†
H. V.
Jain
a,
D.
Verthelyi
b and
S. L.
Beaucage
*a
aDivision of Biotechnology Review and Research IV, CDER, FDA, 10903 New Hampshire Avenue, Silver Spring, Maryland 20933, USA. E-mail: Serge.Beaucage@fda.hhs.gov; Harsh.Jain@fda.hhs.gov
bDivision of Biotechnology Review and Research III, CDER, FDA, 10903 New Hampshire Avenue, Silver Spring, Maryland 20933, USA
First published on 24th July 2015
Abstract
A convenient approach to the delivery of uncharged peptide nucleic acids (PNA) or phosphorodiamidate morpholino (PMO) oligomers in mammalian cells is presented and consists of extending the sequence of these oligomers with a short (6-mer) PNA-polyA or PMO-polyA tail. Recognition of the polyA-tailed PNA or PMO oligomers by an 8-mer amphipathic trans-acting polythymidylic thiophosphate triester element (dTtaPS) results in efficient internalization of these oligomers in several cell lines. Our findings indicate that internalization of the oligomers occurs through an energy-dependent mechanism and macropinocytosis appears to be the prevailing endocytic pathway used for cellular uptake. The functionality of the internalized oligomers is demonstrated by alternate splicing of the pre-mRNA encoding luciferase in HeLa pLuc 705 cells.
Introduction
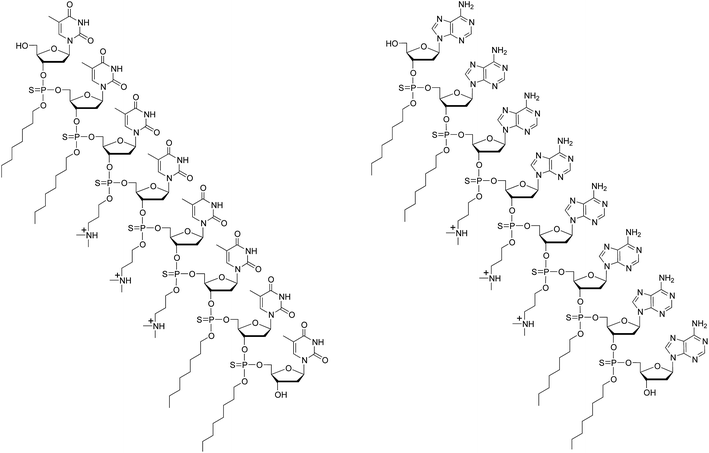
Most human genes undergo alternative splicing events, which are triggered by intricate and highly regulated machinery requiring the sequence-specific binding of several proteins to nuclear pre-mRNAs. Steric interference imparted by RNase H-incompetent oligonucleotide analogues, complementary to specific pre-mRNA splice sites, has been shown to be efficient at redirecting the splicing machinery during assembly of mature mRNAs.1,2 Indeed, skipping the mutated exons in dystrophin pre-mRNA as an approach to the clinical treatment of Duchenne muscular dystrophy, underscores the biomedical significance of alternative splicing events.3–5 Negatively charged 2′-O-methyl RNA phosphorothioate sequences6,7 and uncharged peptide nucleic acid (PNA)8,9 or phosphorodiamidate morpholino (PMO)10–14 oligomers are nuclease-resistant and RNase-H incompetent oligonucleotide analogues that have been found potent at inducing splice redirecting events.6–14 While cationic lipids are efficient carriers for in vitro cellular transfection of negatively charged DNA/RNA sequences and their analogues, these carriers could not be successfully used for cellular internalization of uncharged PNA or PMO oligomers.15 The conjugation of cationic cell-penetrating peptides (CPP) to PNAs or PMOs resulted in improved cellular uptake of these oligomers and led to pre-mRNA splicing correction activities in mammalian cells and animal models.16–21 Interestingly, the use of PNA-CPP conjugates was shown to internalize PNA oligomers in HeLa pLuc 705 cells by virtue of sequence complementarity.15 An inherent limitation of this approach to cellular internalization is the requirement for a PNA-CPP conjugate to be complementary to each PNA targeting a specific mRNA sequence.During the course of our studies on the cellular uptake of an uncharged thermosensitive DNA oligonucleotide prodrug (15-mer) in mammalian cells, we discovered that replacement of four neutral thiophosphate triester functions with four positively charged ones greatly improved the uptake of the modified DNA prodrug in Vero, GC-2 and HeLa cells.22 We then hypothesized that extending PNA or PMO oligomers with a short PNA-polyA or PMO-polyA tail could provide an affinity recognition site for a positively charged, trans-acting polythymidylic thiophosphate triester element (dTtaPS) through sequence complementarity. In principle, the affinity recognition of polyA-tailed PNA or PMO oligomers by dTtaPS could facilitate the uptake of these oligomers in live cells while averting the shortcomings associated with the cellular delivery of cationic CPP conjugates. With the intent of validating our hypothesis, the synthesis of dTtaPS, a polythymidylic acid (8-mer) with three N,N-dimethylaminopropyl and four octyl thiophosphate triester functions (Fig. 1) was performed. At physiological pH, the N,N-dimethylaminopropyl functions are positively charged while the hydrophobic octyl groups are necessary to impart amphiphilicity, which has been reported23,24 to facilitate in vitro cellular delivery of nucleic acids. Our objectives are to: (i) demonstrate the efficiency of dTtaPS-mediated internalization of polyA-tailed PNA and PMO oligomers in various cell lines, and (ii) confirm the functionality of these oligomers through the well-studied alternate splicing of the pre-mRNA encoding luciferase in HeLa pLuc 705 cells.
