Open Access Article
Open Access Article‘Clickable’ polymeric coatings: from antibacterial surfaces to interfaces with cellular and biomolecular affinity
Aysun
Degirmenci
a,
Rana
Sanyal
ab and
Amitav
Sanyal
 *ab
*ab
aDepartment of Chemistry, Bogazici University, Bebek, Istanbul 34342, Türkiye
bCenter for Life Sciences and Technologies, Bogazici University, Bebek, Istanbul 34342, Türkiye. E-mail: amitav.sanyal@bogazici.edu.tr
First published on 19th August 2024
Abstract
Functional polymeric coatings have become indispensable in various biomedical devices since they provide tailored interfaces with desirable properties that enable such applications. For finding an optimal system with the best performance, adopting a modular approach to interface engineering is essential for practical applications. Efficient functionalization of interfaces with specific (bio)molecules, probes, and bioactive ligands endows these interfaces with desirable properties such as biological sensing, adhesion, wettability, and anti-biofouling. In this context, ‘click’ reactions, including copper-catalyzed azide–alkyne cycloaddition (CuAAC), strain-promoted azide–alkyne cycloaddition (SPAAC), nucleophilic and radical thiol–ene, and Diels–Alder (DA) reactions, emerge as pivotal methods for effective modification of polymer-coated surfaces. This review provides an in-depth overview of utilizing ‘clickable’ group-containing polymeric coatings to create functional interfaces for biomedical applications, with a particular emphasis on antimicrobial surfaces and interfaces conducive to cellular and biomolecular immobilizations. Leveraging the versatility and modularity of surface modifications via ‘click’ chemistry, this review aims to inspire researchers to explore this promising approach for engineering functional polymeric interfaces.
1. Introduction
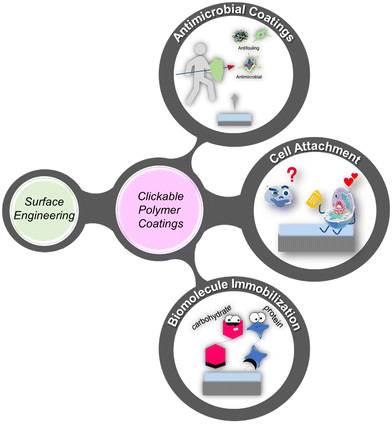
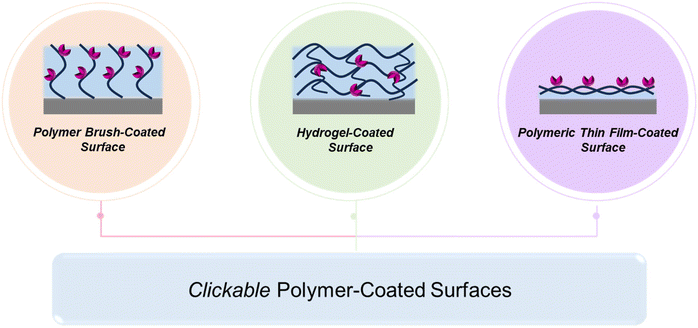
In recent years, polymer-based coatings have transcended their conventional role of material protection, emerging as pivotal components in diverse biomedical applications, spanning biosensing, imaging, and materials science.1–3 Notably, the fabrication of ‘clickable’ polymeric coatings has garnered significant attention for their indispensable role in these fields. Specifically, the demand for ‘clickable’ polymeric coatings offering facile functionalization under mild conditions has surged, facilitating rapid modifications with functional molecules tailored to specific applications.The versatility of chemical transformations encompassed by ‘click’ chemistry presents a powerful toolset for imparting functional attributes to interfaces, enabling a broad spectrum of applications ranging from biomolecule attachment and sensing to the creation of antibacterial coatings and cell-adhesive surfaces, both in specific and nonspecific ways (Scheme 1). Deploying ‘clickable’ polymeric interfaces allows for the functionalization of planar or spherical surfaces, contingent upon the application requirements. Modifications commonly include polymeric thin film coatings, polymer brushes, and polymeric hydrogel coatings (Fig. 1).
In this comprehensive review, we commence with a succinct overview of ‘click’ chemistry as a potent tool for efficient functionalization, followed by dedicated sections exploring its diverse applications. Through the analysis of recent literature examples, we underscore the efficacy and versatility of the ‘click’ chemistry-based approach. Our review aims to provide early-stage researchers with insightful guidance toward potential research avenues tailored to their specific applications while also offering seasoned researchers a comprehensive perspective to integrate related approaches into their work.
2. Click chemistry as a tool for polymer functionalization
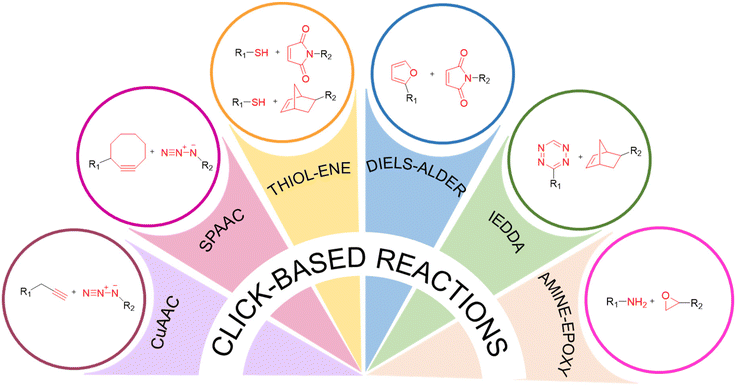
Since its inception in 2001, the ‘click’ chemistry paradigm has revolutionized the post-polymerization modification of polymeric materials,4 offering an attractive method for imparting functional attributes to polymer-coated surfaces. These reactions exhibit high efficiency under relatively mild conditions, facilitating the effective functionalization of soluble polymers and the attachment of biologically relevant moieties at the interface despite the heterogeneous reaction environment. Among the various ‘click’ reactions, copper-catalyzed azide–alkyne cycloaddition (CuAAC),5–10 strain-promoted alkyne–azide cycloaddition (SPAAC),11 Diels–Alder (DA) reactions,12 inverse electron-demand Diels–Alder (IEDDA) reactions,13 radical thiol–ene/thiol–yne reactions,14,15 and thiol–maleimide Michael addition16 reactions have gained popularity due to their fast reaction kinetics, high efficiency, specificity, and workability under mild conditions.To date, these ‘click’ reactions have found extensive utility in engineering materials for various biomedical applications, including biosensing, drug delivery, imaging, rendering surfaces antibacterial, and promoting cellular attachment in specific or nonspecific ways. Essential factors that govern the selection of a particular ‘click’ reaction may depend on factors such as (a) ready incorporation of the complementary reactive groups on the polymer and the (bio)molecule of interest, (b) compatibility of the (bio)molecule of interest to the reaction conditions; (c) stability/reversibility of attachment, and (d) desired spatiotemporal control over the functionalization process. One should weigh the advantages and limitations of the chemical transformations in light of the surface and the (bio)molecule of interest, as well as how the chemistry at the interface will affect the performance of the functionalized interface (Fig. 2) (Table 1).
| ‘Click’ reactions | Advantages | Limitations |
|---|---|---|
| Cu(I)-catalyzed azide–alkyne cycloaddition (CuAAC) reaction | • Fast reaction rate | • Toxicity of copper |
| • Regioselectivity | ||
| Strain-promoted azide–alkyne cycloaddition (SPAAC) reaction | • No metal catalyst | • Not regiospecific |
| • No byproduct | • Multistep synthesis | |
| • Bioorthogonality | • Expensive strained alkynes | |
| Radical thiol–ene reaction | • Fast reaction rate | • Needs heat or UV/Vis irradiation |
| • Well tolerable for many functional groups | ||
| Thiol-Michael addition | • Fast reaction kinetics | • Reactivity toward thiols in vivo |
| • No byproduct | ||
| Diels–Alder (DA) reaction | • Catalyst-free | • Spatial restrictions due to the adsorption of the reactants on the surface |
| • No byproduct | ||
| • Reversible with temperature | ||
| Inverse electron demand Diels–Alder (IEDDA) reaction | • Bioorthogonality | • Multistep synthesis of clickable tetrazine |
| • High reaction rate | ||
| • Chemoselectivity | ||
| Amine-epoxy reaction | • Catalyst-free | • Needs moderate to high temperatures |
3. Antibacterial/antimicrobial surfaces
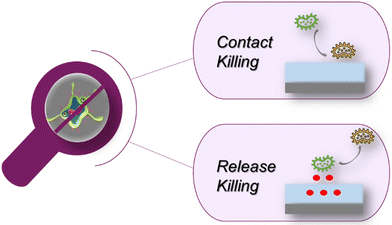
A multitude of biomedical devices, including implants, catheters, endotracheal tubes, and hernia nets, are susceptible to bacterial contamination, posing a significant risk of infections. Treatment modalities for such infections range from antibiotic therapy to surgical removal of the contaminated implant as a last resort. Bacterial adhesion to material surfaces often culminates in the formation of biofilms, which not only amplify bacterial load but also confer resistance to eradication, facilitated by the protective extracellular polymeric matrix exuded by the bacteria. Given that bacterial adhesion and subsequent biofilm formation occur predominantly at material interfaces, substantial research efforts have been directed towards developing polymeric coatings capable of rendering these interfaces both antiadhesive and bactericidal.17–19Polymeric coatings endowed with anti-biofouling properties have garnered considerable attention for enhancing in vivo compatibility and inhibiting microbial attachment. Among these, poly(ethylene glycol) (PEG) stands out as one of the most extensively studied polymers for achieving antifouling coatings. PEG-based polymers are renowned for their innate resistance to protein adhesion,20 cellular attachment,21 and bacterial colonization.22 Additionally, zwitterionic polymers have emerged as promising candidates for antifouling materials due to their superior hydration properties, which effectively deter bacterial adhesion.23,24 In addition to functioning as anti-adhesive surfaces, these coatings are designed to eradicate pathogens. This is typically achieved through either a ‘contact killing’ mechanism or a ‘release killing’ platform (Fig. 3).
Cationic materials have been explored for several decades as promising candidates for fabricating antimicrobial agents. Surfaces, particles, and soluble polymers carrying positive charges have been used as antimicrobial agents for a long time. In particular, quaternary ammonium salts (QASs) are among the most widely used low-molecular-weight antimicrobial agents because of their excellent cell membrane penetration ability, low toxicity, and environmental stability.25 A range of antibacterial polymers and hydrogel-based coatings containing QASs have been reported.26–29 Thus, surface modification using a QAS-containing polymer is an obvious choice for rendering a surface antibacterial.
As a surface coating, an interesting example was reported by Rittschof and coworkers, who fabricated a QAS-containing polymeric coating using layer-by-layer (LbL) ‘click’ deposition to combat biofouling.26 In this example, the CuAAC reaction was employed to obtain a multilayer coating rather than attaching an antibacterial component. Firstly, they prepared polydopamine-coated stainless steel (SS-PDA) surfaces. After that, alkyne-functionalized SS surfaces were fabricated by adding propargylamine to the polydopamine-modified surface. The alkyne-functionalized surface was reacted with an azide-containing azido-poly(PEGMA) polymer, whereby while some of the azide groups reacted with the alkyne groups on the modified surface to anchor the polymer, the residual azide groups on the polymer-coated surface were available for further modification. The ‘clickable’ polymer coating (SS-PPEGMA) obtained was subsequently reacted with a quaternary trimethyl ammonium chloride-based alkynyl-poly(META) polymer in the presence of CuSO4/sodium ascorbate. Subsequent layers were built using the CuAAC reaction with the azido-poly(PEGMA) copolymer. A multilayer coating was thus obtained using a layer-by-layer deposition approach. The polymer coatings exhibited good resistance to bacterial adhesion and efficient bactericidal activity against marine Pseudomonas sp.NCIMB 2021. In addition to the antifouling and bactericidal effect against bacteria, adhesion of barnacle (Amphibalanus (= Balanus) amphitrite) cyprids was reduced by up to 80% on the QAS group containing the polymer multilayer coatings. Notably, the authors showed that efficacy against bacterial fouling and adhesion of barnacle cyprids increased with the number of deposited polymer layers (Fig. 4).
