 Open Access Article
Open Access ArticleRecent advances in plasmonic Prussian blue-based SERS nanotags for biological application
Ya-Qin
Liu
a,
Wei
Zhu
*b,
Ji-Ming
Hu
a and
Ai-Guo
Shen
 *b
*b
aCollege of Chemistry and Molecular Sciences, Wuhan University, Wuhan 430072, China
bSchool of Printing and Packaging, Wuhan University, Wuhan 430079, China. E-mail: zwmaterials@whu.edu.cn; agshen@whu.edu.cn
First published on 21st October 2021
Abstract
The reliability and reproducibility of surface-enhanced Raman scattering (SERS) technology is still a great challenge in bio-related analysis. Prussian blue (PB)-based SERS tags have attracted increasing interest for improving these deficiencies due to its unique Raman band (near 2156 cm−1) in the Raman-silent region, providing zero-background bio-Raman labels without interference from endogenous biomolecules. Moreover, the stable PB shell consisting of multiple layers of CN− reporters ensure a stable and strong Raman signal output, avoiding the desorption of the Raman reporter from the plasmonic region by the competitive adsorption of the analyte. More importantly, they possess outstanding multiplexing potential in biological analysis owing to the adjustable Raman shift with unique narrow spectral widths. Despite more attention having been attracted to the structure and preparation of PB-based SERS tags for their better biological applications over the past five years, there is still a great challenge for SERS suitable for applications in the actual environment. The biological applications of PB-based SERS tags are comprehensively recounted in this minireview, mainly focusing on quantification analysis, multiple-spectral analysis and cell-imaging joint phototherapy. The prospects of PB-based SERS tags in clinical diagnosis and treatment are also discussed. This review aims to draw attention to the importance of SERS tags and provide a reference for the design and application of PB-based SERS tags in future bio-applications.
1. Introduction
Since the surface-enhanced Raman scattering (SERS) phenomenon was first confirmed to perform the great capacity of improvement in the sensitivity of Raman spectroscopy in the 1970s,1–3 the million-fold enhancement of the Raman signal on metal surfaces, and even the detection at the single-molecule level has been achieved by SERS.4–6 In the past few decades, SERS has been demonstrated in numerous important applications in various fields, such as material sciences,7,8 analytical chemistry,9–11 physical chemistry,12,13 and biomedical research.2,14,15 Admittedly, SERS has become an ideal alternative for biological analysis because of its inherent advantages, including the weak Raman signal of water, fingerprinting capabilities, ultra-sensitivity, high specificity, resistance to photobleaching and photodegradation.16,17In general, SERS biological analysis can be classified into direct (label-free)18–20 and indirect (labelled) SERS detection.21–23 Direct SERS detection depends on capturing bio-analytes in the plasmonic region, thus acquiring the fingerprint information of bio-analyte. However, it is limited in the biosystem due to the lack of standard SERS spectra for numerous biomolecules, complicated analysis of Raman peaks and complex biological environments. In contrast, indirect SERS detection relies on the Raman signals provided by SERS tags, circumventing the toilsome analysis of Raman spectra and providing a strategy to study some physical properties in the biosystem, which are impossible to be reflected directly by inherently related molecules. Thus, indirect SERS detection has been widely applied to monitor the dynamic changes in biomolecules24,25 and intracellular environment properties.26,27 Notably, the SERS tag plays a vital role in indirect SERS detection,28,29 consisting of an enhanced substrate, Raman reporters and targeting molecules. The commonly-used Raman reporters are shown in Table 1, including some nitrogen-containing cationic dyes,30,31 sulfur-containing dyes,32,33 thiol-small molecules34–36 and triple-bond-containing molecules.37,38 These Raman reporters have contributed to the preeminent SERS application in life sciences by binding with the enhanced substrates through the electrostatic forces or S-Au(Ag) interactions. Nevertheless, these Raman reporters can fall off due to the competitive adsorption of the analyte on the metal surface, resulting in unstable Raman signals. Although protection strategies with a carbon shell39 or polymers40 have been proposed to avoid the desorption of the Raman reporters, they are still limited due to the strict synthesis process of the protective shell, the biocompatibility, the background signal interference and the production efficiency of the hot spots. Therefore, effective Raman reporters are still a key to improving the sensitivity and stability of the indirect SERS detection.
| Type | Example | Specific Raman scattering | Linking mode | Ref. |
|---|---|---|---|---|
| Traditional Raman reporters | Crystal violet | Fingerprint region (400–1800 cm−1) | Electrostatic force | 30 and 41 |
| 3,3′-Diethylthiadicarbocyanine iodide | S–Au(Ag) interaction | 32 and 33 | ||
| 4-Methylbenzenethiol | 36 and 42 | |||
| Triple-bond containing Raman reporters | 4-Mercaptobenzonitrile | Raman silent region (1800–2800 cm−1) | S–Au(Ag) interaction | 43 and 44 |
| 4-Ethynylbenzenethiol derivatives | 37 |
Prussian Blue (PB), a dark blue pigment discovered in the 1706s, has been demonstrated as a promising coordination polymer as the Raman reporter with the formula of FeIII4[FeII(CN)6]3·nH2O.45–47 Interestingly, it was discovered that PB only possesses a strong and sharp Raman peak in the Raman-silent region (1800–2800 cm−1), where the Raman signal interference of biological endogenous molecules is effectively circumvented.48,49 Based on the unique characteristics of PB, Liu's group achieved a high-sensitivity immunoassay for antigen detection with the limit of detection (LOD) of 8.0 fg mL−1 using a PB-based SERS tag.50 Background-free cancer cell imaging has also been achieved by them. They demonstrated the potential of PB as a high signal-to-background ratio (SBR) and zero-background resonant Raman reporter. Since then, the unique Raman scattering of PB and its analogues (PBA) have made them rising stars in optical labels with the properties of ultra-sensitivity, super-stability, and high-specificity. Recently, PB-based SERS tags with different morphologies have been successfully prepared for indirect SERS detection and imaging in biological systems. The wide application of the PB-based SERS tags will facilitate the investigation of complex intracellular biological events without the interference of endogenous signals and provide new possibilities for precise disease diagnosis.