Materials and methods
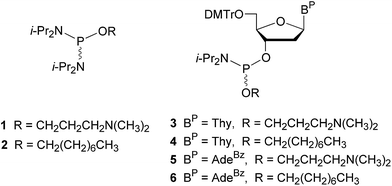
Solid-phase synthesis of dTtaPS and dAtaPS
Solid-phase synthesis of dTtaPS and dAtaPS was performed on a 0.2 μmole scale using a succinylated long chain alkylamine controlled-pore glass support loaded with either 5′-O-(4,4′-dimethoxytrityl)-2′-deoxythymidine or N6-benzoyl-5′-O-(4,4′-dimethoxytrityl)-2′-deoxyadenosine as the leader nucleoside. The syntheses were carried out using an ABI 394 DNA/RNA synthesizer and appropriate phosphoramidites 3–6 as 0.1 M solutions in dry MeCN. The coupling reactions were effected in the presence of 1H-tetrazole over a period of 600 s. All ancillary reagents necessary for the preparation of the DNA sequences were purchased and utilized as recommended by the instrument's manufacturer. The oxidation step of the synthesis cycle was performed using 3-(dimethylaminomethylidene)amino-3H-1,2,4-dithiazole-3-thione [0.05 M in pyridine–MeCN (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 v/v)]. This sulfurization reaction (600 s) was performed before the capping reaction (120 s), which was followed by the iterative cleavage of the 5′-DMTr group under acidic conditions (3% TCA in CH2Cl2, 60 s). Upon complete assembly of dTtaPS or dAtaPS, spectrophotometric measurements of the released DMTr cation at 498 nm revealed an overall yield in the range of 90 ± 5% for dTtaPS or dAtaPS. The dTtaPS synthesis column was placed into a stainless steel pressure vessel and exposed to pressurized methylamine gas (∼2.5 bar, 25 °C, 3 min), whereas the dAtaPS column was subjected to pressurized ammonia gas (∼10 bar, 25 °C, 12 h). Upon removal of residual methylamine or ammonia gas from the pressure container, dTtaPS or dAtaPS was eluted off the synthesis column using a solution (0.5 ml) of Et3N
3 v/v)]. This sulfurization reaction (600 s) was performed before the capping reaction (120 s), which was followed by the iterative cleavage of the 5′-DMTr group under acidic conditions (3% TCA in CH2Cl2, 60 s). Upon complete assembly of dTtaPS or dAtaPS, spectrophotometric measurements of the released DMTr cation at 498 nm revealed an overall yield in the range of 90 ± 5% for dTtaPS or dAtaPS. The dTtaPS synthesis column was placed into a stainless steel pressure vessel and exposed to pressurized methylamine gas (∼2.5 bar, 25 °C, 3 min), whereas the dAtaPS column was subjected to pressurized ammonia gas (∼10 bar, 25 °C, 12 h). Upon removal of residual methylamine or ammonia gas from the pressure container, dTtaPS or dAtaPS was eluted off the synthesis column using a solution (0.5 ml) of Et3N![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeCN
MeCN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (1
H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60
60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 39 v/v/v). Each dTtaPS and dAtaPS solution was evaporated to dryness using a stream of air. dTtaPS or dAtaPS were then stored frozen at −20 °C.
39 v/v/v). Each dTtaPS and dAtaPS solution was evaporated to dryness using a stream of air. dTtaPS or dAtaPS were then stored frozen at −20 °C.
dTtaPS: +MALDI-TOF: calcd for C127H202N19O47P7S7 [M + H]+ 3187, found 3184.
dAtaPS: +MALDI-TOF: calcd for C127H194N43O31P7S7 [M + H]+ 3259, found 3256.
Thermal denaturation protocol
dTtaPS is mixed with the polyA-tailed PNA oligomer 10 in either 1× PBS buffer (pH 7.4) or 1.0 M NaCl in 1× PBS buffer while keeping the total strand concentration at ca. 2 μM. Thermal denaturation profiles (absorbance vs. temperature) of the dTtaPS:10 complex was measured at 260 nm using a UV/Vis spectrophotometer equipped with a Peltier temperature controller and interfaced with a personal computer. Denaturation of the complex was performed over a temperature range of −5 °C to 60 °C. The temperature was ramped at the rate of 1°C min−1; temperature measurements were recorded at every 1 °C with an equilibration time of 30 s between each measurement. Heating and cooling profiles were recorded.Cell culture
HEK293 (ATCC® CRL-1573™), HeLa (ATCC® CCL-2™) MCF7 (ATCC® HTB-22™) and SK-N-SH (ATCC® HTB-11™) cell lines were purchased from ATCC. All cell lines were exponentially grown in ATCC recommended media containing 100 U penicillin and 100 μg ml−1 streptomycin. The HeLa pLuc 705 cell line was exponentially grown in Dulbecco's Minimum Essential Medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FBS), 1.5 mM L-glutamine, 1.0 mM sodium pyruvate, 100 U penicillin, 100 μg ml−1 streptomycin and 200 μg ml−1 hygromycin; the cell culture was maintained at 37 °C in a humidified incubator under 5% CO2.Complex formation between PNA or PMO oligomers and dTtaPS or dAtaPS
Appropriate amounts of PNA or PMO oligomers were solubilized in serum-free OptiMEM (20 μl); dTtaPS or dAtaPS was added to the solution to achieve a final concentration that is twice that of the oligomer. After a 30 min incubation at 37 °C, the PNA or PMO oligomer:dTtaPS or dAtaPS complexes were stored at 4 °C for 15 minutes or until used. Stock solutions (2×) of the complexes were made by adding OptiMEM to appropriate volumes so that the extracellular concentration of the PNA/PMO oligomer and dTtaPS/dAtaPS is 1.0 μM and 2.0 μM, respectively, in all cellular uptake experiments.Flow cytometry analysis of cellular uptake in live cells
The internalization of fluorescently-labelled PNA or PMO sequences (Table 1) in HEK 293, HeLa, MCF7 and SK-N-SH cells was assessed by flow cytometry. Cells were seeded in a 96-well plate (2 × 104 cells per well) and allowed to grow at 37 °C for 24 h in 10% FBS-MEM. The culture medium of each well was then replaced with fresh medium (100 μl) containing any of the fluorescently-labelled PNA or PMO sequences, the final concentration of which was 1.0 μM. After an incubation of 18 h at 37 °C, the medium was removed from each well by suction and 0.25% trypsin (50 μl) was added. Upon complete cell detachment, a solution (100 μl) of ice-cold 2% FBS in phosphate-buffered saline (PBS, pH 7.4) was added to each well. The cells were collected into tubes, to which was added 0.4% trypan blue (50 μl) and 200 μM monensin (20 μl), and analyzed by fluorescence-activated cell sorting (FACS) using a BD FACScan™ flow cytometer. A total of 5000 events were counted and the results were reported as a percentage of the cells that had internalized the fluorescently-labelled oligomers.| Oligomer | Type | Sequencea |