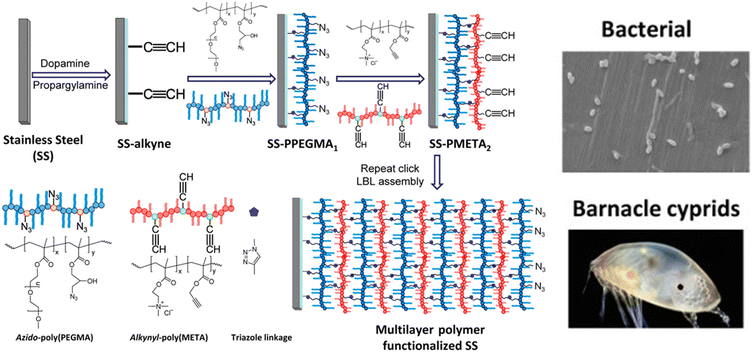 | ||
| Fig. 4 Schematic illustration of antifouling and antibacterial polymer multilayer coating fabrication via layer-by-layer ‘click’ deposition. Adopted with permission from Yang et al.26 Copyright © 2012 American Chemical Society. | ||
Another example of a multilayered antibacterial film was recently reported by Zhu and coworkers, who utilized a metal-catalyst-free thiol–yne ‘click’ reaction to install a QAS-containing coating (Fig. 5).27 The authors synthesized poly[oligo(ethylene glycol)fumarate]-co-poly[dodecyl bis(2-hydroxyethyl)methylammoniumfumarate] (POEGDMAM) containing multi-enes and poly[oligo(ethylene glycol)mercaptosuccinate] (POEGMS) containing multi-thiols. Subsequently, 3-mercaptopropyl trimethoxysilane (MPTMS) modified silicon or quartz surfaces were treated with a solution of POEGDMAM and POEGMS (at 1% concentration) to obtain a film via thiol–ene conjugate reaction, and multilayer hydrogel films were obtained by repeating this treatment step. Due to ammonium groups in POEGDMAM, the obtained hydrogel films demonstrated excellent antibacterial activity against Staphylococcus aureus (S. aureus) and Escherichia coli (E. coli). Similar to the observations by Rittschof and coworkers, in the abovementioned example, the authors observed that the antibacterial activity of the hydrogel film was enhanced by increasing the number of layers.
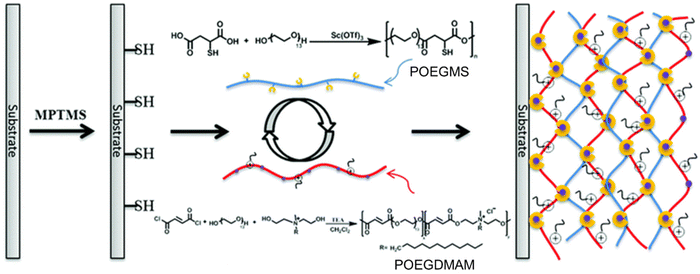 | ||
| Fig. 5 The fabrication of ultrathin hydrogel films by LbL thiol–ene ‘click’ chemistry using POEGMS and POEGDMAM polymers. Adopted with permission from Zhu et al.27 Copyright © 2014 The Royal Society of Chemistry. | ||
Apart from QAS-containing polymers, ε-poly(lysine) (EPL) and poly(hexamethylene biguanide) (PHMB) polymers are cationic polymers that exhibit excellent antimicrobial activities.30,31 Truong and coworkers reported robust, anti-infective coatings containing EPL and PHMB cationic polymers.32 First, they coated a stainless steel surface with furan-containing polyethyleneimine (PEI) and furan-containing poly(acrylic acid) using the LbL technique. After that, these multilayer polymer coatings were crosslinked using bismaleimide-PEG via the DA reaction to fabricate a stable crosslinked structure. Finally, the oxanorbornene unit in the DA cycloadduct was used for the conjugation of a poly(hexamethylene biguanide)-thiol (PHMB-SH) or with ε-poly(lysine) (EPL-SH) using the thiol–ene reaction to impart antimicrobial properties. They demonstrated that the obtained crosslinked resultant multilayer coatings modified with cationic PHMB-SH and EPL-SH polymers not only showed fast self-healing ability but also displayed efficient antibacterial activity against E. coli and S. aureus.
In recent years, as a viable alternative to PEG-based coatings, the utilization of zwitterionic polymers to combat bacterial adhesion has been on the rise due to their excellent non-fouling property. Therefore, several antimicrobial and antifouling zwitterionic polymeric coatings have been reported in recent years.23,24,33–36 Rittschof and coworkers reported the fabrication of antifouling and antimicrobial polymer brushes onto stainless steel using barnacle cement as surface anchor groups.33 In their work, barnacle cement was used as a surface anchor to introduce ‘clickable’ groups on stainless steel surfaces for further modification. The barnacle cement-coated surface was reacted with ethylene sulfide, propargyl carbonylimidazole, and azidoethyl carbonylimidazole to obtain a thiol, alkyne, and azide group-containing stainless-steel surface, respectively. An antifouling zwitterionic SS-PMPC surface was prepared using thiol–ene photopolymerization of 2-methacryloyloxyethylphosphorylcholine (MPC) from the thiol-containing SS surface. Likewise, protein-resistant SS-PPEGMA and protein-adsorbing SS-PPFS polymer surfaces were prepared by clicking an azide-terminated poly(poly(ethylene glycol)methyl ether methacrylate) (azido-PPEGMA) and poly(2,3,4,5,6-pentafluorostyrene) (azido-PPFS) via the CuAAC reaction based graft-to approach. In addition, antifouling alkyne-bearing poly(N-hydroxyethylacrylamide) (alkynyl-PHEAA) and antibacterial alkyne-modified poly(2-(methacryloyloxy)ethyl trimethylammonium chloride) (alkynyl-PMETA) were clicked onto the azide-containing SS surface. The zwitterionic SS-PMPC, non-ionic SS-PHEAA, and antibacterial SS-PMETA polymer surfaces inhibited the adhesion of Gram-negative E. coli and Gram-positive S. epidermidis, with viable adherent bacterial fractions being significantly low compared to that on the pristine SS surface.
Kang and coworkers reported antifouling and antimicrobial coatings using zwitterionic and cationic binary polymer brushes via a ‘click’ reaction34 (Fig. 6). An effective method to fabricate antifouling and antimicrobial surfaces using zwitterionic and cationic binary polymer brushes onto poly-dopamine-coated stainless steel (SS) surfaces was reported. Firstly, the SS surface was deposited with polydopamine (PDA) and reacted with ethylene sulfide and 3-azidopropyl carbonylimidazole to introduce thiol and azide groups onto the PDA-coated surface. Zwitterionic 2-methacryloyloxyethyl phosphorylcholine was grafted and polymerized from a thiol-containing PDA-coated surface via thiol–ene photopolymerization to obtain PMPC-grafted surface (SS-g-PMPC). After that, an alkyne-bearing poly(2-(methacryloyloxy)ethyl trimethylammonium chloride) (PMETA) obtained using atom transfer radical polymerization (ATRP) was conjugated onto the azide groups on the surface to obtain a bifunctional zwitterionic and cationic polymer brush coating. The resulting polymer brush coatings exhibited good resistance to the bacterial adhesion of S. aureus and Pseudomonas sp., as well as toward the microalgal attachment of Amphora coffeaeformis. This interesting dual-functionalization approach provides an exciting approach to impart multifunctionality to surfaces.
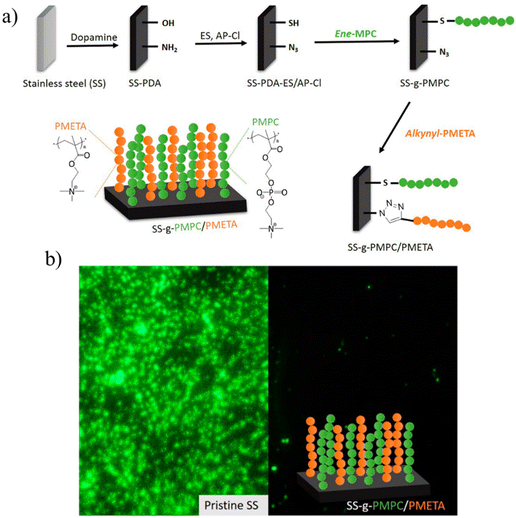 | ||
| Fig. 6 (a) Schematic illustration of fabricating binary polymer brush coatings on stainless steel surfaces via ‘click’ reaction; (b) fluorescence microscopy images of pristine SS and SS-g-PMPC/PMETA brushes after exposure to S. aureus. Adopted with permission from Xu et al.34 Copyright © 2017 American Chemical Society. | ||
Another example employing a PDA-based initial coating followed by the attachment of antifouling and antibacterial polymers was reported by Yang and coworkers.36 The authors synthesized a series of multifunctional antimicrobial polymeric coatings by using diethylene glycol diglycidyl ether (PEGDGE), polylysine, and poly[glycidylmethacrylate-co-3-(dimethyl(4-vinylbenzyl)ammonium)propyl sulfonate] (poly(GMA-co-DVBAPS)) to benefit from their antifouling, bactericidal, and release properties, respectively. The polymers were also tightly anchored onto various substrates (glass, polypropylene, polyethylene terephthalate, and catheter), presumably because of Michael-addition and ring-opening reactions during the co-deposition process. Three polymeric interfaces containing a polydopamine/poly(GMA-co-DVBAPS) (PDA/PDV) coating, a polydopamine/polylysine/poly(GMA-co-DVBAPS) (PDA/L-PDV) coating, and a polydopamine/polylysine/poly(GMA-co-DVBAPS)/PEGDGE (PDA/l-PDV-PEGDGE) coating were fabricated using this approach. The PDA/PDV coating showed a distinct salt-responsive conformation change, which released 95.8% of adhered E. coli. In addition, the PDA/L-PDV coating also provided salt-responsive bacterial release (∼91.4/∼95.1% for E. coli/S. aureus) and killed the attached bacteria (up to ∼92.8/∼90.2%). Moreover, the zwitterionic PDA/L-PDV-PEGDGE coatings with multiple antimicrobial mechanisms demonstrated long-term surface resistance for E. coli (∼96 h)/S. aureus (∼60 h), antibacterial activity (∼44.7/∼54.7%), and very efficient salt-responsive surface regeneration ability (∼94.4/∼95.4%). As the authors suggested, the present strategy offers a promising approach for fabricating antibacterial coatings where components acting simultaneously with different mechanisms can be incorporated onto a surface to obtain a multifunctional coating (Fig. 7).