In this review, we give a detailed discussion on the most recent advances of PB-based SERS tags. At first, we will emphatically discuss the rational design of the PB-based SERS tag to provide credible guidance for extending the biological application of the PB-based SERS tag. Secondly, the applications of PB-based SERS tags in the biological field will be summarized, such as the monitoring of the intracellular environment, tumour imaging, bacterial coding and cancer cell classification (Fig. 1). Finally, the challenges and the prospects of PB-based SERS tags in biological applications are also discussed.
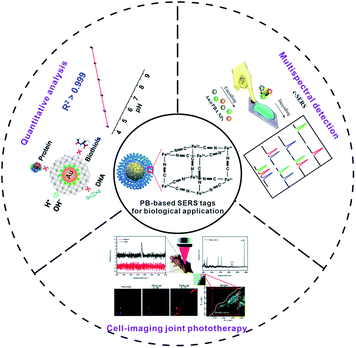 | ||
| Fig. 1 Multifunctional PB-based SERS tags used for biological applications. PB-based SERS nanotags for quantitative analysis, adapted from ref. 88 with permission from American Chemical Society. Copyright 2020 American Chemical Society. Multispectral detection, adapted from ref. 89 with permission from American Chemical Society. Copyright 2020 American Chemical Society. Cell-imaging joint phototherapy, adapted from ref. 65 with permission from Royal Society of Chemistry. Copyright 2020 Royal Society of Chemistry. | ||
2. Rational design of PB-based SERS tags for bioanalysis
The SERS tags are usually the first-mentioned part in the indirect SERS detection, generally consisting of a SERS-enhanced substrate, Raman reporters and targeting biomolecules28,51 (Fig. 2A). Protection shells are also introduced into some SERS tags to obtain a more stable Raman signal output. Constructing PB-based SERS tags should take the following principles into account: (1) a proper metal nanostructure will provide strongly enhanced Raman signals because of the local surface plasmonic resonance (LSPR) on the metal surface,1,52 which lays a solid foundation for SERS tags. The SERS properties of the tags can also be greatly affected by the chemical composition,53,54 size,55,56 and structure of the enhanced substrate.57,58 (2) In general, the Raman reporters are required to attach to the surface of a SERS-enhanced substrate because the most widely accepted mechanisms, electromagnetic enhancement (EM) and chemical enhancement (CE), are strongly distance-dependent or chemical bonding-dependent, respectively. In addition, a strong interaction between the Raman reporters and substrate is indispensable for preventing reporter desorption during further modification or use. (3) A dense, uniform Raman reporter layer formed on the substrates will contribute to a strong and stable signal. (4) Furthermore, the detection specificity in bioanalysis is decided by the biorecognition element, e.g. aptamer, which is used to target the analyte.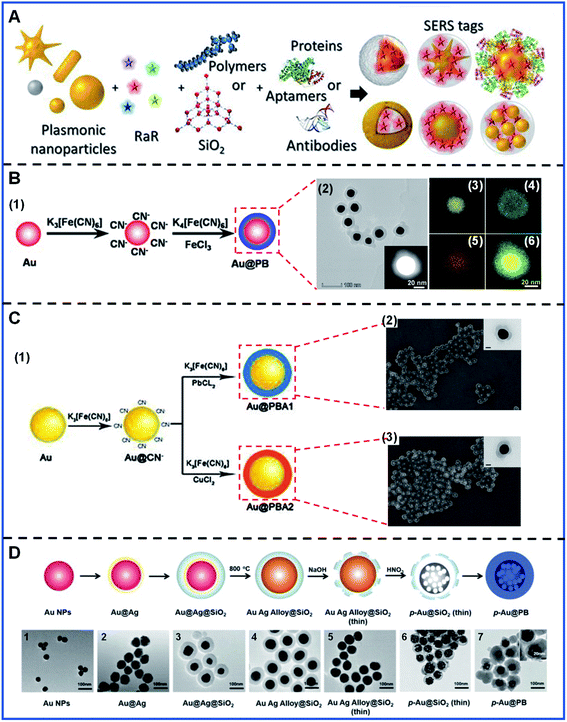 | ||
| Fig. 2 The basic constituents of SERS tags and the synthesis of PB-based nanomaterials. (A) The representation of the basic elements of SERS tags. Adapted with permission from ref. 51. Copyright 2019 American Chemical Society. (B) The synthesis and characterization of Au@PB NPs: (B.1) A schematic illustrating the synthesis of Au@PB NPs; (B.2) TEM images, the inset is the corresponding high-angle annular darkfield scanning TEM image; (B.3–B.6) EDX mapping of Au, Fe, O and the overlay images. Reproduced with permission from ref. 61. Copyright 2021 American Chemical Society. (C) The synthesis and characterization of Au@PBA NPs: (C.1) Schematic illustrating the synthesis procedure of Au@PBA NPs; (C.2) SEM image of Au@PBA (Cu) NPs, the inset is the TEM image of a single Au@PBA (Cu) NP; (C.3) SEM image of Au@PBA (Pb) NPs, the inset is the TEM image of a single Au@PBA (Pb) NP. Reproduced with permission from ref. 62. Copyright 2021 Elsevier. (D) The synthesis and characterization of p-Au@PB NPs: (D.1–D.7) TEM images of the formed process of p-Au@PB NPs, the inset is the magnified image of p-Au@PB NPs. Adapted with permission from ref. 63. Copyright 2019 John Wiley & Sons, Inc. | ||
2.1. Component of PB-based SERS tags
PB-based SERS tags can be simply divided into two parts: one is PB-based nanomaterials, another is targeting molecules. To date, the PB-based nanomaterials commonly comprise the gold (Au) nanosphere as the core and the PB or the PBA as the shell. Au nanoparticles (Au NPs) are the most widely used enhanced substrate for PB-based SERS tags in biological applications due to their established synthetic methods, super excellent biocompatibility and low toxicity. Although silver is recommended as the most efficient plasmonic metal, it is unsuitable for PB-based SERS tags due to its extreme instability caused by the etching of CN− and the toxicity to living systems.59,60 In addition, the spherical particle is relatively easy to control during the fabrication and has better long-term stability. Meanwhile, the PB or PBA shell as the Raman reporter can provide a single stable signal output in the bio-silent region. The core–shell nanostructure ensures the strong Raman signal of PB-based nanomaterials, effectively circumventing the desorption of the Raman reporter caused by the competitive adsorption of the analyte.Apart from spherical Au NPs as the core, another attempt has also been made for the fabrication of porous gold (p-Au) NPs as the core. PB-coated p-Au NPs with abundant Raman hot spots have been put forward by Liu's group to amplify the Raman signals of the PB shell. PB is not only used as the Raman reporter but can also be used as the protective shell for other Raman reporters. For instance, PB-caged 4-mercaptopyridine (4-MPy)-Au NPs have been successfully synthesized by Liu's group, in which the PB shell is both used as the internal standard and a cage-like protective layer, as well as a carrier that only allows the entry of H+ or OH−.