|---|---|---|
| a Fl, fluorescein; • phosphorothioate diester. | ||
| 7 | PNA | Fl-CCTCTTACCTCAGTTACA-AAAAAA-NH2 |
| 8 | PNA | Fl-GTGGCCGTTTACGTCGCC-AAAAAA-NH2 |
| 9 | PNA | Fl-CCTCTTACCTCAGTTACA-NH2 |
| 10 | PNA | H-CCTCTTACCTCAGTTACA-AAAAAA-NH2 |
| 11 | PNA | H-GTGGCCGTTTACGTCGCC-AAAAAA-NH2 |
| 12 | PNA | H-CCTCTTACCTCAGTTACA-NH2 |
| 13 | 2′-OMe RNA | 5′-C•C•T•C•T•T•A•C•C•T•C•A•G•T•T•A•C•A |
| 14 | 2′-OMe RNA | 5′-G•T•G•G•C•C•G•T•T•T•A•C•G•T•C•G•C |
| 15 | PNA | H-CCTCTTACCTCAGTTACA-TTTTTT-NH2 |
| 16 | PNA | H-CCTCTTACCTCAGTTACA-CCCCCC-NH2 |
| 17 | PNA | H-AAAAAA-CCTCTTACCTCAGTTACA-NH2 |
| 18 | PMO | CCTCTTACCTCAGTTACA-AAAAAA-Fl |
| 19 | PMO | GTGGCCGTTTACGTCGCC-AAAAAA-Fl |
| 20 | PMO | CCTCTTACCTCAGTTACA-Fl |
| 21 | PMO | CCTCTTACCTCAGTTACA-AAAAAA |
| 22 | PMO | GTGGCCGTTTACGTCGCC-AAAAAA |
| 23 | PMO | CCTCTTACCTCAGTTACA |
| 24 | PMO | CCTCTTACCTCAGTTACA-AATAAA |
Luciferase assay protocol
HeLa pLuc 705 cells were seeded in a 96-well plate (2 × 104 cells per well) and allowed to grow at 37 °C for 18 h in 10% FBS-DMEM (100 μl). For each dose-dependence experiment, the culture medium of each well was replaced with either fresh serum-free (50 μl) or 20% serum-containing OptiMEM (50 μl) for experiments intended to be performed in serum-free or in 10% serum-containing medium. A 2× solution of PNA or PMO oligomer:dTtaPS or dAtaPS complexes in OptiMEM (50 μl) was added to the cells in order to achieve pre-determined complex concentrations, as indicated in the legends of relevant Figures. After a 4 hour incubation at 37 °C, 20% FBS in OptiMEM (100 μl) was added to serum-free experiments and cells were incubated for an additional 18 h at 37 °C. The culture medium was then removed by suction and the cells were lysed upon addition of the Pierce luciferase cell lysis buffer (50 μl) and mechanical agitation for 10 minutes at ambient temperature. The cell lysate (30 μl) was placed in a white 96-well plate and was followed by the addition of Bright-Glow reagent (20 μl); luciferase activity was then measured using a microplate reader. For each well, luminescence was integrated over a period of 1 s and recorded as relative light units (RLU). Luminescence measurements are reported based on the amount of total protein present in the test sample.Protein concentration measurements
Protein concentrations were determined from a fraction (5 μl) of the above luciferase assay cell lysate using the Pierce Coomassie (Bradford) protein assay kit as per the manufacturer's instructions.RT-PCR analysis of splicing correction events
Total RNA was isolated from a fraction of the above luciferase assay cell lysate using TRIzol (Life Technologies) as per the manufacturer's recommendations. The total RNA (1 μg) was then subjected to RT-PCR using the high capacity cDNA reverse transcription kit and 2× SYBR green (Applied Biosystems) as per the manufacturer's instructions using the forward and reverse primers 5′-TTGATATGTGGATTTCGAGTCGTC and 5′-TGTCAATCAGAGTGCTTTTGGCG, respectively. The cDNA products were analysed on 3% agarose gels, which were stained with ethidium bromide. Fluorescence was measured using a GE ImageQuant LAS 4000 scanner.Energy-dependence of the dTtaPS-assisted internalization of PNA oligomer 10 in HeLa pLuc 705 cells
HeLa pLuc 705 cells were seeded in a 96-well plate (2 × 104 cells per well) and allowed to grow at 37 °C for 18 h in 10% FBS-DMEM as described above in the luciferase assay protocol. While the cell culture was maintained at 37 °C or 4 °C, a 2× solution of PNA oligomer 10:dTtaPS complex was prepared as described above. The cell culture medium was replaced with the PNA oligomer:dTtaPS complex solution to provide an oligomer and dTtaPS final concentration of 1.0 μM and 2.0 μM, respectively. After an incubation of 1 h at 37 °C or 4 °C, the medium was removed by suction and luciferase activity was measured as described above in the luciferase assay protocol. Luminescence measurements were normalized to the amounts of protein present in the test samples and reported as percent luminescence remaining relative to that of the experiment performed at 37 °C.Endocytic pathway assays
HeLa pLuc 705 cells were seeded in a 96-well plate and grown in 10% FBS-DMEM as described above. The cell culture was then pre-incubated for 30 min at 37 °C in the presence of chlorpromazine, nystatin or 5-(N-ethyl-N-isopropyl)amiloride (EIPA), the final concentration of which was 30 μM, 50 μM or 1 mM, respectively. A 2× solution of PNA oligomer 10:dTtaPS or PMO oligomer 21:dTtaPS complexes was added to the cells to provide an oligomer and dTtaPS final concentration of 1.0 μM and 2.0 μM, respectively. The cells were then incubated for an additional 30 min at 37 °C, whereupon luciferase activity was measured as described above in the luciferase assay protocol. Luminescence measurements were normalized to the amounts of protein present in the test samples and reported as percent luminescence remaining relative to that of the experiment performed in the absence of endocytic pathway inhibitors.Transfection inhibition assay
Appropriate amounts of PNA or PMO oligomers were solubilized in serum-free OptiMEM (10 μl); dTtaPS was added to the solution to achieve a final concentration that is twice that of the oligomer. A solution of 4 M urea in serum-free OptiMEM (10 μl) was added to produce a final urea concentration of 2 M. After a 30 min incubation at 37 °C, the PNA or PMO oligomer:dTtaPS complexes were stored at 4 °C for 15 minutes or until used. Stock solutions (2×) of the complexes were made by adding OptiMEM to appropriate volumes (300 μl). HeLa pLuc 705 cells were seeded in a 96-well plate (2 × 104 cells per well) and allowed to grow at 37 °C for 18 h in 10% FBS-DMEM, as described above. The cell culture of each well was replaced with fresh 20% serum-containing OptiMEM (50 μl) to which was added 50 μl of the 2× stock solution of the PNA or PMO oligomer:dTtaPS complexes to produce an extracellular urea concentration of 67 mM. After a 4 hour incubation at 37 °C, the medium was replaced with fresh 10% FBS-DMEM. The cell culture was then processed as described above in the luciferase assay protocol. Luminescence measurements are reported as a percentage of the relative light units (RLU) measured in the absence of urea, which is arbitrarily set to 100%.Confocal microscopy analysis of dTtaPS-assisted internalization of PNA or PMO oligomers in HeLa pLuc 705 cells