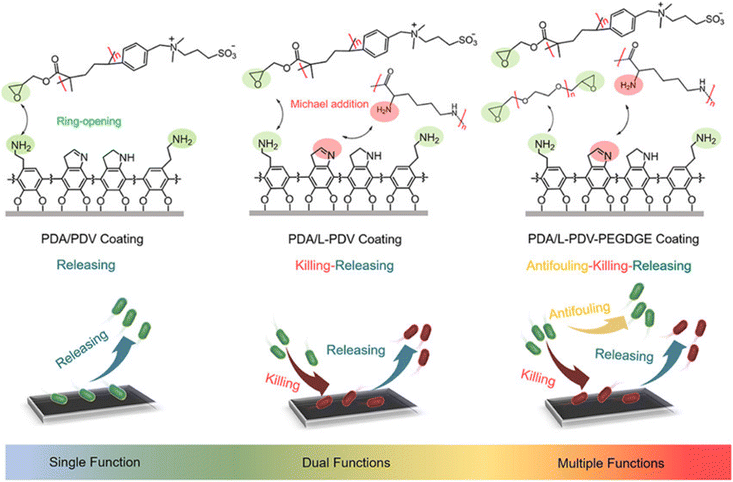 | ||
| Fig. 7 Schematic representation of mussel-inspired polymeric coatings realizing functions from single to multiple antimicrobial mechanisms. Three functional polymers, i.e., polylysine, diethylene glycol diglycidyl ether (PEGDGE), and poly[glycidylmethacrylate-co-3-(dimethyl(4-vinylbenzyl)ammonium)propyl sulfonate] (poly(GMA-co-DVBAPS)), were selected. Adapted with permission from Mao et al.36 Copyright © 2021 American Chemical Society. | ||
Another approach to inhibit microbial attachment is the employment of antimicrobial coatings where polymers are conjugated with cationic biocides, antibiotics, or antimicrobial peptides (AMPs). Among them, AMPs provide an effective antimicrobial activity due to their potent activity.37 AMPs can be incorporated into the polymeric coating physically38 or through chemical conjugation.37,39 The chemical immobilization of AMPs provides a stable antimicrobial coating by decreasing toxicity and unwanted release from the surface.40 AMPs can be chemoselectively conjugated to an appropriately functionalized surface containing ‘clickable’ groups.41–57 Kizhakkedathu and coworkers have reported several AMP-conjugated antimicrobial coatings.42,44,51 In one of their studies, an AMP-conjugated polymer brush was fabricated on a titanium surface.42 Using surface-initiated ATRP (SI-ATRP), an amine-containing polymer brush was obtained, which was modified to install thiol-reactive maleimide functional groups through post-polymerization modification. Cysteine-functionalized cationic antimicrobial peptide Tet213 (KRWWKWWRRC) was conjugated to a polymer brush via the thiol–maleimide ‘click’ reaction. The authors demonstrated that the AMP density and the grafting density of the polymer brush significantly influenced the antimicrobial activity of AMP-conjugated polymer brushes against Pseudomonas aeruginosa (Fig. 8).
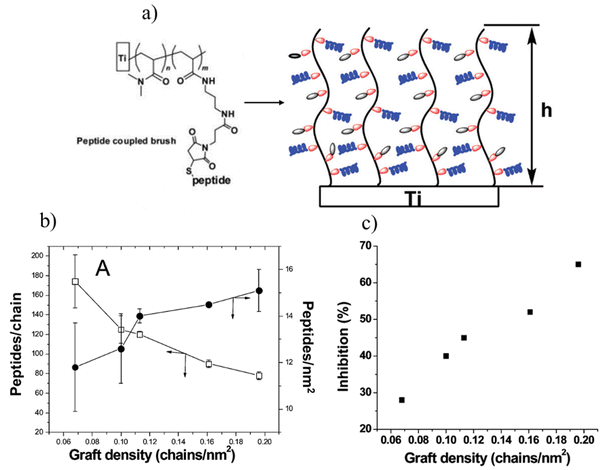 | ||
| Fig. 8 (a) The schematic illustration of a cysteine-functionalized cationic antimicrobial peptide Tet213 (KRWWKWWRRC) conjugated polymer brush via thiol–maleimide ‘click’ reaction; (b) effect of graft density on the number of peptides grafted per chain and peptide density on the surface; (c) effect of graft density and thickness of the peptide grafted brush on bacterial growth inhibition. Reprinted with permission from Gao et al.42 Copyright © 2011 American Chemical Society. | ||
In a subsequent study, Kizhakkedathu and coworkers used the maleimide-containing polymer brush coating for in vivo application,44 and they showed that the AMP-tethered brush-coated surfaces maintained excellent antibacterial activity in vitro and in vivo. The coating was effective in resisting biofilm formation, and the biofilm resistance depended on the nature of modified AMPs. They also demonstrated that the coatings could protect against bacterial infection in vivo. In an alternative approach, Sanyal, Kizhakkedathu, and coworkers reported a thiol-reactive polymeric coating on a TiO2 surface to achieve an antimicrobial polymeric thin film.51 Using reversible addition–fragmentation chain transfer (RAFT) polymerization, a surface-anchorable copolymer containing dopamine side chain residues as surface attachment groups and furan-protected maleimide groups as masked maleimide groups were synthesized. A robust coating on TiO2 surfaces was obtained using a DOPA-containing copolymer. The protected maleimide groups on the surface were activated by removing the furan group using the retro-Diels–Alder (rDA) reaction to get the thiol-reactive maleimide units. After obtaining a ‘clickable’ maleimide-containing polymeric film, the antimicrobial peptide E6 (RRWRIVVIRVRRC) was conjugated onto the polymer-coated surface via the thiol–maleimide Michael addition. The authors investigated the antimicrobial activity of peptide-conjugated polymer-coated surfaces against Gram-positive and Gram-negative bacteria. They showed that the hydrophilic polymer coatings considerably decreased the adhesion level of bacteria compared to the pristine inorganic surface and killed >80% of the adhered bacteria on the surface (Fig. 9). As a different approach, Wang and coworkers reported the preparation of an antimicrobial titanium surface by conjugating peptide via CuAAC chemistry.52 First, a ‘clickable’ titanium surface was obtained using a silane-based coupling agent bearing an alkynyl group. Subsequently, the alkyne group containing surface was coated with azide-functionalized PEG, which was directly conjugated with antimicrobial peptide (PEG-HHC36:N3-PEG12-KRWWKWWRR) via the CuAAC reaction to furnish an AMP conjugated PEG-based antimicrobial coating with an AMP density of 897.4 ± 67.3 ng cm−2 (2.5 ± 0.2 molecules per nm2) on the surface. The authors showed that this construct inhibited 90.2% of Staphylococcus aureus and 88.1% of Escherichia coli after 2.5 h of incubation. They applied the CCK-8 assay using mouse bone mesenchymal stem cells, and it indicated that this AMP-conjugated titanium coating had negligible cytotoxicity. Moreover, an in vivo assay showed that this coating could kill 78.8% of S. aureus after 7 days. In 2022, Chen and coworkers reported an elegant example of a mussel-inspired antibacterial peptide coating on ureteral stents for encrustation prevention.56 A polypeptide consisting of four catecholic dopamine (DOPA) units and one Ac-(DA)-Gly-(DA)-Lys[(PEG5)-(Mpa)-(Mal-DBCO)]-(DA)-Gly-(DA) (DBCO) group, abbreviated as (DA)4-DBCO and DBCO-containing modified polydopamine, was obtained. The amount of DOPA was increased to achieve more stable substrate adhesion onto the surface. They also prepared a specific antibacterial peptide, namely, (3-azido)-RWRWRW-NH2, including the azido group. After synthesis, the stent surface was coated with (DA)4-DBCO. After that, the azide-containing antibacterial peptide (3-azido)-RWRWRW-NH2 was conjugated to the stent surface via ‘click’ chemistry, where copper (Cu) ions were used to coordinate the mussel-inspired antibacterial peptide to improve the stability. They reported that the Cu2+ coordinated antibacterial coating could improve stability and antibacterial ability with low or no cytotoxicity in vivo and in vitro. In addition, they demonstrated that AMP-conjugated stents could resist infection and reduce encrustation formation in vivo for a long time. Another important example of a mussel-inspired biomimetic surface for rational integration of anti-infectivity and osteo-inductivity onto PEEK (polyetheretherketone) implants was reported by Geng and coworkers.57 They coated PEEK surfaces with PEG-based DOPA-rich peptide with a ‘clickable’ azide group. Subsequently, coated PEEK surfaces were conjugated with dibenzocyclooctyne (DBCO)-modified AMP (RWRWRW) peptide and the osteogenic growth peptide (OGP) through bioorthogonal SPAAC reaction. They showed an excellent anti-infection effect of the dual peptide conjugated surface by implanting S. aureus-contaminated PEEK rods into the femurs of rats in vivo. Furthermore, they reported that a newly formed bone surrounding the implant was found after six weeks of implantation, which was much denser than bare PEEK and PEEK coated with either peptide.
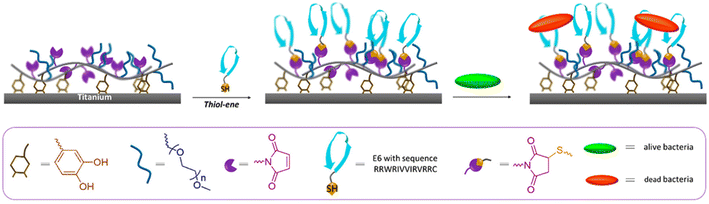 | ||
| Fig. 9 Schematic illustration of an AMP-conjugated thiol-reactive polymeric coating on a TiO2 surface to fabricate an antimicrobial coating. Adapted with permission from Gevrek et al.51 Copyright © 2019 American Chemical Society. | ||
In addition to polymer coating examples, ‘clickable’ hydrogel coatings have also been reported to obtain antimicrobial films.45,46,48 Liskamp and coworkers reported the fabrication of a hydrogel coating on a PET surface to achieve an antimicrobial coating.45 They obtained the hydrogel coating on the PET surface using a PEGDA crosslinker, pentaerythritol tetrakis(3-mercaptopropionate) (PTMP), in the presence of DMPA as a photoinitiator via thiol–ene reaction. Moreover, the antimicrobial peptide (AMP) HHC10 (H-KRWWKWIRW-NH2) was incorporated into a PEG-based hydrogel coating using a radical thiol–ene ‘click’ reaction in a single step. The authors demonstrated that the resulting AMP-conjugated hydrogels displayed bactericidal activity against Gram-positive S. aureus and S. epidermidis and Gram-negative E. coli in in vitro studies.
Antimicrobial surfaces via the incorporation of bactericides is another way to decrease bacterial attachment. While antibiotics are widely utilized to treat and manage bacterial infections, there are concerns such as the burst release of drugs, low efficacy, and the evolution of bacteria with antibiotic resistance.58 Thus, approaches where antibiotics are loaded into the polymeric coatings through chemical conjugation are of interest, and ‘click’ reactions provide an avenue to design release-killing antibacterial coatings.59–62 For example, Song and coworkers fabricated vancomycin-conjugated polymer brushes to suppress S. aureus colonization (Fig. 10).60 They coated a Ti6Al4V alloy surface with an ATRP initiator; subsequently, azide-bearing polymer brushes were synthesized using an azide-containing TEG-based methacrylate monomer. After that, alkyne-functionalized vancomycin was conjugated onto the azide-containing polymer brush via the CuAAC reaction. They observed that covalently tethered vancomycin suppressed S. aureus colonization on polymer brush-coated metallic surfaces. In addition, in vivo, animal studies revealed that vancomycin-conjugated polymer-coated titanium pins inserted in femoral canals infected with S. aureus significantly decreased bacterial colonization. The authors suggest that the disclosed surface functionalization strategy may be extended to conjugate any antibacterial agent to mitigate periprosthetic infections.