2.2. Preparation and advantages of PB-based nanomaterials
2.3. Specific targeting of analytes
The recognition of PB-based SERS tags toward a specific cellular region or specific molecule in the biosystem is typically attained with the specific targeting ligands, which generally consist of specific proteins, peptides, aptamers, polysaccharides, or antibodies. For instance, some proteins or peptides containing free sulfhydryl groups, such as ovalbumin, were coated on the NPs due to the inherent capacity of proteins to attach to the NPs via S–Au bonds.64 Stable covalent bonds between the aptamers62 or polysaccharides65 and the PB-based nanomaterials can also be formed with the aid of poly-L-lysine (PLL) as a bridge, the 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) and N-hydroxysuccinimides (NHS) as the coupling reagent. The activated PLL by EDC and NHS provides abundant amine moieties to react with the carboxylic acid groups in the aptamers or polysaccharides. Similarly, the positively charged PLL is also used as the bridge for the connection of the PB-based nanomaterials and the negative antibody50 by electrostatic adsorption. In addition, some special molecules, e.g., folic acid (FAs)63 receptors, are overexpressed in various cancer cells, thus FAs can be treated as the specific targeting ligands. Even though the functional PB-based nanomaterials through specific biorecognition molecules have been widely used as SERS tags for both in vitro and in vivo cell targeting and imaging, a variety of bio-applications are still constantly emerging.3. Biological applications of Prussian blue-based SERS nanotags
Many organic molecules, such as proteins,66,67 DNA,68,69 and metabolites,70,71 perform essential functions in multiple biological processes, including signal transduction, cell adhesion and migration, metabolism, etc. The visible spatial distributions and reliable quantification detection of these important molecules present in biological systems provide solid foundations to further understand their important function and will drive advances in various fields such as early disease diagnostics. Moreover, the intracellular environment is also a critical physiological parameter that is an indicator to reveal the abnormal phenomenon in vivo.72,73 The comprehensive detection and imaging of these endogenous molecules have attracted more and more attention with the increased rate of various diseases. PB-based SERS tags are particularly helpful for long-term imaging applications in biosystems due to their ability to resist photobleaching and photodegradation, which overcomes the shortcoming of fluorophore labels. Furthermore, the low cytotoxicity of SERS tags is the prerequisite to enter the cell, increasing the requirements for the selection of the enhanced substrate and the Raman reporter. Taking this into account, PB-based SERS tags exhibit powerful advantages because PB was allowed to be an antidote for the treatment of thallium and cesium intoxication by the Food and Drug Administration (FDA) in 2003, showing the outstanding biocompatibility of PB. We present below some typical examples that elaborate the current interest in using PB-based SERS tags, as compared to previously reported methods.3.1. Reliable SERS quantitative analysis
Signal reproducibility of SERS tags is decisive for reliable quantitative SERS bioanalysis. Quantitative SERS analysis is by far plagued due to the uneven distribution of hot spots on SERS-enhanced substrates and the possible desorption of Raman reporters. The environmental conditions also have a crucial impact and can cause the fluctuation of the Raman signal. Various attempts, such as protective coating,29,74 internal standards75,76 and ratiometric SERS77,78 assays, have been carried out to alleviate the present dilemma in quantitative SERS analysis techniques. The introduction of a protective coating effectively avoids the leaching of Raman reporter molecules and prevents potential signal fluctuation caused by the surrounding medium. However, the increased shell thickness probably results in a decrease in the detection sensitivity due to the distance dependence of EM, and the complex synthesis process increases the time and labour costs. Although ratiometric quantitative SERS analysis offers an alternative way to improve the reliability of the quantification SERS analysis through deformation of spectral bands, which are provided by the reaction between the target analyte and Raman reporters, it is tricky and unrealistic to construct numerous appropriate SERS probes for every analyte of interest. The use of an internal standard (IS) can solve the above problem. Ren's group developed core-IS molecule-shell (CMS) nanoparticles to correct the signal fluctuation and protect the IS molecule from the interference of the external environment.79 Nevertheless, the introduction of IS itself will bring about the inaccurate signal when the IS molecules immobilized on the surface of enhanced substrates by coordinate or electrostatic interactions do not enter the hot spot during signal acquisition. Furthermore, CMS nanostructures are complicated to synthesize, and controlling the IS molecules is not a facile process.Based on the present deficiencies in SERS detection and quantification, Li and colleagues developed ultrathin core–shell Au@PB NPs with the characteristic Raman vibrational band at 2155 cm−1, in which PB is used as the IS to improve the accuracy and reliability of quantification SERS analysis.80 The characteristic PB spectral band in the Raman silent region significantly reduced the signal overlap and interference caused by analyte signals, and is thus suitable for use as an IS for