HeLa pLuc 705 cells (5 × 104 cells per well) were seeded in 8-well chambered slides and cultured for 24 h at 37 °C in 10% FBS-MEM (200 μl). The culture medium of each well was then replaced with 20% FBS containing fresh medium (200 μl) and the fluoresceinated PNA or PMO oligomer:dTtaPS complex that was prepared as described above to provide an oligomer and dTtaPS final concentration of 1.0 μM and 2.0 μM, respectively. After a 12 hour incubation at 37 °C, the medium was removed and a 0.4% trypan Blue solution (200 μl) was added to quench the fluorescence stemming from extracellular fluoresceinated oligonucleotides. After a period of 5 minutes at ambient temperature, the cells were washed twice with PBS (pH 7.4, 200 μl) followed by the addition of fresh phenol red-free DMEM (200 μl) supplemented with 10% FBS. The intracellular distribution of fluorescence was determined without fixation using a Leica SP8 confocal microscope system and analysed using the Leica LASAF software. Hoechst 33342 was used, in accordance with the manufacturer's staining protocols, for the detection of nuclei. Images were acquired at a 63× magnification.Results and discussion
The chemical synthesis of dTtaPS and dAtaPS
The solid-phase synthesis of dTtaPS and dAtaPS begins with the preparation of the phosphordiamidites 1 and 2, which are required for the preparation of the deoxyribonucleoside phosphoramidites 3–6 (Fig. 2).Thus, the reaction of an equimolar amount of commercial bis(N,N-diisopropylamino)chlorophosphine with either 3-(N,N-dimethylamino)propan-1-ol or 1-octanol in the presence of excess triethylamine in anhydrous benzene, produced the phosphordiamidite 1 or 2 in a yield exceeding 85% after purification on silica gel. Treatment of commercially available 4,4′-dimethoxytriyl deoxythymidine or N6-benzoyl-4,4′-dimethoxytrityl 2′-deoxyadenosine with the phosphordiamidite 1 or 2 and 1H-tetrazole in a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 : 1, respectively, in anhydrous MeCN afforded the deoxyribonucleoside phosphoramidites 3–6. These phosphoramidites were isolated as white powders in yields of 75–85% after purification on silica gel and lyophilization from dry benzene. The identity of phosphoramidites 3–6 was confirmed by 31P-NMR spectroscopy and high-resolution mass spectrometry (data shown as ESI†).
2 : 1, respectively, in anhydrous MeCN afforded the deoxyribonucleoside phosphoramidites 3–6. These phosphoramidites were isolated as white powders in yields of 75–85% after purification on silica gel and lyophilization from dry benzene. The identity of phosphoramidites 3–6 was confirmed by 31P-NMR spectroscopy and high-resolution mass spectrometry (data shown as ESI†).
The synthesis of dTtaPS and dAtaPS on appropriately functionalized controlled pore glass (CPG) supports was easily achieved using the deoxyribonucleoside phosphoramidites 3–6 as 0.1 M solutions in MeCN. The coupling time of the 1H-tetrazole-activated phosphoramidites 3–6 was extended to 10 min to maximize stepwise coupling efficiencies, which were determined to be in the order of 98–99% based on the 4,4′-dimethoxytrityl cation assay.25 Upon exposure to pressurized gaseous amines and release from the CPG support, the purity of diastereomeric dTtaPS and dAtaPS was assessed by C4-RP-HPLC analysis, which revealed broad product peaks consistent with the diastereomeric and amphiphilic nature of these compounds. The purity of dTtaPS and dAtaPS was nonetheless found adequate to support our investigations.
A short PNA-polyA stretch is necessary and sufficient for dTtaPS-mediated internalization and bioactivity of PNA oligomers in live mammalian cells
The dTtaPS-mediated cellular internalization of fluorescently-labelled PNA oligomers (Table 1) in HEK 293, HeLa, HeLa pLuc 705, MCF7 and SK-N-SH live cells was evaluated by FACS analysis. As shown in Fig. 3, polyA-tailed PNA oligomers 7 and 8 were efficiently internalized in the presence of dTtaPS in all the cell lines under study. In sharp contrast, the PNA oligomer 9 lacking the polyA stretch was not substantially internalized under similar conditions in any of the cell lines.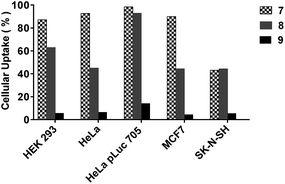 | ||
| Fig. 3 Internalization of the dTtaPS:PNA oligomer complexes in live mammalian cells. The extracellular concentration of dTtaPS and each PNA oligomer (7–9) is 2.0 μM and 1.0 μM, respectively. | ||
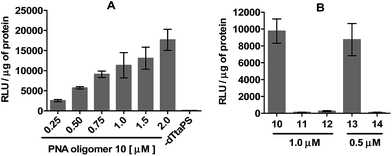
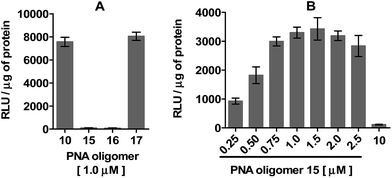
The efficiency of dTtaPS in mediating the cellular uptake of polyA-tailed PNA oligomers was assessed through a splice correction assay performed in HeLa pLuc 705 cells. The concentration-dependence of luciferase activity resulting from the dTtaPS-mediated internalization of PNA oligomer 10 in the proposed cell line is shown in Fig. 4A and compared with the luciferase activities measured either from the dTtaPS-assisted deliveries of PNA oligomers 11 and 12 or 2′-OMe RNA sequences 13 and 14 (Fig. 4B). As clearly demonstrated in Fig. 4A, the PNA oligomer 10 induced splice correction of the pre-mRNA encoding luciferase, in a dose-dependent manner, in the nucleus of the cells. When dTtaPS was absent, no luciferase activity was detected. It should be noted that the lowest concentration of oligomer 10 required for exhibiting luciferase activity has not been investigated to further support the efficiency of the dTtaPS-mediated internalization process. Fig. 4B shows that the polyA-tailed PNA oligomer 11 failed, as expected for a negative control, to restore luciferase activity. The successful dTtaPS-assisted delivery of the fluorescently-labelled PNA oligomers 7 and 8 in HeLa pLuc 705 cells (Fig. 3) suggests that the dTtaPS-mediated delivery of 11 should have been as successful as that of 10 under similar conditions. The PNA oligomer 12, which is 10 lacking the polyA stretch, did not restore luciferase activity, because dTtaPS failed to efficiently internalize its fluoresceinated congener 9 in HeLa pLuc 705 cells (Fig. 3).