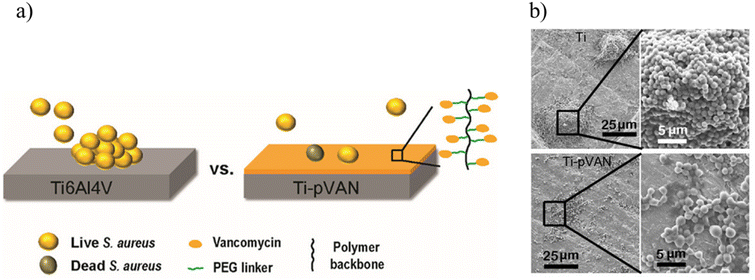 | ||
| Fig. 10 (a) Fabrication of a vancomycin conjugated polymer brush on a titanium alloy for suppression of S. aureus colonization; (b) SEM of S. aureus adhered on unmodified Ti6Al4V and Ti-pVAN plates after 5 h incubation with 1.2 × 105 CFU per mL S. aureus. Adapted with permission from Zhang et al.60 Copyright © 2019 American Chemical Society. | ||
Although the covalent conjugation of incorporating antibiotics is a promising strategy to minimize burst release, alternative design approaches to fabricate drug-loaded coatings may also minimize unwanted release. Segura and coworkers reported a study along these lines,62 where poly(allyl mercaptan) (PAM) was synthesized using the thiol–ene radical copolymerization of 1,3-propanedithiol and allyl sulfide. Titanium wires were submerged in chloroform in a solution consisting of PAM and tetra-PEG-thiol. Subsequent irradiation under UV light for 5 min induced the formation of a tetra-PEG-PAM coating directly on the implant surface. Vancomycin was introduced into the coating by mixing with tetra-PEG and PAM before UV irradiation. Vancomycin release in a PBS bath showed that the drug concentration in the solution increased over time and was above the minimum inhibitory concentration (MIC) (0.5 μg mL−1 for S. aureus Xen 36) for 14 days. The authors showed that titanium pins coated with three layers of PEG-PAM + Vanc decreased the burst release of Vanc and resulted in concentrations above the MIC for 17 days. They performed a zone of inhibition (ZOI) assay to measure in vitro antimicrobial activity against S. aureus and observed distinctive dead zones on the plate. Moreover, they incorporated a variety of clinically relevant antibiotics such as cefazolin, cefepime, ceftriaxone, piperacillin, rifampin, tazobactam, clindamycin, tobramycin, and linezolid, and reported that the thus-obtained antibiotic-loaded titanium pins possessed the ability to inhibit S. aureus growth.
4. Cell adhesive coatings
The specific functionalization of interfaces with bioactive molecules like cell adhesive peptides and growth factors is highly important in investigating cell–material interactions, which can provide control over the response of a cell to its surroundings. Because of the complex interaction of the cell with the extracellular matrix (ECM) and crosstalk between different signaling molecules, there is a demand for the fabrication of substrates that can be modified explicitly with desired ligands. One of the most widely employed surface modifications entails the attachment of the cell adhesive peptide containing the arginine–glycine–aspartic acid (RGD) sequence. This specific sequence is present in many of the ECM proteins, and it is known to specifically bind to the integrin receptors on the cell's surface, promoting cellular attachment to the underlying substrate.63,64 In this regard, efforts have been devoted to fabricating RGD-conjugated polymer-based coatings to obtain cell adhesive surfaces,63,65,66 and many of those have been achieved using ‘click’ reaction-based conjugation.67–87Coating surfaces of titanium-based biomedical implants are of high interest to facilitate their integration within the surrounding tissue. To date, various cell adhesive peptide-conjugated polymeric coatings have been investigated.69,71,80 In a recent study, Sanyal and coworkers reported the fabrication of a ‘clickable’ hydrogel coating on a titanium surface.80 A furan-protected maleimide group containing a hydrogel layer on a titanium surface was obtained. Using photopolymerization, an adlayer of dopamine-containing methacrylate was utilized to anchor the hydrogel coating during hydrogel formation. The initial coating containing the strained oxabicyclonorbornene group is amenable for functionalization with radical thiol–ene and the IEDDA ‘click’ reactions (Fig. 11a). After thermal treatment, the protected maleimide units were converted to thiol-reactive maleimide groups, allowing functionalization of hydrogel coatings via the thiol–maleimide Michael addition (Fig. 11b). The maleimide group containing hydrogel coating could be readily functionalized with a cell adhesive RGD peptide (RGDC), which promotes the attachment and proliferation of fibroblast NIH-3T3 cells (Fig. 11c).
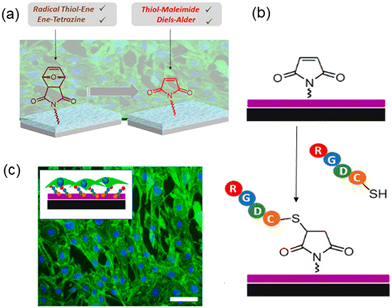 | ||
| Fig. 11 (a) Fabrication of a maleimide containing hydrogel coating on a titanium surface; (b) schematic representation of the RGDC conjugated hydrogel coating for cellular attachment and proliferation; (c) NIH-3T3 cell proliferation on the RGD-conjugated hydrogel coating. Reprinted with permission from Gevrek et al.80 Copyright © 2020 by the authors. Licensee MDPI, Basel, Switzerland. | ||
While most studies have investigated the irreversible attachment of cells onto polymeric substrates, several reports have shown the study of reversible binding platforms. The reversible binding can be useful for catch and release applications,88–91 where a particular target cell can be caught on the substrate and then released for downstream genetic analysis, thus providing more information on the spread of malignant cells in a disease like cancer. Another advantage of a reversible system could be that it facilitates obtaining an interface with patterned cells. The latter was recently demonstrated by Koga and coworkers, who reported the fabrication of a photocleavable peptide-poly(2-hydroxyethyl methacrylate) (PHEMA) hybrid graft polymer to modulate the cell binding affinities of 2D and 3D coatings (Fig. 12).77 They synthesized a ‘clickable’ poly(HEMA-co-PgA) copolymer using HEMA and propargyl acrylate. To prepare a 2D film coating, glass plates (1 cm2) were coated with poly(HEMA-co-PgA) using the spin coating method (Fig. 12a). They demonstrated that the presence of the photocleavable linker enabled spatially controlled cleavage of conjugated materials from the surface (Fig. 12b). A red dye conjugate could be cleaved from the surface by spatially controlled exposure to UV irradiation, while the unexposed area remains intact, as is evident from the fluorescence image of the substrate after the UV exposure. The alkyne-bearing polymer-coated materials were conjugated with a photo-cleavable azide-terminated oligopeptide [Arg-Gly-Asp-Ser (RGDS)] with a photolabile 3-amino-3-(2-nitrophenyl)-propanoic acid moiety, using the CuAAC reaction. The authors showed that the RGDS conjugated thin film acted as an effective interface for successful attachment and proliferation of NIH/3T3 fibroblasts and MC3T3-E1 osteoblast-like cells in vitro. Furthermore, they showed that UV exposure to the surface led to the detachment of cells from the surface due to the photo-cleavage of RGDS grafts (Fig. 12c). Moreover, the applicability of this system to 3D materials was examined, and the authors reported that cell adhesion was remarkably enhanced on RGDS-conjugated 3D-printed PLA objects.
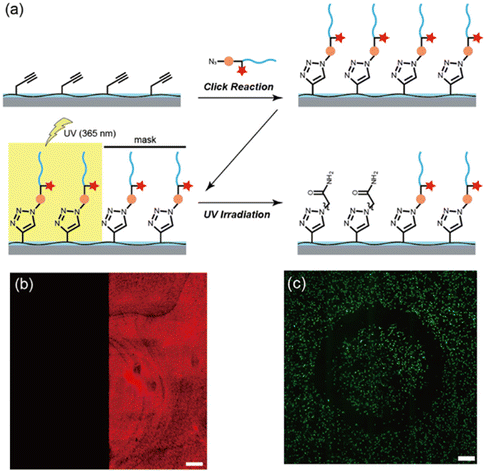 | ||
| Fig. 12 (a) Fabrication of a photocleavable peptide-poly(2-hydroxyethyl methacrylate) hybrid graft copolymer via ‘click’ reaction to modulate the cell affinities of 2D and 3D substrates; (b) confocal microscopy image of the RGDS-patterned film (left: UV-irradiated region, right: nonirradiated region); (c) phototriggered cell release from the hybrid thin film by using a circular ring-patterned photomask. Adapted with permission from Nishimura et al.77 Copyright © 2019 American Chemical Society. | ||
Minko and coworkers reported an alternative approach to obtain patterned domains of cells without resorting to the metal-catalyzed CuAAC reaction.87 A stimuli-responsive non-enzymatic cell harvesting was performed using nanostructured thermoresponsive polymer films made of cell-adhesive epoxy photoresist domains and cell-adhesion resistive poly(N-isopropylacrylamide) (pNIPAm) brush regions. The cell adhesive RGD peptide was conjugated to a polymer film via the amine-epoxy ‘click’ reaction. They reported that while RGD peptide conjugated domains enable cell binding, pNIPAm-based soft thermoresponsive polymer brush domains generate pressure, which allows the detachment of cells at a temperature below the lower critical solution temperature (LCST) of the polymer.
While most of the work reported to date involves functionalizing the interface with a single peptide motif such as RGD, it is known that the cell surface possesses multiple receptors and thus often utilizes many different interactions with their environment. Strategies to obtain polymeric surfaces capable of installing multiple bioactive ligands would be valuable in introducing further control over the cell material interface. An interesting approach was reported by Wegner and coworkers, who reported the preparation of a dual-functionalized nanostructured surface via a ‘click’ reaction to examine the crosstalk and relationship between different signaling molecules and the clustering effect in ligand–receptor interactions.73 The authors fabricated gold nanoparticles on the glass surface using block polymer micelle nanolithography. After obtaining gold nanoparticles on the glass surface, the surface was coated with a silane- and alkyne-bearing PEG polymer (CH3CH2O)3Si-PEG3000-alkyne, and thus a ‘clickable’ alkyne-bearing gold nanoparticle containing polymer coating was constructed. A thiol-containing adhesive cRGD peptide was introduced to gold nanoparticles on the polymer coating using gold–thiol chemistry. Further conjugation of the polymer coating with an azide-bearing PHSRN synergy peptide, which enhances cell spreading in the presence of cRGD, was undertaken. The work established that the presence of the synergy peptide PHSRN affects the integrin-mediated adhesion of rat embryonic fibroblast cells, wild type (RFT WT) via cRGD units. REF-WT cells were seeded onto polymer coatings, and the density of attached cells on the surface and their spreading area were quantified. Focal cell adhesions were probed using vinculin and paxillin staining to examine the cell attachment to the surfaces. While the REF WT cells did not adhere to surfaces modified solely with the synergy peptide PHSRN and only a few cells showed attachment on surfaces functionalized with cRGD peptide, the cells adhered well on surfaces functionalized with both PHSRN and cRGD peptides. In addition, they observed that the cells had a smaller spreading area on only cRGD-functionalized surfaces compared to surfaces modified with both cRGD and PHSRN. Moreover, the cells on surfaces functionalized with the two signaling molecules developed mature focal adhesion and well-developed actin fibers characteristic of adhered fibroblasts. The authors suggested that such dual-functionalized surfaces can also be used to examine the crosstalk and spatial requirements for processes with cRGD and PHSRN signaling molecules (Fig. 13).