quantitative SERS analysis in complex systems. These core–shell Au@PB NPs were applied to detect the concentrations of crystal violet (CV) in lake water and the concentrations of dopamine (DA) in blood serum without any pretreatment. It was demonstrated that the accuracy of CV and DA quantification detection was effectively improved with the coefficient of determination of CV quantification detection increasing from 0.9738 to 0.9968 and that of DA quantification detection increasing from 0.9690 to 0.9950. Li's group also demonstrated that Au@PB NPs remain stable in different pH and temperature conditions, and are thus feasible for the detection of practical samples80 (Fig. 3). Lu's group synthesized stable Au@PB@Au NPs and employed them for trace detection of Interleukin-6 (IL-6) in plasma. PB is used as IS molecules and is further protected by Au shell, significantly improving the linearity of IL-6 detection.81
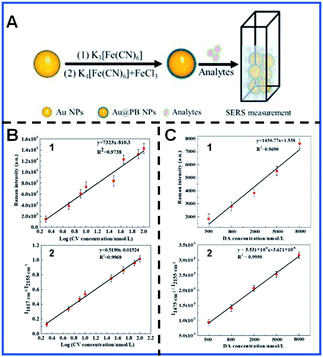 | ||
| Fig. 3 Schematic showing the synthesis of Au@PB NPs for SERS quantitative analysis (A) and the calibration curves of CV (B) and DA (C). (B.1 and B.2) Calibration plot based on the I1617 cm−1vs. log[CV] and I1617 cm−1/I2155 cm−1vs. log[CV], respectively. (C.1 and C.2) Calibration plot based on I1475 cm−1vs. [DA] and I1475 cm−1/I2155 cm−1vs. [DA]. Reproduced with permission from ref. 80. Copyright 2019 American Chemical Society. | ||
Dynamic quantification detection of intracellular local pH has been successfully achieved by several groups based on the pH-dependent SERS spectra of some Raman reporters, such as MBA,82,83 2-aminobenzenethiol (2-ABT),84,85 or 4-mercaptopyridine (4-MPy).86,87 It was refreshing that a PB-caged pH-responsive SERS probe was designed by Liu's group with 4-MPy as the pH-responsive molecule.88 This pH-responsive probe can provide an excellent response toward pH, even in the presence of many disturbances, such as human serum albumin (HSA), glutathione (GSH) and cell culture medium. Fig. 4 shows the typical pH imaging for intracellular pH monitoring under the diverse conditions in living HeLa cells using this PB-caged pH-responsive SERS probe. The obtained Raman band at 2138 cm−1 of PB can be employed to ascertain the probe distribution with high accuracy. Furthermore, the changes in the intensity at 1594 cm−1 and 1614 cm−1 (I1594 cm−1/I1614 cm−1) were employed for the pH variation quantification, showing the ability of this novel probe for the statistical analyses of intracellular pH. More importantly, the PB-caged strategy also has potential for simultaneous nanodrug transport and tumour detection in biomedical applications. Even though great progress has been made for improving the reliability of quantitative SERS analysis, further advances in quantitative SERS bioanalysis will be promoted by better design strategies, such as the combination of the solid SERS substrates and PB-based nanoparticles, which guarantee a reproducible number of Raman reporters in the hot spots. The batch preparation route with good repeatability for synthesizing NPs with uniform size and morphology will also be a creative endeavour in the future.
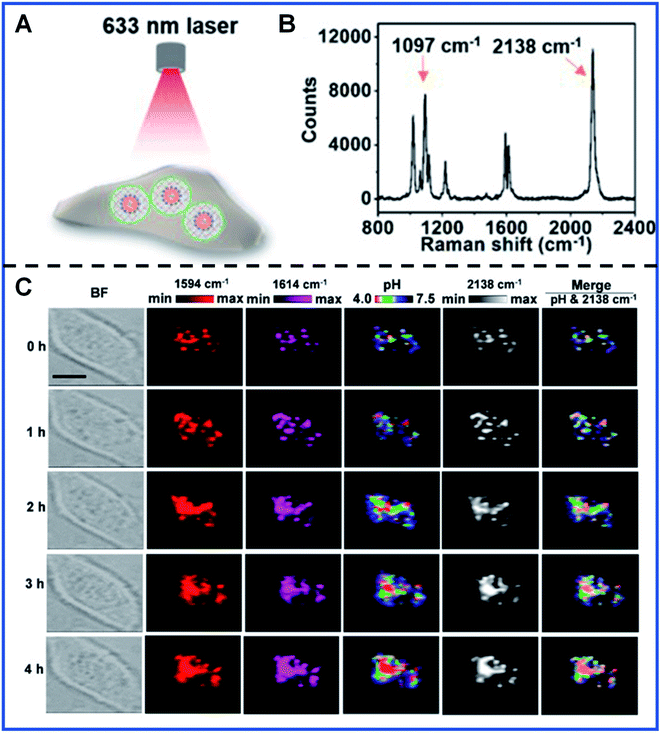 | ||
| Fig. 4 Intracellular pH monitoring using PB-caged 4-MPy-Au NPs as the pH probe. (A) A schematic illustrating the profiling of intracellular probes carried out with a 633 nm laser. (B) Raman spectrum showing the characteristic Raman band of pH probes in PBS. (C) Real-time imaging of a single cell at different times using PB-caged 4-MPy-Au NPs as the pH probe. Reproduced with permission from ref. 88. Copyright 2020 American Chemical Society. | ||
3.2. Multispectral bioanalysis
Multi-colour imaging has attracted more and more attention for satisfying the increased demand for intracellular multi-component analysis with the rapid development of life analytical chemistry. Developing novel multi-colour imaging methods and probes is essential to the better understanding of the various intracellular activities such as intracellular molecule interactions, intercellular communication, and the molecular changes during the production process of cancer. PB-based SERS tags have the potential to be used as multi-signal output due to their narrow peak width and adjustable Raman shift. Therefore, PB-based SERS tags have aroused extensive interest for their employment in biological multi-colour analysis and imaging.Shen's group performed a series of pioneering studies on encoding the Raman intensities and Raman shifts of PB-based SERS tags in multispectral bioanalysis. They designed three Au@PBA NPs with distinguishable Raman shifts in the bio-silent region by replacing the Fe2+/Fe3+ with other metal ions, including Au@PBA (Pb) NPs, Au@PBA (Co) NPs and Au@PBA (Cu) NPs. Based on three Au@PBA nanomaterials, they proposed combined SERS (c-SERS) and applied it in the high-throughput detection of micrometre-scale objects, such as bacteria.89 The c-SERS output involves the various combination of independent SERS signal outputs from three PBA-encoded SERS tags, which were obtained when three Au@PBA NPs were confined in a micron-scale object. As shown in Fig. 5A, seven kinds of SERS signal outputs can be obtained with this encoding strategy, thus seven diverse micro-scale analytes can be recognized in a single detection. They demonstrated the simultaneous recognition of seven types of bacteria based on the “ID” information provided by the diverse combination of SERS tags (Fig. 5B). Through primary decoding, C-SERS could simultaneously realize the high-throughput labelling of (2n − 1) various micro-scale targets with only n distinguishing SERS tags, which greatly expanded the capacity of optical labels.