For comparability purposes, the positive control 2′-OMe RNA sequences 13 was transfected in HeLa pLuc 705 cells using Lipofectamine® 2000 as the carrier in serum-free medium (Fig. 4B). Under these conditions, restoration of luciferase activity was comparable to that obtained by the dTtaPS-assisted transfection of the polyA-tailed PNA oligomer 10 in serum-containing medium. No significant luciferase activity was detected when the negative control 2′-OMe RNA sequences 14 was transfected under identical conditions (Fig. 4B).
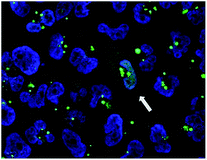
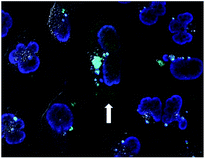
We then investigated the relationship between the concentration of dTtaPS and the extent of luminescence production upon transfection of the polyA-tailed PNA oligomer 10 in HeLa pLuc 705 cells. At a PNA oligomer concentration of 1.0 μM, luciferase activity was found optimal when the dTtaPS concentration was in the range of 1.5–3.0 μM (Fig. S1†). Confocal microscopy analysis of the dTtaPS-assisted uptake of the fluorescently-labelled PNA oligomers 7 in HeLa pLuc 705 cells revealed, as shown in Fig. 5, that endosomal sequestration of 7 is predominant after a contact time of 12 h at 37 °C. Fig. 5 also shows that PNA oligomer 7 escaped endosomal sequestration to some extent, as evidenced by nuclear fluorescence.
Recognition of the PNA-polyA stretch on the cellular internalization and bioactivity of PNA oligomers
The spatial arrangement and criticality of the polyA stretch of PNA oligomer 10 in terms of requirements for recognition by dTtaPS and cellular internalization were evaluated. This was achieved by moving the polyA stretch from the N-terminus of 10 to its C-terminus or replacing the polyA stretch with a polyT or a polyC stretch at the N-terminus. The consequences of these structural changes were monitored by measuring luciferase activity upon dTtaPS-mediated transfection of these modified PNA oligomers (15–17) in HeLa pLuc 705 cells. Fig. 6A shows that moving the polyA stretch from the N-terminus of 10 to its C-terminus did not significantly affect the dTtaPS-assisted transfection efficiency of PNA oligomer 17 under similar conditions. However, replacing the polyA stretch of 10 with a polyT or a polyC stretch at the N-terminus did not result in substantial expression of luciferase activity, presumably due to poor dTtaPS-mediated internalization of PNA oligomers 15 and 16. These findings suggest that recognition of the polyA stretch of PNA oligomer 10 by dTtaPS is necessary for its efficient cellular uptake. Interestingly, when the polyT-tailed PNA oligomer 15 is transfected in HeLa pLuc 705 cells using the dAtaPS DNA element, luciferase activity is restored to a level that is dependent on the concentration of 15 (Fig. 6B). Under these conditions, about half the level of luciferase activity was obtained when compared to that of the dTtaPS-assisted delivery of 10 (Fig. 6A). Although we cannot rule out the possibility that the dAtaPS-mediated delivery of 15 may be less efficient than the dTtaPS-assisted delivery of 10, one might argue that once internalized, cellular factors may competitively interact with the positively charged dAtaPS and allow the polyT stretch of uncharged PNA oligomer 15 to interact, by sequence complementarity, with the polyadenylated tail of cellular mRNAs. Such an interference may potentially lead to an appreciably decreased concentration of 15 at the aberrant splice site of the luciferase pre-mRNA, when compared to the concentration of 10 under similar conditions. No significant level of luciferase activity was detected when dAtaPS was used for the delivery of the polyA-tailed PNA oligomer 10 (Fig. 6B). These results are consistent with the recognition of the polyT stretch of PNA oligomer 15 by dAtaPS, as a critical requirement for efficient cellular uptake. As anticipated, dAtaPS failed to recognize and internalize the polyC-tailed or the polyA-tailed PNA oligomers 16 or 17, respectively, in HeLa pLuc 705 cells (data not shown).A short PMO-polyA stretch is necessary and sufficient for dTtaPS-mediated internalization and bioactivity of PMO oligomers in live mammalian cells
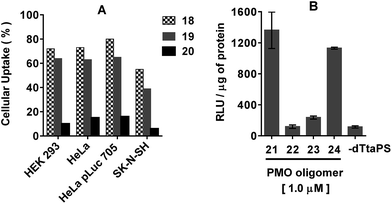
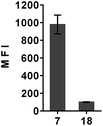
The applicability of dTtaPS to the delivery of a different class of uncharged nucleic acid sequences in several mammalian cell lines was carried out using PMO oligomers under experimental conditions similar to those used for PNA oligomers. As shown in Fig. 7A, results of the FACS analyses confirmed the dTtaPS-assisted delivery of the polyA-tailed PMO oligomers 18 and 19 in HEK 293, HeLa, HeLa pLuc 705, and SK-N-SH cell lines.As anticipated from the outcome of the cellular uptake of PNA oligomer 9 (Fig. 3), dTtaPS failed to efficiently internalize the PMO oligomer 20 lacking the polyA tail, in all the cell lines under investigation. Although dTtaPS has been comparably efficient at internalizing the fluoresceinated polyA-tailed PNA and PMO oligomers 7 and 18 in HeLa pLuc 705 cells in terms of percentage of transfected cells (see Fig. 3 and 7a), dTtaPS did not deliver comparable amounts of 7 and 18 to a comparable number of transfected cells. Indeed, FACS analysis of the dTtaPS-mediated uptake of PNA and PMO oligomers 7 and 18, when performed in the mean fluorescence intensity mode, revealed that the internalization of PNA oligomer 7 is ∼10-fold greater than that of the PMO oligomer 18 (Fig. 8). These findings are in agreement with those obtained from the dTtaPS-assisted internalization of PMO oligomer 21 in HeLa pLuc 705 cells (Fig. 7B), indicating that the restoration of luciferase activity is about 5- to 8-fold less than that obtained with the PNA oligomer 10 under conditions of similar oligomer concentrations (Fig. 4A and B).