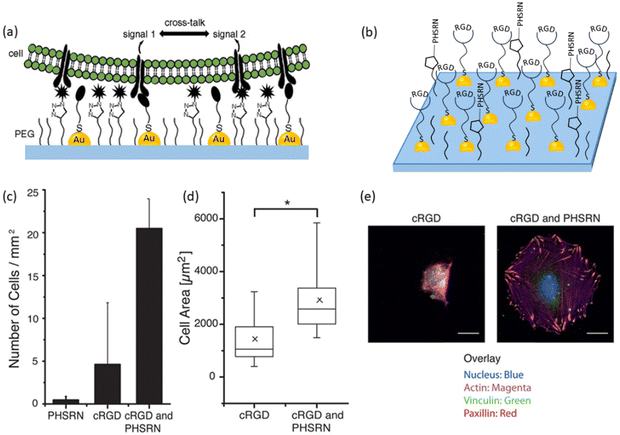 | ||
| Fig. 13 (a) The fabrication of a dual-functionalized nanostructured polymer coating by ‘click’ chemistry for cellular adhesion; (b) scheme for dual functionalization with adhesion peptide cRGD and synergy peptide PHSRN; (c) density of adherent REF WT cells on substrates with gold nanoparticles and PEG-alkyne (10 mol%) modified with cRGD and/or PHSRN; (d) the spreading area of cells on cRGD- or cRGD- and PHSRN-conjugated interfaces; (e) fluorescence images of adherent cells on substrates functionalized with cRGD or cRGD and PHSRN. The nucleus is shown in blue, actin in magenta, vinculin in green, and paxillin in red. Adapted with permission from Schenk et al.73 Copyright © 2014 American Chemical Society. | ||
5. Interfaces for biosensing and biomolecular immobilization
Microarray platforms are attracting increasing attention due to their indispensable role in various areas of chemical, biological, and biomedical sciences. It is anticipated that microarrays would enable the selection and assessment of drug candidates and disease diagnosis and allow one to evaluate treatment outcomes.92 The underlying approach for microarray fabrication involves surface modification of a substrate to immobilize probe molecules, where surface science plays an important role.93 An early example of a microarray was composed of recombinant DNA plasmids deposited on filter paper, and then it was hybridized with specific cDNA sequences.94 Today, in addition to DNA microarrays, there are many examples of microarrays for proteins,95 peptides,96 antibodies,97 carbohydrates,98 and cells.99 Microarrays’ performance strongly depends on the spatial localization of spots on the surface, spot density, and morphology.100 In addition to these factors, the physical and chemical properties of the surface are crucial to determine the robustness of the array technology.101 The specific and nonspecific binding of molecules is affected by the surface, density, and orientation of immobilized biomolecules, which play a crucial role in the sensitivity and selectivity of the array technique.102 In this regard, polymeric coatings have attracted attention in fabricating homogeneous coatings with increased binding capacity, control over probe localization, and enabling selective biomolecular recognition with the target molecule. One of the main advantages of using a polymeric platform is that the polymer can be designed to tune the coating for a specific application.103Commonly employed fabrication of polymeric coatings for sensing biological targets entails immobilizing biological probes such as carbohydrates, proteins, and oligonucleotides. Effective conjugation chemistry is vital, and the heterogeneous nature of the interface makes this challenging. Thus, highly effective reactions, such as the ‘click’ reactions, have emerged as valuable methods for selective and directional functionalization with high efficiency, selectivity, and fast reaction kinetics.4 Several examples of ‘clickable’ coatings have been reported for the past two decades.104–115 Hawker and coworkers reported an example of microarrays covalently attached at the hydrogel-coated PEG-based surface using thiol–ene chemistry.104 These hydrogel-coated microarrays provide many advantages, such as robustness, tunable mechanical properties, and the ability to incorporate orthogonal functional groups using thiol and alkene pendant groups. Fabrication of high-throughput spotted microarrays on hydrogel coating containing thiol-functional peptides for cell adhesion was demonstrated. Furthermore, functional groups such as azide, aldehyde, and NHS-ester were incorporated using thiol–ene chemistry to immobilize biomolecules and/or dye.
Seminal contributions in using reactive polymer coatings with reactive functional groups were reported almost two decades ago. For example, Pirri et al. reported the fabrication of DNA microarrays using a copolymer composed of N,N-dimethylacrylamide, N-acryloyloxysuccinimide, and 3-(trimethoxysilyl)propyl methacrylate, namely copoly(DMA-NAS-MAPS).105 Amine-containing oligonucleotides were conjugated onto the reactive copolymer coating to obtain a high-density array capable of undergoing hybridization with cDNA targets. In subsequent studies, the same group employed the polymer coating to obtain protein microarrays.106 In an approach incorporating ‘click’ reactions with the abovementioned copolymer, Cretich and coworkers reported the fabrication of peptide microarrays to screen Burkholderia cepacia complex infection among cystic fibrosis patients.108 Microarrays of Burkholderia-derived peptide probes that were selectively oriented on polymeric coatings were obtained by ‘click’ reactions such as thiol–maleimide, copper-catalyzed azide–alkyne cycloaddition, and copper-free strain-promoted azide–alkyne cycloaddition reactions (Fig. 14). The authors compared the immobilization efficiency of microarrays constructed with the different chemoselective reactions and evaluated diagnostic performances in contrast to random immobilization techniques. Notably, while random immobilization was not feasible for reliable diagnosis and showed poor binding efficiency and reproducibility, correct peptide orientation due to chemoselective ‘click’ reactions demonstrated successful diagnostic performance. Further performance improvement was observed when the peptide probes were spaced from the chip surfaces using small PEG units. Interestingly, all the evaluated immobilization strategies displayed good efficiency in the final diagnostic performance.
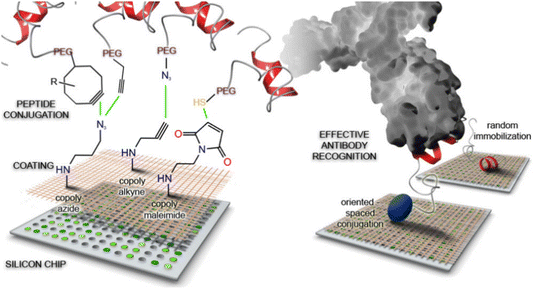 | ||
| Fig. 14 Schematic illustration of peptide conjugation onto a ‘clickable’ polymer-coated silicon chip for effective antibody recognition. Adapted with permission from Gori et al.108 Copyright © 2016 American Chemical Society. | ||
Around a similar time, Chiari and coworkers also reported the fabrication of a ‘clickable’ polymer coating using post-polymerization modification for DNA microarray applications.109 The surface attachable copolymer copoly(DMA-NAS-MAPS), which they had earlier reported was utilized to fabricate the reactive polymeric coating on the substrate, and groups such as azide, alkyne, thiol, and maleimide groups were introduced to the surface. The authors note that the degradation of the active NHS-ester could be problematic when large volumes are used for protein immobilization, such as in microchannel derivatization. The authors showed the improved stability and benefits of using the ‘click’ reactions. Also, they showed the applicability of these functionalized surfaces in a highly effective solid-phase PCR for the genotyping of KRAS G12D mutation (Fig. 15).
 | ||
| Fig. 15 (a) Fabrication of clickable polymer coatings via post-polymerization modification for microarray technology and (b) schematic representation of the oligonucleotides immobilized on the surfaces, coated with the ‘click’ copolymers. Adapted with permission from Sola et al.109 Copyright © 2016 American Chemical Society. | ||
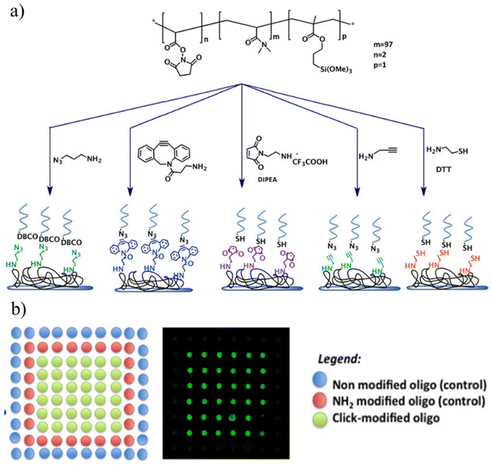
Apart from the derivatization of reactive polymeric coatings with ‘clickable’ groups, several reports utilize polymeric coatings that directly incorporate these reactive handles.116–136 Thus, the parent copolymers fabricating the coatings possess a ‘clickable’ functional group, thus avoiding any intermediate surface modification step before bioimmobilization. Hoven and coworkers reported fabricating a ‘clickable’ and antifouling platform using a polymer containing a propargyl group and 2-methacryloyloxyethyl phosphorylcholine for biosensing application.123 They synthesized a poly-[(propargyl methacrylate)-ran-(2-methacryloyloxyethyl phosphorylcholine)] (PPgMAMPC) polymer using RAFT polymerization. Subsequently, the PPgMAMPC polymer was converted into a thiol-terminated PPgMAMPC-SH polymer by aminolysis of the phenyl-dithioester group. After that, the PPgMAMPC-SH polymer was immobilized on the gold-coated surface using a “grafting to” approach, and an alkynyl group containing polymer coating was obtained. Azide-containing biotin and peptide nucleic acid (PNA) were conjugated to the polymeric surface via the CuAAC reaction, and biosensing application was demonstrated by detecting specific target molecules, such as streptavidin and DNA. Using this direct approach, Sanyal and coworkers have reported many examples of ‘clickable’ polymer coatings for biomolecular immobilization and sensing.120,126,127,130,133 For example, Sanyal, Klok, and coworkers reported the facile fabrication of ‘clickable’ maleimide-containing polymer brushes via “grafting-from” surface-initiated atom transfer radical copolymerization (SI-ATRP).120 They polymerized poly(ethylene glycol) methyl ether methacrylate (PEGMEMA) and a furan-protected maleimide-containing monomer (FuMaMA) via SI-ATRP. Subsequently, a thermally induced retro Diels–Alder reaction gave thiol-reactive maleimide-including copolymer brushes. After the fabrication and characterization of copolymer brushes, they explored post-polymerization functionalization of polymer brushes using thiol-containing dyes and a biotin-derivative via the thiol–maleimide conjugation reaction. The authors immobilized streptavidin-coated quantum dots onto biotin-conjugated copolymer brushes. In another study, Sanyal and coworkers reported the fabrication of a thiol-reactive polymeric thin film coating for biomolecule immobilization (Fig. 16).127 The authors synthesized poly(ethylene glycol) (PEG)-based copolymer containing alkoxylsilyl groups to provide surface binding and furan-protected maleimide groups for post-polymerization modification. Si/SiO2 surfaces were coated with this copolymer, and after a thermal treatment, a maleimide group containing a thin polymeric film was obtained. The attachment of a thiol-containing fluorescent BODIPY-SH dye demonstrated surface modification via a thiol-maleimide reaction. Moreover, they conjugated the polymeric film with biotin-SH using the microcontact printing method. After biotin-SH conjugation, the residual maleimide units on the printed surface were neutralized using thiol-containing PEG to eliminate nonspecific binding. Streptavidin was immobilized on the biotinylated polymeric coating, and the authors showed that the amount of the immobilized protein could be tuned by varying the surface composition.
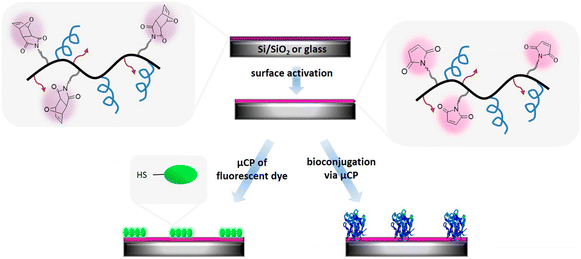 | ||
| Fig. 16 Fabrication of a thiol-reactive polymer-based thin film coating for sensing and biomolecule immobilization. Adapted with permission from Gevrek et al.127 Copyright © 2017 American Chemical Society. | ||
Apart from functionalization through side-chain modifications, Sanyal and coworkers reported the fabrication of polymer brushes with ‘clickable’ end groups and utilized them for sensing or biomolecule immobilization.133 DEGMA-based polymer brushes were obtained using surface-initiated RAFT polymerization. The dithioester groups at the end of polymer chains enabled the installation of ‘clickable’ azide, maleimide, and alkene groups via a radical cross-coupling reaction with appropriately functionalized azo-containing molecules. The authors showed that modified polymer brushes can be functionalized using alkyne or thiol-containing dyes and bioactive ligands using CuAAC reaction, Michael addition, and radical thiol–ene reaction. Moreover, the approach allows the installation of polymer brushes conjugated with multivalent motifs using dendritic azo-containing molecules. Terminal alkene-containing dendrons were functionalized with thiol-containing mannose ligands. The dendritic presentation of the protein binding sugar ligand enabled efficient sensing of concanavalin A (ConA) lectin (Fig. 17).