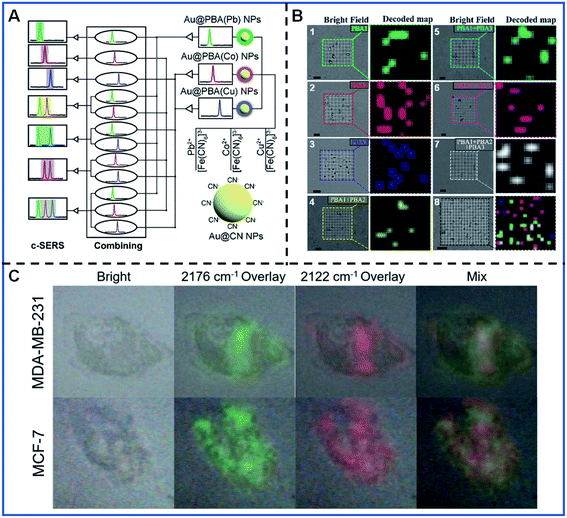 | ||
| Fig. 5 The multispectral analysis of bacteria recognition and cancer cell subtyping. (A) Schematic illustrating the conception of the c-SERS. Adapted with permission from ref. 89. Copyright 2019 American Chemical Society. (B) Decoding and spatial visualization of various bacteria based on the c-SERS. B.1–B.7 corresponds to four types of bacteria, including Escherichia coli, Staphylococcus aureus, Pseudomonas aeruginosa, and Bacillus cereus. B.8 represents the simultaneous detection of various bacteria. Adapted with permission from ref. 89. Copyright 2019 American Chemical Society. (C) Simultaneous Raman imaging of two kinds of cancer cells. Adapted with permission from ref. 62. Copyright 2021 Elsevier. | ||
Apart from the c-SERS strategy, the Raman intensity difference of PB-based SERS tags caused by the marker expression difference is also an essential indicator for the accurate recognition of cell subtyping. Shen's group substantiated the conclusion by the fine synthesis of Au@PBA (Pb) NPs and Au@PBA (Cu) NPs.62Fig. 5C shows the different cell marker imaging of MCF-7 and MDA-MB-231 cell lines by simultaneously incubating two kinds of cell lines with two kinds of Au@PBA NPS. MCF-7 and MDA-MB-231 were able to be discriminated by the ratio of the average Raman intensities at 2176 cm−1 and the average Raman intensities at 2122 cm−1 (I2176 cm−1/I2122 cm−1), which correspond to the expression of the epidermal growth factor receptor (EGFR) and epithelial cell adhesion molecule (EpCAM), respectively. This kind of multi-colour imaging has great potential for cancer cell diagnosis and is likely to promote profiling advances in cancer cell subtypes.
3.3. Cell-imaging joint phototherapy
The SERS technique is advantageous in live-cell imaging due to the following. First, high sensitivity can be achieved with low laser power, thus avoiding light-induced injury to the cells. Second, the Raman signal of water is fairly weak, satisfying the detection requirement of the bio-environment. Third, the production process of Raman scattering and the data acquisition are relatively short, enabling real-time and long-time monitoring of biological processes. PB-based SERS tags are increasingly being used in living cell studies due to their low toxicity, excellent biocompatibility, strong and sharp Raman bands.A vital application of PB-based SERS tags in living cells is the detection of cancer biomarkers or bacteria, demonstrating their excellent capacity for high-throughput identification of cancer cells and bacteria. Yin et al. synthesized Au@PB NPs in which PB was used as a background-free Raman reporter. After surface modification with targeting molecules, the FA-conjugated Au@PB NPs were used to recognize the FA receptor overexpressed in HeLa cells.50
Similarly, Li and colleagues developed FA-conjugated p-Au@PB NPs to recognize the FA receptor overexpressed in HeLa cells and downregulation in HepG2 cells (Fig. 6A).63 The Raman intensity of p-Au@PB NPs is largely stronger than that of Au@PB NPs and PB NPs under the same conditions (Fig. 6B), showing the potential of p-Au@PB NPs for higher detection sensitivity.