Of interest, the replacement of one PMO-A residue with a PMO-T in the polyA tail of PMO oligomer 21 did not appreciably decrease the ability of dTtaPS to internalize 24 in HeLa pLuc 705 cells on the basis of luciferase activity (Fig. 7B). Thus, one A → T modification in the polyA tail of 24 is not sufficient to prevent its recognition and cellular internalization by dTtaPS. In the absence of dTtaPS, the internalization of PMO oligomer 21 is reduced to that of the negative control 22 or that of the positive control 23 lacking the polyA tail.
The effect of dTtaPS concentration on the cellular uptake of PMO oligomer 21 in HeLa pLuc 705 cells was also investigated. Similar to the study performed with the PNA oligomer 10, the dTtaPS-mediated delivery of PMO oligomer 21 was found optimal, in terms of luciferase activity expression, at a dTtaPS concentration in the range of 2.0–3.0 μM (Fig. S2†). Confocal microscopy analysis of the dTtaPS-assisted internalization of the fluorescently-labelled PMO oligomer 18 in HeLa pLuc 705 cells revealed endosomal sequestration of 18 and nuclear fluorescence (Fig. 9), which is consistent with the level of luminescence measured upon splicing correction activity effected by dTtaPS-mediated transfection of the polyA-tailed PMO oligomer 21 (Fig. 7B).
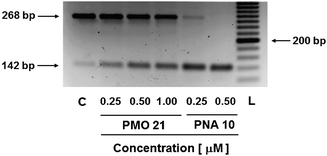
Although the results obtained from the luciferase assays and confocal microscopy analysis of the dTtaPS-assisted delivery of PNA oligomers 7 and 10 and PMO oligomers 18 and 21 provided evidence that the uncharged PNA and PMO sequences reached the nuclei of HeLa pLuc 705 cells and induced splice correction of the luciferase pre-mRNA, those results did not provide information on the extent splice correction had occurred. Reverse transcription of the aberrantly and correctly spliced luciferase pre-mRNAs and PCR amplification of the corresponding cDNAs revealed, after separation of the PCR products by agarose electrophoresis, the presence of the 142 bp cDNA fragment corresponding to the correctly spliced luciferase pre-mRNA along with the 268 bp cDNA fragment corresponding to the aberrantly spliced pre-mRNA (Fig. 10). This assessment is supported by the basal expression of luciferase in the untreated HeLa pLuc 705 cell control experiment,18 which shows the presence of both cDNA fragments. The dTtaPS-mediated delivery of the PNA oligomer 10 led to a high level of splicing correction (∼90%) at an oligomer concentration of 250 nM, whereas the delivery of PMO oligomer 21 by dTtaPS induced splicing redirection to the extent of ∼50% at an oligomer concentration of 1.0 μM. These results correlate quite well with luciferase activity measurements as shown in Fig. 4B and 7B.
Mechanism of the dTtaPS-assisted internalization of PNA or PMO oligomers
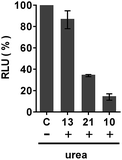
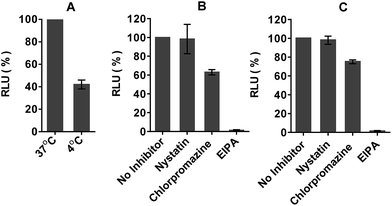
The mechanism by which the polyA tail of PNA or PMO oligomers is recognized by dTtaPS has been investigated. Although recognition specificity appears to occur through Watson–Crick base-pairing, thermal denaturation of the dTtaPS:10 complex in PBS buffer (pH 7.4) did not produce a typical sigmoidal denaturation profile from which a melting temperature could be determined. With the objective of conclusively demonstrating that the polyA tail of PNA oligomer 10 or PMO oligomer 21 is required for its dTtaPS-mediated internalization in HeLa pLuc 705 cells, we designed an experiment under which, the internalization of 10 or 21 by dTtaPS is conducted in the presence of urea. As a denaturant, urea is expected to interfere with the ability of dTtaPS to recognize the polyA tail of 10 or 21 and inhibit the cellular internalization of these sequences, thereby resulting in poorer luciferase activity. As shown in Fig. 11, the dTtaPS-mediated internalization of the PNA sequence 10 or PMO sequence 21 resulted in a decreased production of luciferase activity by more than 80% or 65%, respectively, relative to that of the control experiment performed in the absence of urea. As anticipated, the Lipofectamine® 2000-mediated internalization of the negatively charged 2′-OMe RNA sequence 13 is essentially unaffected by the presence of urea. Our findings strongly suggest that the recognition of polyA-tailed PNA or PMO oligomers by dTtaPS is specific and proceeds through weak base-pairing interactions due to the relatively bulky diastereomeric phosphorothioate triester functions of the short dTtaPS DNA transfection reagent.In order to determine whether the dTtaPS-mediated internalization of PNA oligomer 10 or PMO oligomer 21 in HeLa pLuc 705 cells proceeded through an energy dependent mechanism, a cellular uptake experiment was carried out at 37 °C while another experiment was similarly conducted at 4 °C. Fig. 12A shows that when the cellular uptake experiment is carried out at 4 °C, restoration of luciferase activity is decreased by at least 50% relative to that measured from the experiment carried out at 37 °C. These results indicate that the dTtaPS-assisted internalization of PNA oligomer 10 is in agreement with an endocytosis uptake mechanism.26 We then set out to identify the most probable endocytic pathway leading to the dTtaPS-mediated uptake of PNA oligomer 10 and PMO oligomer 21. Well-documented endocytic pathway inhibitors were employed for this purpose, namely, chlorpromazine for inhibition of clathrin-coated pits-mediated endocytosis, nystatin for inhibition of caveolae-mediated endocytosis and 5-(N-ethyl-N-isopropyl)amiloride (EIPA) for inhibition of macropinocytosis.24,25 These endocytic pathway inhibitors were used at concentrations that are known to not significantly cause cell cytotoxicity.27Fig. 12B and C shows that nystatin did not inhibit luciferase expression, whereas chlorpromazine and EIPA inhibited restoration of luciferase activity to the extent of ∼35% and ∼95%, respectively.