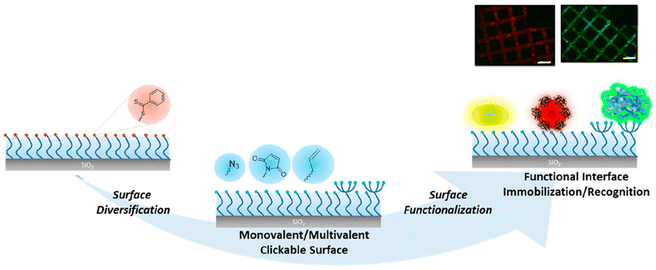 | ||
| Fig. 17 Schematic representation of the fabrication of ‘clickable’ monovalent and multivalent ligand-containing polymer brushes for sensing and biomolecule immobilization. Adapted with permission from Degirmenci et al.133 Copyright © 2022 American Chemical Society. | ||
Rühe and coworkers reported an elegant example of ‘CHicable’ and ‘clickable’ copolymers for network formation and surface modification (Fig. 18).134 In this study, they presented the fabrication of multifunctional polymer networks via a combination of C,H-insertion crosslinking (CHic), and ‘click’ chemistry. Multifunctional P(DMAA)-based copolymers bearing reactive ‘CHicable’ and ‘clickable’ groups were synthesized via free-radical polymerization. An N,N-dimethylacrylamide monomer was utilized to obtain a hydrophilic matrix. 4-Methacryloyloxybenzophenone (MABP) or 2-acryloxyanthraquinone (AOAQ) was used to serve as a ‘CHicable’ photoreactive group. N-(3-Azidopropyl)methacrylamide (AzMA) was responsible for further functionalization via the CuAAC reaction and taking apart in C,H insertion crosslinking (CHic) reactions. Network formation and surface binding can be achieved by activating benzophenone/anthraquinone groups, which have been built into the prepolymers, with UV light (λirr = 365 nm) while selectively preserving the reactive binding sites. The authors suggest that the azide groups remained after the crosslinking step, and therefore, alkyne-modified bio(molecules) can be conjugated via ‘click’ reactions. They showed Cy5-alkyne attachment onto the surface via the CuAAC reaction. Moreover, cyclooctyne-based streptavidin protein was immobilized onto the azide-containing polymeric coating via the metal-free SPAAC reaction. After that, immobilization of biotin-conjugated anti-IL-6 antibodies onto the streptavidin-modified surface was performed. The authors showed that the recombinant human IL6 analyte was successfully detected using this anti-IL-6 antibody immobilized surface.
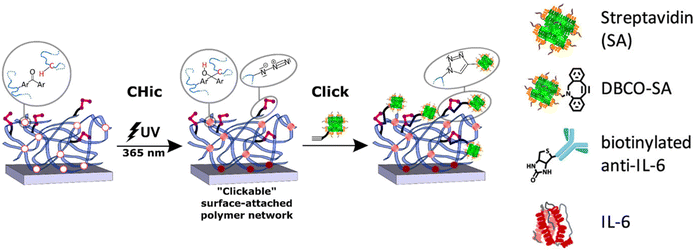 | ||
| Fig. 18 Schematic illustration of the preparation and functionalization of surface-attached photo-cross-linked ‘clickable’ polymer networks (CHic–Click) and ‘click’ conjugation with an alkyne-bearing bio(molecule) for further immobilization. Reprinted with permission from Straub et al.134 Copyright © 2021 The Authors. Published by American Chemical Society. | ||
Apart from the patterning of biomolecules, there is growing interest in the fabrication of cellular microarrays.137–139 Sha, Ma, and coworkers reported the fabrication of PEG-derived polymer brush micropatterns for obtaining a cellular microarray platform.140 The authors constructed a homogeneous hyperbranched poly(ethylene glycol) (PEG)-based brush layer as an anti-biofouling background and an extension layer of square-grid poly(2-(2-azido-2-methyl-1-oxopropoxy)ethyl methacrylate) (PAMEMA) polymer brush micropatterns on silicon surfaces. The ‘clickable’ azide groups on the side chains of the PAMEMA polymer brush enabled the covalent immobilization of alkyne-bearing target biomolecules such as RGD-peptide, fibronectin, and biotin for cell binding and protein immobilization. The authors showed that culture of human bone-derived marrow stromal cells (BMSCs) and mouse L929 cells could be conducted on the hierarchical microarray structure on the polymer coating in a high-throughput manner (Fig. 19).
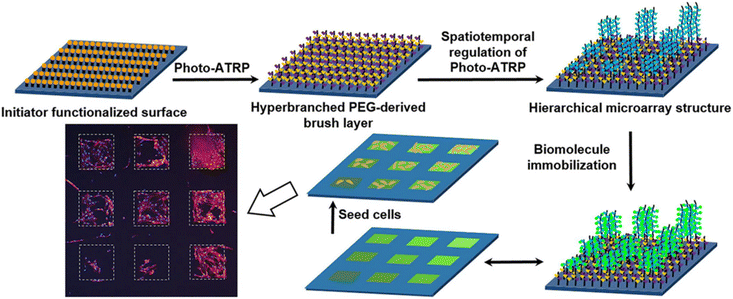 | ||
| Fig. 19 Fabrication of PEG-derived brush micropatterns for cellular microarray platforms. Adapted with permission from Zhao et al.140 Copyright © 2020 Elsevier. | ||
6. Conclusion
Facile and pragmatic approaches to obtaining functional interfaces are highly useful for several biomedical applications. This focused review highlights that employing ‘click’ reaction-based strategies provides a modular and effective approach to fabricating polymer-coated planar surfaces. Successful post-polymerization surface functionalization strategies have been demonstrated using ‘click’ reactions such as the CuAAC reaction, SPAAC reaction, thiol–ene reaction, DA cycloaddition, and IEDDA reaction. While all these approaches are quite effective for the fabrication of functional surfaces, depending on the final application, one may need to compare the performance of functionalized interfaces obtained using different chemistries. The properties of coatings such as their biocompatibility, long-term stability, and compatibility with sterilization processes required for final application would be important parameters. Also, the scalability of fabrication would be relevant when large scale clinical tests and in-field performance evaluations are required.Eventually, it is the performance of the fabricated materials in a real-life scenario that puts the device to the actual test. While many studies demonstrate in vitro performance for many antibacterial surfaces, reports with in vivo tests are limited. This is understandable due to the high costs, as well as the experience and availability of animal facilities required for the task. As more commercialization is targeted, one can anticipate the industry to move to test such materials into the in vivo stage. For applications where the endpoint is in vitro testing, for example, cellular microarrays or biomolecular sensing, putting the interfaces to real-world tests is relatively simpler. A survey of reports points out that tests related to biological sensing are often done with pure target biomolecules, but most real samples are a complex mixture of many biological entities. Along similar lines, testing the functional interfaces with clinical samples and also comparing the detection outcomes, e.g., sensitivity with conventionally used tests such as Elisa, would provide better performance evaluation and advocate their applicability.
Finally, the area of functional polymeric coatings will drive innovations through interdisciplinary collaborations, and thus the fabrication processes need to be kept simple and modular. Utilization of ‘clickable’ interfaces due to their straightforward and effective utilization certainly helps their wide utilization by researchers in diverse fields. One can envision that the attractive attributes of ‘clickable’ interfaces will continue to increasingly expand their use in addressing many challenges in biomedical sciences.
Data availability
No primary research results or data were generated as part of this review.Conflicts of interest
There are no conflicts to declare.References
- R. M. Arnold, N. E. Huddleston and J. Locklin, J. Mater. Chem., 2012, 22, 19357–19365 RSC.
- M. Krishnamoorthy, S. Hakobyan, M. Ramstedt and J. E. Gautrot, Chem. Rev., 2014, 114, 10976–11026 CrossRef CAS PubMed.
- O. Azzaroni, J. Polym. Sci., Part A: Polym. Chem., 2012, 50, 3225–3258 CrossRef CAS.
- H. C. Kolb, M. G. Finn and K. B. Sharpless, Angew. Chem., Int. Ed., 2001, 40, 2004–2021 CrossRef CAS PubMed.
- C. W. Tornøe, C. Christensen and M. Meldal, J. Org. Chem., 2002, 67, 3057–3064 CrossRef.
- L. A. Canalle, S. S. van Berkel, L. T. de Haan and J. C. M. van Hest, Adv. Funct. Mater., 2009, 19, 3464–3470 CrossRef CAS.
- S. Kantheti, R. Narayan and K. V. S. N. Raju, RSC Adv., 2015, 5, 3687–3708 RSC.
- G. M. Ziarani, Z. Hassanzadeh, P. Gholamzadeh, S. Asadi and A. Badiei, RSC Adv., 2016, 6, 21979–22006 RSC.
- J. M. Spruell, M. Wolffs, F. A. Leibfarth, B. C. Stahl, J. Heo, L. A. Connal, J. Hu and C. J. Hawker, J. Am. Chem. Soc., 2011, 133, 16698–16706 CrossRef CAS PubMed.
- L. Beria, T. N. Gevrek, A. Erdog, R. Sanyal, D. Pasini and A. Sanyal, Biomater. Sci., 2014, 2, 67–75 RSC.
- M. Davydova, A. De Los Santos Pereira, M. Bruns, A. Kromka, E. Ukraintsev, M. Hirtz and C. Rodriguez-Emmenegger, RSC Adv., 2016, 6, 57820–57827 RSC.
- C. M. Preuss, M. M. Zieger, C. Rodriguez-Emmenegger, N. Zydziak, V. Trouillet, A. S. Goldmann and C. Barner-Kowollik, ACS Macro Lett., 2014, 3, 1169–1173 CrossRef CAS PubMed.
- N. Ozbek, E. L. S. Kahveci and M. U. Kahveci, ACS Appl. Polym. Mater., 2021, 3, 3721–3732 CrossRef CAS.
- R. M. Hensarling, V. A. Doughty, J. W. Chan and D. L. Patton, J. Am. Chem. Soc., 2009, 131, 14673–14675 CrossRef CAS PubMed.
- A. S. Quick, A. De Los Santos Pereira, M. Bruns, T. Bückmann, C. Rodriguez-Emmenegger, M. Wegener and C. Barner-Kowollik, Adv. Funct. Mater., 2015, 25, 3735–3744 CrossRef CAS.
- B. Cengiz, N. Ejderyan and A. Sanyal, J. Macromol. Sci., Part A: Pure Appl.Chem., 2022, 59, 443–455 CrossRef CAS.
- K. Bazaka, M. V. Jacob, W. Chrzanowski and K. Ostrikov, RSC Adv., 2015, 5, 48739–48759 RSC.
- H. Chouirfa, H. Bouloussa, V. Migonney and C. Falentin-Daudré, Acta Biomater., 2019, 83, 37–54 CrossRef CAS PubMed.
- T. Peng, Q. Shi, M. Chen, W. Yu and T. Yang, J. Funct. Biomater., 2023, 14, 243 CrossRef CAS PubMed.
- J. L. Dalsin, L. Lin, S. Tosatti, J. Vörös, M. Textor and P. B. Messersmith, Langmuir, 2005, 21, 640–646 CrossRef CAS PubMed.
- X. Fan, L. Lin, J. L. Dalsin and P. B. Messersmith, J. Am. Chem. Soc., 2005, 127, 15843–15847 CrossRef CAS PubMed.
- P. Kingshott, J. Wei, D. Bagge-Ravn, N. Gadegaard and L. Gram, Langmuir, 2003, 19, 6912–6921 CrossRef CAS.
- G. Cheng, Z. Zhang, S. Chen, J. D. Bryers and S. Jiang, Biomaterials, 2007, 28, 4192–4199 CrossRef CAS PubMed.
- Y.-F. Zhao, L.-P. Zhu, Z. Yi, B.-K. Zhu and Y.-Y. Xu, J. Membr. Sci., 2014, 470, 148–158 CrossRef CAS.
- B. Dizman, M. O. Elasri and L. J. Mathias, J. Polym. Sci., Part A: Polym. Chem., 2006, 44, 5965–5973 CrossRef CAS.