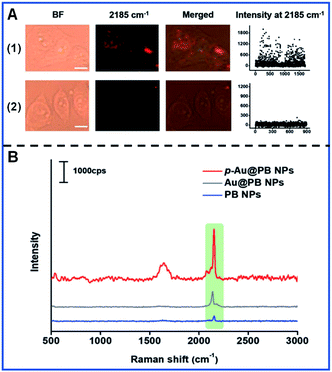 | ||
| Fig. 6 The Raman mapping of multiple cancer cells (A) and the Raman spectra of different NPs (B). (A.1) The Raman mapping of HeLa cells with the upregulation of FA receptors. (A.2) The Raman mapping of HepG2 cells with the downregulation of FA receptors. Reproduced with permission from ref. 63. Copyright 2019 John Wiley & Sons, Inc. | ||
An interesting and novel integrated SERS platform for reliable detection and the photothermal elimination of bacteria was reported.61 As shown in Fig. 7A, a wormlike plasmonic gold film with 4-mercaptophenylboronic acid (4-MPBA) and 4-mercaptobenzonitrile (4-MBN) was prepared, in which 4-MPBA was used to capture the bacteria and 4-MBN was used as the IS molecule. Additionally, vancomycin-modified core–shell Au@PB NPs (Au@PB@Van NPs) were used as the SERS tags. Therefore, both reliable quantification detection based on the statistical analysis of the imaging of bacteria and in situ photothermal elimination of bacteria with the 808 nm laser irradiation can be simultaneously achieved in whole blood samples.
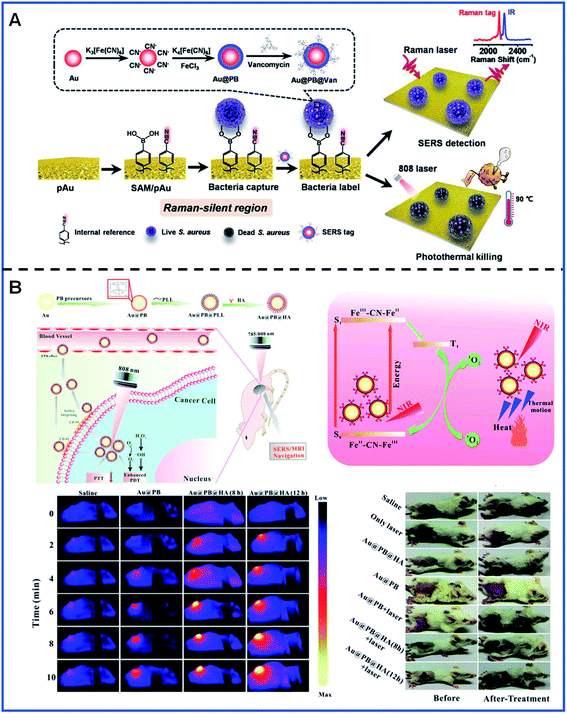 | ||
| Fig. 7 Bacteria detection and tumour location using PB-based SERS tags. (A) A schematic illustrating the detection and elimination process of bacteria. Adapted with permission from ref. 61. Copyright 2021 American Chemical Society. (B) The detection and phototherapy of tumours in vivo. Reproduced from ref. 65 with permission from Royal Society of Chemistry. Copyright 2020 Royal Society of Chemistry. | ||
In vivo SERS imaging plays an essential role in the tracking of the existence and location of tumours and, therefore, has the potential to be used for guiding surgical tumour removal. SERS/MR bimodal imaging has been employed for dendritic cell (DC) activation and tracking by using gadolinium-doped Au@PB NPs as bimodal agents.64 Gd3+ was doped into the PB shells by replacing the FeCl3 solution with the mixture of FeCl3 and GdCl3 in order to obtain a better MR imaging ability. SERS/MR bimodal imaging opens up a promising means for activating and tracking DC. Furthermore, SERS/MR dual-mode imaging has also been used for the guidance of both tumour localization and in situ PT/PD treatment using hyaluronic acid (HA)-conjugated Au@PB NPs as targeting theragnostic agents. During the process of treatment, Au@PB NPs showed a decent photothermal effect. Furthermore, PB can stimulate the generation of 1O2 based on the molecular oxygen present in the surrounding environment using the 808 nm laser irradiation, contributing to the effective treatment of tumours in the mice (Fig. 7B).65
Although PB-based SERS nanotags have exhibited outstanding advantages for photothermal therapy and have improved the quantitative accuracy, it is noted that there is the potential for photodamage to normal cells during the cell imaging process, thus it is essential to improve the specific targeting of the nanotags to the abnormal cells. On the other hand, photodamage is usually caused by high-power lasers and long irradiation times, so it is also urgent to increase the brightness of PB-based SERS tags and design PB-based SERS tags suitable for longer-wavelength lasers. Moreover, further exploration should be carried out to improve the long-term stability of PB-based SERS nanotags for their wide biological applications.
4. Summary and outlook
This review provides a comprehensive summary of PB-based SERS tags and their biological applications. PB-based SERS tags have been synthesized as newly emerging optical nanoprobes consisting of Au NPs, PB or PBA, and functional moieties. Compared with the commonly-used Raman reporter molecules, PB and PBA are powerful competitors as Raman reporters, which provide a distinguished Raman band and avoid the competitive adsorption between analytes and Raman reporters. Moreover, PB possesses the advantages of low toxicity, satisfying the requirement in biological analysis, and PB has been demonstrated as a novel class of MR contrast agent due to the presence of iron ions. PB-based SERS tags have been investigated for the quantification analysis of key molecules highly related to human health as well as the recognition of micro-scale objects such as bacteria and cancer cells. The multifunctional detection platform can be constructed by fine-tuning the various components of the PB-based SERS tag. Even so, all these current studies remain in the early stages for the development of SERS techniques as clinically available means of diagnosis.Previous studies of PB-based SERS tags will help to navigate future development. However, there are still many challenges to be overcome in the field before PB-based SERS tags can be used as commercial nanoprobes to answer significant biomedical questions. The reliability of indirect SERS bioanalysis is decided by both the reproducible preparation and the stability of SERS tags; therefore, the delicate control of the preparation conditions, including a more mature and replicable approach and the further protection with the surfactant, is critical for synthesizing stable and uniform PB-based SERS tags. Moreover, breakthroughs in the femtosecond time scale and nanometre resolution are possible, combined with the application of ultrafast lasers and the development of high spatial resolution techniques such as tip-enhanced Raman scattering (TERS).90,91 With the development and popularity of portable Raman,92,93 SERS techniques with PB-based SERS tags will be a promising tool for point of care (POC) pathogen diagnosis and molecular detection.