Although, the clathrin-coated pits-mediated endocytosis pathway is used to some extent for the dTtaPS-assisted internalization of 10 and 21 in HeLa pLuc 705 cells, macropinocytosis clearly appears to be the prevailing endocytic pathway used for this internalization process.
The efficiency of the dTtaPS-mediated transfection of PMO oligomer 21 in HeLa pLuc 705 cells relative to that of other transfection reagents
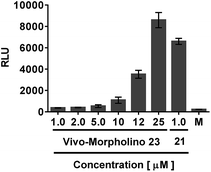
A meaningful comparison of transfection efficiency between dTtaPS and Lipofectamine® 2000 could not be established for the internalization of PMO oligomer 21 in HeLa pLuc 705 cells because Lipofectamine® 2000 cannot internalize uncharged nucleic acids in the absence of a complementary negatively charged DNA sequence28 and its molar concentration is unknown. Although CPP-PNA or PMO conjugates have been shown to be efficient in the delivery of uncharged nucleic acids in mammalian cells and animal tissues,29–31 the preparation and purification of those conjugates are tedious and laborious.15 Moreover, CPP-conjugates have been reported to convey unpredictable physico-chemical32 and functional properties to uncharged nucleic acids.33 In order to circumvent these drawbacks, we decided to use the commercially available Vivo-Morpholino conjugate of PMO oligomer 23, which is arginine-rich like many CPP conjugates, for comparing its transfection efficiency with that of the trans-acting DNA element dTtaPS. As shown in Fig. 13, the use of dTtaPS as a transfecting reagent led to the production of luciferase activity in HeLa pLuc 705 cells that is 18-fold greater than that produced by Vivo-Morpholino, based on relative light units (RLU) measured at an oligomer concentration of 1.0 μM. These results strongly demonstrate the efficiency of dTtaPS as a transfection reagent.Cytotoxicity studies
The cytotoxicity of PNA oligomer 10:dTtaPS and PMO oligomer 21:dTtaPS complexes in HeLa pLuc 705 cells was evaluated over a period of 18 h using a commercial cell-counting kit. Increasing the concentration of 10 or 21 from 1.0 μM to 2.5 μM while keeping the concentration of dTtaPS at 2.0 μM did not induce significant cytotoxicity when compared to that of the medium or in the absence of dTtaPS (ESI data Fig. S3A and B†). Similarly, the cytotoxicity of dTtaPS and dAtaPS to HeLa pLuc 705 cells was found to be minimal at concentrations optimal for transfection efficiency (Fig. S4†).Conclusions
The amphipathic trans-acting phosphorothioate DNA elements dTtaPS and dAtaPS are the first DNA-based transporters ever used for the delivery of uncharged nucleic acid sequences in mammalian cells. These DNA elements were easily prepared through solid-phase DNA synthesis protocols using appropriate deoxyribonucleoside phosphoramidites. dTtaPS was found to efficiently internalize polyA-tailed PNA or PMO oligomers in HEK 293, HeLa and SK-N-SH cells. The polyA stretch of either PNA or PMO oligomers is necessary and sufficient for the dTtaPS-assisted delivery of these oligomers in live cells. Our findings indicate that internalization of the oligomers occurs through an energy-dependent mechanism and macropinocytosis appears to be the prevailing endocytic pathway used for cellular uptake. The recognition of polyA-tailed PNA or PMO oligomers by dTtaPS is specific and appears to occur through weak base-pairing interactions. The dTtaPS-assisted delivery of PNA oligomer 10 or PMO oligomer 21 in HeLa pLuc 705 cells enabled these oligomers to travel across the cytosol and nuclear membrane to bind to the pre-mRNA encoding luciferase and induce its correct splicing to restore luciferase activity. In the absence of the polyA stretch or when the polyA stretch is replaced with a polyT or a polyC stretch, dTtaPS failed to internalize PNA or PMO oligomers in live HeLa pLuc 705 cells and prevented splice redirection of the luciferase pre-mRNA. dAtaPS was also found competent in internalizing specifically the polyT-tailed PNA oligomer 15 in HeLa pLuc 705 cells and reinstating luciferase activity. Unlike commercial cationic lipids, which preferably require serum-free media for optimal transfection performance, the phosphorothioate DNA elements dTtaPS and dAtaPS performed comparably in both serum-free and serum-containing media (Fig. S5†) while exhibiting little cytotoxicity at optimal transfection concentrations. Having demonstrated the proof of concept for this simple and efficient procedure for cellular internalization of uncharged nucleic acid analogues, future work will focus on broadening the use of amphipathic trans-acting phosphorothioate DNA elements for in vitro and in vivo delivery of therapeutically relevant uncharged and negatively charged nucleic acid-based drugs.Acknowledgements
This research is supported in part by an appointment to the Postgraduate Research Participation Program at the Center for Drug Evaluation and Research administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the U.S. Food and Drug Administration. The authors thank Prof. Rudolph L. Juliano (University of North Carolina, Chapel Hill) for kindly providing the HeLa pLuc 705 cell line and Dr Kazuyo Takeda of the Confocal Microscopy core facility at FDA-CBER for image acquisitions.Notes and references
- A. Aartsma-Rus and G. J. B. van Ommen, RNA, 2007, 13, 1609–1624 CrossRef CAS PubMed.
- P. Järver, L. O'Donovan and M. J. Gait, Nucleic Acid Ther., 2014, 24, 37–47 CrossRef PubMed.
- Q. L. Lu, A. Rabinowitz, Y. C. Chen, T. Yokota, H. Yin, J. Alter, A. Jadoon, G. Bou-Gharios and T. Partridge, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 198–203 CrossRef CAS PubMed.