- W. J. Yang, D. Pranantyo, K.-G. Neoh, E.-T. Kang, S. Lay-Ming Teo and D. Rittschof, Biomacromolecules, 2012, 13, 2769–2780 CrossRef CAS PubMed.
- H. Wang, G. Zha, H. Du, L. Gao, X. Li, Z. Shen and W. Zhu, Polym. Chem., 2014, 5, 6489–6494 RSC.
- W. J. Yang, X. Tao, T. Zhao, L. Weng, E.-T. Kang and L. Wang, Polym. Chem., 2015, 6, 7027–7035 RSC.
- M. R. Kazemian, L. Wang and S. Liu, ACS Appl. Bio Mater., 2019, 2, 5021–5031 CrossRef CAS PubMed.
- W.-Y. Wang, H.-W. Hu, J.-C. Chiou, K.-F. Yung and C.-W. Kan, Polym. Chem., 2023, 14, 5226–5252 RSC.
- S. Chen, S. Huang, Y. Li and C. Zhou, Front. Chem., 2021, 9, 659304 CrossRef CAS PubMed.
- C. Zhou, J. Zhou, X. Ma, D. Pranantyo, J. Li, L. Xu and V. X. Truong, Mater. Sci. Eng., C, 2021, 121, 111828 CrossRef CAS PubMed.
- W. J. Yang, T. Cai, K.-G. Neoh, E.-T. Hang, S. Lay-Ming Teo and D. Rittschof, Biomacromolecules, 2013, 14, 2041–2051 CrossRef CAS PubMed.
- G. Xu, P. Liu, D. Pranantyo, L. Xu, K.-G. Neoh and E.-T. Kang, Ind. Eng. Chem. Res., 2017, 56, 14479–14488 CrossRef CAS.
- P. Sae-ung, A. Wijitamornloet, Y. Iwasaki, P. Thanyasrisung and V. P. Hoven, Macromol. Mater. Eng., 2019, 304, 1900286 CrossRef.
- S. Mao, D. Zhang, X. He, Y. Yang, I. Protsak, Y. Li, J. Wang, C. Ma, J. Tan and J. Yang, ACS Appl. Mater. Interfaces, 2021, 13, 3089–3097 CrossRef CAS PubMed.
- F. Costa, I. F. Carvalho, R. C. Montelaro, P. Gomes and M. C. L. Martins, Acta Biomater., 2011, 7, 1431–1440 CrossRef CAS PubMed.
- A. S. Kathleen, E. F. Helen, F. C. Tanguy, M. C. Cristopher, R. Loose, G. N. Stephanopoulos and P. T. Hammond, Biomaterials, 2010, 31, 2348–2357 CrossRef PubMed.
- M. K. Chug and E. J. Brisbois, ACS Mater. Au, 2022, 2, 525–551 CrossRef CAS PubMed.
- L. Ferreira and A. Zumbuehl, J. Mater. Chem., 2009, 19, 7796–7806 RSC.
- K. Glinel, A. M. Jonas, T. Jouenne, J. Leprince, L. Galas and W. T. S. Huck, Bioconjugate Chem., 2009, 20(1), 71–77 CrossRef CAS PubMed.
- G. Gao, K. Yu, J. Kindrachuk, D. E. Brooks, R. E. W. Hancock and J. N. Kizhakkedathu, Biomacromolecules, 2011, 12, 3715–3727 CrossRef CAS PubMed.
- Y. Li, C. M. Santos, A. Kumar, M. Zhao, A. I. Lopez, G. Qin, A. M. McDermott and C. Cai, Chem. – Eur. J., 2011, 17, 2656–2665 CrossRef CAS PubMed.
- G. Gao, D. Lange, K. Hilpert, J. Kindrachuk, Y. Zou, J. T. J. Cheng, M. Kazemzadeh-Narbat, K. Yu, R. Wang, S. K. Straus, D. E. Brooks, B. H. Chew, R. E. W. Hancock and J. N. Kizhakkedathu, Biomaterials, 2011, 32, 3899–3909 CrossRef CAS PubMed.
- R. T. C. Cleophas, M. Riool, H. L. C. Q. van Ufford, S. A. J. Zaat, J. A. W. Kruijtzer and R. M. J. Liskamp, ACS Macro Lett., 2014, 3, 477–480 CrossRef CAS PubMed.
- M. A. Cole, T. F. Scott and C. M. Mello, ACS Biomater. Sci. Eng., 2016, 2, 1894–1904 CrossRef CAS PubMed.
- H. P. Felgueiras, L. M. Wang, K. F. Ren, M. M. Querido, Q. Jin, M. A. Barbosa, J. Ji and M. C. L. Martins, ACS Appl. Mater. Interfaces, 2017, 9, 7979–7989 CrossRef CAS PubMed.
- H. Cheng, K. Yue, M. Kazemzadeh-Narbat, Y. Liu, A. Khalilpour, B. Li, Y. S. Zhang, N. Annabi and A. Khademhosseini, ACS Appl. Mater. Interfaces, 2017, 9, 11428–11439 CrossRef CAS PubMed.
- J. He, J. Chen, G. Hu, L. Wang, J. Zheng, J. Zhan, Y. Zhu, C. Zhong, X. Shi, S. Liu, Y. Wang and L. Ren, J. Mater. Chem. B, 2018, 6, 68–74 RSC.
- M. Xiao, J. Jasensky, J. Gerszberg, J. Chen, J. Tian, T. Lin, T. Lu, J. Lahann and Z. Chen, Langmuir, 2018, 34, 12889–12896 CrossRef CAS PubMed.
- T. N. Gevrek, K. Yu, J. N. Kizhakkedathu and A. Sanyal, ACS Appl. Polym. Mater., 2019, 1, 1308–1316 CrossRef CAS.
- J. Chen, Y. Zhu, M. Xiong, G. Hu, J. Zhan, T. Li, L. Wang and Y. Wang, ACS Biomater. Sci. Eng., 2019, 5, 1034–1044 CrossRef CAS PubMed.
- Y. Xiao, W. Wang, X. Tian, X. Tan, T. Yang, P. Gao, K. Xiong, Q. Tu, M. Wang, M. F. Maitz, N. Huang, G. Pan and Z. Yang, Research, 2020, 2020, 7236946 CAS.
- C. Xuan, L. Hao, X. Liu, Y. Zhu, H. Yang, Y. Ren, L. Wang, T. Fujie, H. Wu, Y. Chen, X. Shi and C. Mao, Biomaterials, 2020, 252, 120018 CrossRef CAS PubMed.
- K. Li, J. Chen, Y. Xue, T. Ding, S. Zhu, M. Mao, L. Zhang and Y. Han, Chem. Eng. J., 2021, 423, 130133 CrossRef CAS.
- Q. Yao, J. Zhang, G. Pan and B. Chen, ACS Appl. Mater. Interfaces, 2022, 14, 36473–36486 CrossRef CAS PubMed.
- M. Li, J. Bai, H. Tao, L. Hao, W. Yin, X. Ren, A. Gao, N. Li, M. Wang, S. Fang, Y. Xu, L. Chen, H. Yang, H. Wang, G. Pan and D. Geng, Bioact. Mater., 2022, 8, 309–324 CAS.
- Y. Wang, Y. Yang, Y. Shi, H. Song and C. Yu, Adv. Mater., 2019, 32, 1904106 CrossRef PubMed.
- L. Hu, P. Zhao, H. Deng, L. Xiao, C. Qin, Y. Du and X. Shi, RSC Adv., 2014, 4, 13477–13480 RSC.
- B. Zhang, B. M. Braun, J. D. Skelly, D. C. Ayers and J. Song, ACS Appl. Mater. Interfaces, 2019, 11, 28641–28647 CrossRef CAS PubMed.
- M. Czuban, M. W. Kulka, L. Wang, A. Koliszak, K. Achazi, C. Schlaich, I. S. Donskyi, M. D. Luca, J. M. M. Oneto, M. Royzen, R. Haag and A. Trampuz, Mater. Sci. Eng., C, 2020, 116, 111109 CrossRef CAS PubMed.
- W. Xi, V. Hegde, S. D. Zoller, H. Y. Park, C. M. Hart, T. Kondo, C. D. Hamad, Y. Hu, A. H. Loftin, D. O. Johansen, Z. Burke, S. Clarkson, C. Ishmael, K. Hori, Z. Mamouei, H. Okawa, I. Nishimura, N. M. Bernthal and T. Segura, Nat. Commun., 2021, 12, 5473 CrossRef CAS PubMed.
- U. Hersel, C. Dahmen and H. Kessler, Biomaterials, 2003, 24, 4385–4415 CrossRef CAS PubMed.
- S. L. Bellis, Biomaterials, 2011, 32, 4205–4210 CrossRef CAS PubMed.
- H. Ma, J. Hyun, P. Stiller and A. Chilkoti, Adv. Mater., 2004, 16, 338–341 CrossRef CAS.
- S. Tugulu, P. Silacci, N. Stergiopulos and H.-A. Klok, Biomaterials, 2007, 28, 2536–2546 CrossRef CAS PubMed.
- G. A. Hudalla and W. L. Murphy, Langmuir, 2010, 26, 6449–6456 CrossRef CAS PubMed.
- R. Wang, W. Chen, F. Meng, R. Cheng, C. Deng, J. Feijen and Z. Zhong, Macromolecules, 2011, 44, 6009–6016 CrossRef CAS.
- Y. Li, M. Zhao, J. Wang, K. Liu and C. Cai, Langmuir, 2011, 27, 4848–4856 CrossRef CAS PubMed.
- K. A. Kilian and M. Mrksich, Angew. Chem., Int. Ed., 2012, 51, 4891–4895 CrossRef CAS PubMed.
- J.-T. Wu, C.-H. Huang, W.-C. Liang, Y.-L. Wu, J. Yu and H.-Y. Chen, Macromol. Rapid Commun., 2012, 33, 922–927 CrossRef CAS PubMed.
- F. Lin, J. Zheng, J. Yu, J. Zhou and M. L. Becker, Biomacromolecules, 2013, 14, 2857–2865 CrossRef CAS PubMed.
- F. C. Schenk, H. Boehm, J. P. Spatz and S. V. Wegner, Langmuir, 2014, 30, 6897–6905 CrossRef CAS PubMed.
- A. K. Muszanska, E. T. J. Rochword, A. Gruszka, A. A. Bastian, H. J. Busscher, W. Norde, H. C. van der Mei and A. Herrmann, Biomacromolecules, 2014, 15, 2019–2026 CrossRef CAS PubMed.
- F. He, B. Luo, S. Yuan, B. Liang, C. Choong and S. O. Pehkonen, RSC Adv., 2014, 4, 105–117 RSC.
- T.-P. Sun, C.-H. Tai, J.-T. Wu, C.-Y. Wu, W.-C. Liang and H.-Y. Chen, Biomater. Sci., 2016, 4, 265–271 RSC.
- S.-N. Nishimura, N. Hokazono, Y. Taki, H. Motoda, Y. Morita, K. Yamamoto, N. Higashi and T. Koga, ACS Appl. Mater. Interfaces, 2019, 11, 24577–24587 CrossRef CAS PubMed.
- Z.-Y. Guan, C.-Y. Wu, T.-Y. Chen, S.-T. Huang, Y.-C. Chiang and H.-Y. Chen, ACS Biomater. Sci. Eng., 2019, 5, 1753–1761 CrossRef CAS PubMed.
- Q. Chen, S. Yu, D. Zhang, W. Zhang, H. Zhang, J. Zou, Z. Mao, Y. Yuan, C. Gao and R. Liu, J. Am. Chem. Soc., 2019, 141, 16772–16780 CrossRef CAS PubMed.
- T. N. Gevrek, A. Degirmenci, R. Sanyal and A. Sanyal, Polymers, 2020, 12, 1211 CrossRef CAS PubMed.