Diversified design is another developing orientation of PB-based SERS tags. The gadolinium-doped strategies provide a new perspective for the biomedical application of PB-based SERS tags. For instance, the PB-based SERS tags have the potential to promote wound healing by doping with calcium ions or zinc ions. The fine-tuning of the morphology of the PB shell, such as the caged PB shell, can also be used as the nanodrug carrier for the treatment of tumours and cancer. Additionally, PB-based SERS tags provide much information for in vivo tumour location, thereby assisting with the selective removal of tumour tissue from the damaged tissue. When combined with the 808 nm laser irradiation, the prognosis after surgery will be significantly improved. Although there is still much work to be done using PB-based SERS tags for the reliable quantitative measurements in complex biosystems and the surgery guidance combined with the treatment in the biomedical area, we can still look forward to the increased SERS-based practical applications along with further development of portable instruments and the preparation method of PB-based SERS tags.
Notably, PB is regarded as one of the metal–organic frameworks (MOFs), indicating the potential of MOF for constructing SERS nanotags used in SERS detection. MOFs, as highly porous crystalline materials, have rapidly attracted great interest in the emerging MOF-SERS field due to their nanoporous functionality, large surface area, excellent dispersibility, and designable functions. Recently, different MOFs have been used to stabilize noble metal nanoparticles and adsorb analyte molecules in close proximity to the “hotspots”, improving the detection reproducibility and sensitivity of SERS application. MOFs have been applied in SERS detection in multiple areas, such as environmental analysis,94–96 food safety,97–99 bio-medicine100,101 and real-time monitoring.102,103 In the future, the introduction of more novel MOF materials or the modification of MOF materials for constructing SERS tags will further extend the reliable application of SERS.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (No. 21874102 and 22074109).Notes and references
- P. A. Mosier-Boss, Nanomaterials, 2017, 7, 142 CrossRef PubMed.
- J. Perumal, Y. Wang, A. B. E. Attia, U. S. Dinish and M. Olivo, Nanoscale, 2021, 13, 553 RSC.
- J. Sun, L. Gong, W. Wang, Z. Gong, D. Wang and M. Fan, Luminescence, 2020, 35, 808 CrossRef PubMed.
- K. Kneipp, Y. Wang, H. Kneipp, L. T. Pe, I. Itzkan, R. R. Dasarirelman and M. S. Feld, Phys. Rev. Lett., 1997, 78, 1667 CrossRef CAS.
- S. M. Stranahan and K. A. Willets, Nano Lett., 2010, 10, 3777 CrossRef CAS PubMed.
- J. P. Camden, J. A. Dieringer, Y. Wang, D. J. Masiello, L. D. Marks, G. C. Schatz and R. P. Van Duyne, J. Am. Chem. Soc., 2008, 130, 12616 CrossRef CAS PubMed.
- Z. Zheng, S. Cong, W. Gong, J. Xuan, G. Li, W. Lu, F. Geng and Z. Zhao, Nat. Commun., 2017, 8, 1993 CrossRef PubMed.
- P. K. Kannan, P. Shankar, C. Blackman and C. H. Chung, Adv. Mater., 2019, 31, 1803432 CrossRef PubMed.
- R. A. Alvarez-Puebla and L. M. Liz-Marzan, Chem. Soc. Rev., 2012, 41, 43 RSC.
- D. Li, D. M. Yao, C. N. Li, Y. H. Luo, A. H. Liang, G. Q. Wen and Z. l. Jiang, TrAC, Trends Anal. Chem., 2020, 127, 115885 CrossRef CAS.
- M. Fan, G. F. S. Andrade and A. G. Brolo, Anal. Chim. Acta, 2020, 1097, 1 CrossRef CAS PubMed.
- S. A. Ogundare and W. E. van Zyl, Cellulose, 2019, 26, 6489 CrossRef CAS.
- H. K. Lee, Y. H. Lee, C. S. L. Koh, G. C. Phan-Quang, X. Han, C. L. Lay, H. Y. F. Sim, Y. C. Kao, Q. An and X. Y. Ling, Chem. Soc. Rev., 2019, 48, 731 RSC.
- F. S. Flores, E. Méndez, M. L. Castaños, A. C. Lima, K. A. L. Castaños, M. A. G. Fuentes and A. M. Albores, Biosensors, 2021, 11, 66 CrossRef PubMed.
- M. Sarah, F. Karen and G. Duncan, J. Photochem. Photobiol., C, 2014, 21, 40 CrossRef.
- Y. Zeng, K. M. Koo, M. Trau, A. G. Shen and J. M. Hu, Appl. Mater. Today, 2019, 15, 431 CrossRef.
- X. S. Zheng, I. J. Jahn, K. Weber, D. Cialla-May and J. Popp, Spectrochim. Acta, Part A, 2018, 197, 56 CrossRef CAS PubMed.
- Y. C. Song, T. L. Xu, L. P. Xu and X. J. Zhang, Nanoscale, 2018, 10, 20990 RSC.
- B. Han, Y. L. Zhang, L. Zhu, X. H. Chen, Z. C. Ma, X. L. Zhang, J. N. Wang, W. Wang, Y. Q. Liu, Q. D. Chen and H. B. Sun, Sens. Actuators, B, 2018, 270, 500 CrossRef CAS.
- E. Garcia-Rico, R. A. Alvarez-Puebla and L. Guerrini, Chem. Soc. Rev., 2018, 47, 4909 RSC.