- P. L. Harding, A. M. Fall, K. Honeyman, S. Fletcher and S. D. Wilton, Mol. Ther., 2007, 15, 157–166 CrossRef CAS PubMed.
- H. M. Moulton and J. D. Moulton, Biochim. Biophys. Acta, 2010, 1798, 2296–2303 CrossRef CAS PubMed.
- Q. L. Lu, C. J. Mann, F. Lou, G. Bou-Gharios, G. E. Morris, S. A. Xue, S. Fletcher, T. A. Partridge and S. D. Wilton, Nat. Med., 2003, 9, 1009–1014 CrossRef CAS PubMed.
- C. J. Mann, K. Honeyman, G. McClorey, S. Fletcher and S. D. Wilton, J. Gene Med., 2002, 4, 644–654 CrossRef CAS PubMed.
- H. Yin, Q. Lu and M. Wood, Mol. Ther., 2008, 16, 38–45 CrossRef CAS PubMed.
- T. Shiraishi, S. Pankratova and P. E. Nielsen, Chem. Biol., 2005, 12, 923–929 CrossRef CAS PubMed.
- S. Abes, H. M. Moulton, P. Clair, P. Prevot, D. S. Youngblood, R. P. Wu, P. L. Iversen and B. Lebleu, J. Controlled Release, 2006, 116, 304–313 CrossRef CAS PubMed.
- H. M. Moulton, M. C. Hase, K. M. Smith and P. L. Iversen, Antisense Nucleic Acid Drug Dev., 2003, 13, 31–43 CrossRef CAS PubMed.
- H. M. Moulton, M. H. Nelson, S. A. Hatlevig, M. T. Reddy and P. L. Iversen, Bioconjugate Chem., 2004, 15, 290–299 CrossRef CAS PubMed.
- S. Fletcher, K. Honeyman, A. M. Fall, P. L. Harding, R. D. Johnsen and S. D. Wilton, J. Gene Med., 2006, 8, 207–216 CrossRef CAS PubMed.
- S. Cirak, V. Arechavala-Gomeza, M. Guglieri, L. Feng, S. Torelli, K. Anthony, S. Abbs, M. A. Garralda, J. Bourke and D. J. Wells, et al. , Lancet, 2011, 378, 595–605 CrossRef CAS.
- T. Shiraishi and P. E. Nielsen, Artif. DNA: PNA XNA, 2011, 2, 90–99 CrossRef.
- T. S. Zatsepin, J. J. Turner, T. S. Oretskaya and M. J. Gait, Curr. Pharm. Des., 2005, 11, 3639–3654 CrossRef CAS.
- M. Zorko and Ü. Langel, Adv. Drug Delivery Rev., 2005, 57, 529–545 CrossRef CAS PubMed.
- N. Bendifallah, F. W. Rasmussen, V. Zachar, P. Ebbesen, P. E. Nielsen and U. Koppelhus, Bioconjugate Chem., 2006, 17, 750–758 CrossRef CAS PubMed.
- S. Abes, J. J. Turner, G. D. Ivanova, D. Owen, D. Williams, A. Arzumanov, P. Clair, M. J. Gait and B. Lebleu, Nucleic Acids Res., 2007, 35, 4495–4502 CrossRef CAS PubMed.
- J. J. Turner, S. Jones, M. M. Fabani, G. Ivanova, A. A. Arzumanov and M. J. Gait, Blood Cells, Mol., Dis., 2007, 38, 1–7 CrossRef CAS PubMed.
- L. O'Donovan, I. Okamoto, A. A. Arzumanov, D. L. Williams, P. Deuss and M. J. Gait, Nucleic Acid Ther., 2015, 25, 1–10 CrossRef PubMed.
- H. V. Jain, K. Takeda, C. Tami, D. Verthelyi and S. L. Beaucage, Bioorg. Med. Chem., 2013, 21, 6224–6232 CrossRef CAS PubMed.
- M. A. Maslov, T. O. Kabilova, I. A. Petukhov, N. G. Morozova, G. A. Serebrennikova, V. V. Vlassov and M. A. Zenkova, J. Controlled Release, 2012, 160, 182–193 CrossRef CAS PubMed.
- D. Zhi, S. Zhang, B. Wang, Y. Zhao, B. Yang and S. Yu, Bioconjugate Chem., 2010, 21, 563–577 CrossRef CAS PubMed.
- A. Ellington and J. D. Pollard Jr, Introduction to the synthesis and purification of oligonucleotides, in Current Protocols in Nucleic Acid Chemistry, ed. S. L. Beaucage, D. E. Bergstrom, G. D. Glick and R. A. Jones, John Wiley & Sons, Inc., Hoboken, 2000, vol. I, pp. A.3C.1–A.3C.22 Search PubMed.
- I. A. Khalil, K. Kogure, H. Akita and H. Harashima, Pharmacol. Rev., 2006, 58, 32–45 CrossRef CAS PubMed.
- G. D. Ivanova, A. Arzumanov, R. Abes, H. Yin, M. J. A. Wood, B. Lebleu and M. J. Gait, Nucleic Acids Res., 2008, 36, 6418–6428 CrossRef CAS PubMed.
- D. F. Doyle, D. A. Braasch, C. G. Simmons, B. A. Janowski and D. R. Corey, Biochemistry, 2001, 40, 53–64 CrossRef CAS PubMed.
- Y.-F. Li and P. A. Morcos, Bioconjugate Chem., 2008, 19, 1464–1470 CrossRef CAS PubMed.
- P. A. Morcos, Y. Li and S. Jiang, BioTechniques, 2008, 45, 613–623 CrossRef CAS PubMed.
- J. D. Moulton and S. Jiang, Molecules, 2009, 14, 1304–1323 CrossRef CAS PubMed.
- C. E. Prater and P. S. Miller, Bioconjugate Chem., 2004, 15, 498–507 CrossRef CAS PubMed.
- C.-H. Tung and S. Stein, Bioconjugate Chem., 2000, 11, 605–618 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Sources of common reagents and solvents, preparation and characterization of the phosphoramidites 1–6 along with descriptions of routine product purification procedures, high resolution mass spectrometry spectra, and biological data. See DOI: 10.1039/c5ra12038a |
| This journal is © The Royal Society of Chemistry 2015 |