- R. Poreba, A. de los Santos Pereira, R. Pola, S. Jiang, O. Pop-Georgievski, Z. Sedlakova and H. Schönherr, Macromol. Biosci., 2020, 20, 1900354 CrossRef CAS PubMed.
- R. Sivkova, J. Taborska, A. Reparaz, A. de los Santos Pereirra, I. Kotelnikov, V. Proks, J. Kucka, J. Svoboda, T. Riedel and O. Pop-Georgievski, Int. J. Mol. Sci., 2020, 21, 6800 CrossRef CAS.
- Z. Yang, X. Zhao, R. Hao, Q. Tu, X. Tian, Y. Xiao, K. Xiong, M. Wang, Y. Feng, N. Huang and G. Pan, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 16127–16137 CrossRef CAS PubMed.
- Y. Zhang, J. Shen, R. Hu, X. Shi, X. Hu, B. He, A. Qin and B. Z. Tang, Chem. Sci., 2020, 11, 3931–3935 RSC.
- A.-S. Mertgen, A. G. Guex, S. Tosatti, G. Fortunato, R. M. Mossi, M. Rottmar, K. Maniura-Weber and S. Zürcher, Appl. Surf. Sci., 2022, 584, 152525 CrossRef CAS.
- A. Schulte, A. A. de los Santos Pereira, R. Pola, O. Pop-Georgievski, S. Jiang, I. Romanenko, M. Singh, Z. Sedlakova, H. Schönherr and R. Poreba, Macromol. Biosci., 2023, 23, 2200472 CrossRef CAS PubMed.
- Y. Kim, U. M. Jahan, A. P. Deltchev, N. Lavrik, V. Reukov and S. Minko, ACS Appl. Mater. Interfaces, 2023, 15, 49012–49021 CrossRef CAS PubMed.
- S. Li, N. Chen, Z. Zhang and Y. Wang, Biomaterials, 2013, 34, 460–469 CrossRef CAS PubMed.
- F. Karimi, J. Collins, D. E. Heath and L. A. Connal, Bioconjugate Chem., 2017, 28, 2235–2240 CrossRef CAS PubMed.
- T. N. Gevrek, M. Cosar, D. Aydin, E. Kaga, M. Arslan, R. Sanyal and A. Sanyal, ACS Appl. Mater. Interfaces, 2018, 10, 14399–14409 CrossRef CAS PubMed.
- A. Degirmenci, R. Sanyal, M. Arslan and A. Sanyal, Polym. Chem., 2022, 13, 2595–2607 RSC.
- E. F. Petricoin, J. L. Hackett, L. J. Lesko, R. K. Puri, S. I. Gutman, K. Chumakov, J. Woodcock, D. W. Feigal Jr., K. C. Zoon and F. D. Sistare, Nat. Genet., 2002, 32, 474–479 CrossRef CAS PubMed.
- W. J. Brittain, T. Brandsetter, O. Prucker and J. Rühe, ACS Appl. Mater. Interfaces, 2019, 11, 39397–39409 CrossRef CAS.
- J. P. Gergen, R. H. Stern and P. C. Wensink, Nucleic Acids Res., 1979, 7, 2115–2136 CrossRef CAS PubMed.
- M. Dufva and C. B. V. Christensen, Expert Rev. Proteomics, 2005, 2, 41–48 CrossRef CAS PubMed.
- D. Brambilla, M. Chiari, A. Gori and M. Cretich, Analyst, 2019, 144, 5353–5367 RSC.
- S. Song, B. Li, L. Wang, H. Wu, J. Hu, M. Li and C. Fan, Mol. BioSyst., 2007, 3, 151–158 RSC.
- I. Shin, S. Park and M.-R. Lee, Chem. – Eur. J., 2006, 11, 2894–2901 CrossRef PubMed.
- R. Z. Wu, S. N. Bailey and D. M. Sabatini, Trends Cell Biol., 2002, 12, 485–488 CrossRef CAS PubMed.
- M. Dufva, Biomol. Eng., 2005, 22, 173–184 CrossRef CAS PubMed.
- M. D. Sonawane and S. B. Nimse, J. Chem., 2016, 2016, 9241378 Search PubMed.
- S. B. Nimse, K. Song, M. D. Sonawane, D. R. Sayyed and T. Kim, Sensors, 2014, 14, 22208–22229 CrossRef PubMed.
- S. K. Nemani, R. K. Annavarapu, B. Mohammadian, A. Raiyan, J. Heil, A. Haque, A. Abdelaal and H. Sojoudi, Adv. Mater. Interfaces, 2018, 5, 1801247 CrossRef.
- N. Gupta, B. F. Lin, L. M. Campos, M. D. Dimitriou, S. T. Hikita, N. D. Treat, M. V. Tirrell, D. O. Clegg, E. J. Kramer and C. J. Hawker, Nat. Chem., 2010, 2, 138–145 CrossRef CAS.
- G. Pirri, F. Damin, M. Chiari, E. Bontempi and L. E. Depero, Anal. Chem., 2004, 76(5), 1352–1358 CrossRef CAS PubMed.
- M. Cretich, G. Pirri, F. Damin, I. Solinas and M. Chiari, Anal. Biochem., 2004, 332, 67–74 CrossRef CAS PubMed.
- C. Zillio, A. Bernardi, A. Palmioli, M. Salina, G. Tagliabue, M. Buscaglia, R. Consonni and M. Chiari, Sens. Actuators, B, 2015, 215, 412–420 CrossRef.
- A. Gori, L. Sola, P. Gagni, G. Bruni, M. Liprino, C. Peri, G. Colombo, M. Cretich and M. Chiari, Bioconjugate Chem., 2016, 27(11), 2669–2677 CrossRef CAS PubMed.
- L. Sola, F. Damin, P. Gagni, R. Consonni and M. Chiari, Langmuir, 2016, 32, 10284–10295 CrossRef CAS PubMed.
- L. Sola, F. Damin and M. Chiari, Anal. Chim. Acta, 2019, 1047, 188–196 CrossRef CAS PubMed.
- L. Sola, A. Romanato, M. B. Siboni, F. Damin, E. Chiodi, D. Brambilla, M. Cretich, A. Gori and M. Chiari, eXPRESS Polym. Lett., 2019, 13, 1004–1017 CrossRef CAS.
- J. Striebel, M. Vorobii, R. Kumar, H.-Y. Liu, B. Yang, C. Weishaupt, C. Rodriguez-Emmenegger, H. Fuchs, M. Hirtz and K. Riehemann, Adv. NanoBiomed Res., 2020, 1, 2000029 CrossRef.
- L. Sola, D. Brambilla, A. Mussida, R. Consonni, F. Damin, M. Cretich, A. Gori and M. Chiari, Anal. Chim. Acta, 2021, 1187, 339138 CrossRef CAS PubMed.
- R. Kumar, B. Yang, J. Barton, M. Stejfova, A. Schafer, M. Koenig, P. Knittel, P. Cigler and M. Hirtz, Adv. Mater. Interfaces, 2022, 9, 2201453 CrossRef CAS.
- B. Yang, Y. Wang, M. Vorobii, E. Sauter, M. Koenig, R. Kumar, C. Rodriguez-Emmenegger and M. Hirtz, Adv. Mater. Interfaces, 2022, 9, 2270092 CrossRef.
- X.-L. Sun, C. L. Stabler, C. S. Cazalis and E. L. Chaikof, Bioconjugate Chem., 2006, 17, 52–57 CrossRef CAS PubMed.
- L. Q. Xu, K.-G. Neoh, E.-T. Kang and G. D. Fu, Polym. Chem., 2012, 3, 920–927 RSC.
- Y. Li, M. Giesbers, M. Gerth and H. Zuilhof, Langmuir, 2012, 28, 12509–12517 CrossRef CAS PubMed.
- D. Sung, S. Park and S. Jon, Langmuir, 2012, 28, 4507–4514 CrossRef CAS PubMed.
- T. N. Gevrek, T. Bilgic, H.-A. Klok and A. Sanyal, Macromolecules, 2014, 47, 7842–7851 CrossRef CAS.
- A. R. Kuzmyn, A. de los Santos Pereira, O. Pop-Georgievski, M. Bruns, E. Brynda and C. Rodriguez-Emmenegger, Polym. Chem., 2014, 5, 4124 RSC.
- S. C. Lange, E. van Andel, M. M. J. Smulders and H. Zuilhof, Langmuir, 2016, 32, 10199–10205 CrossRef CAS.
- O. Wiarachai, T. Vilaivan, Y. Iwasaki and V. P. Hoven, Langmuir, 2016, 32, 1184–1194 CrossRef CAS PubMed.
- V. Parrillo, A. de los Santos Pereira, T. Riedel and C. Rodriguez-Emmenegger, Anal. Chim. Acta, 2017, 971, 78–87 CrossRef CAS PubMed.
- U. Bog, A. de los Santos Pereira, S. L. Mueller, S. Havenridge, V. Parrillo, M. Bruns, A. E. Holmes, C. Rodriguez-Emmenegger, H. Fuchs and M. Hirtz, ACS Appl. Mater. Interfaces, 2017, 9, 12109–12117 CrossRef CAS PubMed.
- Y. N. Yuksekdag, T. N. Gevrek and A. Sanyal, ACS Macro Lett., 2017, 6, 415–420 CrossRef CAS PubMed.
- T. N. Gevrek, I. Kosif and A. Sanyal, ACS Appl. Mater. Interfaces, 2017, 9, 27946–27954 CrossRef CAS PubMed.
- K. Miyahara, R. Sakai, M. Hara and T. Maruyama, Colloid Polym. Sci., 2019, 297, 927–931 CrossRef CAS.
- D. Di lorio, A. Marti, S. Koeman and J. Huskens, RSC Adv., 2019, 9, 35608–35613 RSC.
- N. Cengiz, T. N. Gevrek, R. Sanyal and A. Sanyal, Bioconjugate Chem., 2020, 31, 1382–1391 CrossRef CAS PubMed.
- Y. Zhang, J. Shen, R. Hu, X. Shi, X. Hu, B. He, A. Qin and B. Z. Tang, Chem. Sci., 2020, 11, 3931–3935 RSC.
- T. S. Svalova, M. V. Medvedeva and A. N. Kozitsina, Electroanalysis, 2021, 33, 2469–2475 CrossRef CAS.
- A. Degirmenci, G. Y. Bas, R. Sanyal and A. Sanyal, Bioconjugate Chem., 2022, 33, 1672–1684 CrossRef CAS PubMed.
- A. J. Straub, F. D. Scherag, H. I. Kim, M.-S. Steiner, T. Brandstetter and J. Rühe, Langmuir, 2021, 37, 6510–6520 CrossRef CAS PubMed.
- G. E. Fenoy, R. Hasler, C. Lorenz, J. Movilli, W. A. Marmisolle, O. Azzaroni, J. Huskens, P. Bauerle and W. Knoll, ACS Appl. Mater. Interfaces, 2023, 15, 10885–10896 CrossRef CAS PubMed.
- V. Damodara and S. Ramakrishnan, Macromolecules, 2023, 56, 7837–7846 CrossRef CAS.
- M. D. Kurkuri, C. Driever, G. Johnson, G. McFarland, H. Thissen and N. H. Voelcker, Biomacromolecules, 2009, 10, 1163–1172 CrossRef CAS PubMed.
- G. Tourniaire, J. Collins, S. Campbell, H. Mizomoto, S. Ogawa, J.-F. Thaburet and M. Bradley, Chem. Commun., 2006, 2118–2120 RSC.
- A. L. Hook, H. Thissen and N. H. Voelcker, Biomacromolecules, 2009, 10, 573–579 CrossRef CAS PubMed.
- H. Zhao, J. Sha, T. Wu, T. Chen, X. Chen, H. Ji, Y. Wang, H. Zhu, L. Xie and Y. Ma, Appl. Surf. Sci., 2020, 529, 147056 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2024 |