- J. Ma, G. Feng, Y. Ying, Y. Shao, Y. She, L. Zheng, A. M. Abd Ei-Aty and J. Wang, Analyst, 2021, 146, 956 RSC.
- W. Fan, S. Yang, Y. Zhang, B. Huang, Z. Gong, D. Wang and M. Fan, ACS Sens., 2020, 5, 3599 CrossRef CAS PubMed.
- A. Teixeira, J. L. Paris, F. Roumani, L. Dieguez, M. Prado, B. Espina, S. Abalde-Cela, A. Garrido-Maestu and L. Rodriguez-Lorenzo, Materials, 2020, 13, 1934 CrossRef CAS PubMed.
- J. Langer, I. Garcia and L. M. Liz-Marzan, Faraday Discuss., 2017, 205, 363 RSC.
- Y. Wang, S. Zong, N. Li, Z. Wang, B. Chen and Y. Cui, Nanoscale, 2019, 11, 2460 RSC.
- Y. Liu, S. Yue, Y. N. Wang, Y. Wang and Z. R. Xu, Sens. Actuators, B, 2020, 310, 127889 CrossRef CAS.
- S. Yue, X. Sun, N. Wang, Y. Wang, Y. Wang, Z. Xu, M. Chen and J. Wang, ACS Appl. Mater. Interfaces, 2017, 9, 39699 CrossRef CAS PubMed.
- Y. Wang, B. Yan and L. Chen, Chem. Rev., 2013, 113, 1391 CrossRef CAS PubMed.
- L. Fabris, J. Opt., 2015, 17, 114002 CrossRef.
- S. Cheng, B. Zheng, D. B. Yao, Y. Wang, J. J. Tian, L. H. Liu, H. J. Liang and Y. S. Ding, Anal. Lett., 2019, 52, 902 CrossRef CAS.
- T. T. B. Quyen, C. C. Chang, W. N. Su, Y. H. Uen, C. J. Pan, J. Y. Liu, J. Rick, K. Y. Lin and B. J. Hwang, J. Mater. Chem. B, 2014, 2, 629 RSC.
- N. Choi, H. Dang, A. Das, M. S. Sim, I. Y. Chung and J. Choo, Biosens. Bioelectron., 2020, 164, 112326 CrossRef CAS PubMed.
- J. He, Y. Qiao, H. Zhang, J. Zhao, W. Li, T. Xie, D. Zhong, Q. Wei, S. Hua, Y. Yu, K. Yao, H. A. Santos and M. Zhou, Biomaterials, 2020, 234, 119763 CrossRef CAS PubMed.
- Y. Feng, L. He, L. Wang, R. Mo, C. Zhou, P. Hong and C. Li, Nanomaterials, 2020, 10, 1000 CrossRef CAS PubMed.
- L. Sun, Z. L. Yu and M. S. Lin, Analyst, 2019, 144, 4820 RSC.
- L. Lin, H. Gu and J. Ye, Chem. Commun., 2015, 51, 17740 RSC.
- Y. Zeng, J. Q. Ren, A. G. Shen and J. M. Hu, J. Am. Chem. Soc., 2018, 140, 10649 CrossRef CAS PubMed.
- Y. T. Shen, L. J. Liang, J. Zhang, Z. Y. Li, J. Yue, J. Q. Wang, W. Q. Xu, W. Shi and S. P. Xu, Sens. Actuators, B, 2019, 285, 84 CrossRef CAS.
- A. G. Shen, L. F. Chen, W. Xie, J. C. Hu, A. Zeng, R. Richards and J. M. Hu, Adv. Funct. Mater., 2010, 20, 969 CrossRef CAS.
- J. Li, H. Liu, P. Rong, W. Zhou, X. Gao and D. Liu, Nanoscale, 2018, 10, 8292 RSC.
- J. L. Gong, J. H. Jiang, H. F. Yang, G. L. Shen, R. Q. Yu and Y. Ozaki, Anal. Chim. Acta, 2006, 564, 151 CrossRef CAS.
- N. G. Khlebtsov, L. Lin, B. N. Khlebtsov and J. Ye, Theranostics, 2020, 10, 2067 CrossRef CAS PubMed.
- S. Sloan-Dennison, M. R. Bevins, B. T. Scarpitti, V. K. Sauve and Z. D. Schultz, Analyst, 2019, 144, 5538 RSC.
- Y. Shen, J. Yue, W. Shi, W. Xu and S. Xu, Biosens. Bioelectron., 2020, 151, 111957 CrossRef CAS PubMed.
- X. W. Wang and L. Cheng, Coord. Chem. Rev., 2020, 419, 213393 CrossRef CAS.
- M. A. Busquets and J. Estelrich, Drug Discovery Today, 2020, 25, 1431 CrossRef CAS PubMed.
- Z. Qin, Y. Li and N. Gu, Adv. Healthcare Mater., 2018, 7, e1800347 CrossRef PubMed.
- S. F. A. Kettle, E. Diana, E. M. C. Marchese, E. Boccaleri and P. L. Stanghellini, J. Raman Spectrosc., 2011, 42, 2006 CrossRef CAS.
- G. Moretti and C. Gervais, J. Raman Spectrosc., 2018, 49, 1198 CrossRef CAS.
- Y. Yin, Q. Li, S. Ma, H. Liu, B. Dong, J. Yang and D. Liu, Anal. Chem., 2017, 89, 1551 CrossRef CAS PubMed.
- E. Lenzi, D. J. D. Aberasturi and L. M. Liz-Marzan, ACS Sens., 2019, 4, 1126 CrossRef CAS PubMed.
- M. Muhammad and Q. Huang, Talanta, 2021, 227, 122188 CrossRef CAS PubMed.
- J. Xu, C. Li, H. Si, X. Zhao, L. Wang, S. Jiang, D. Wei, J. Yu, X. Xiu and C. Zhang, Opt. Express, 2018, 26, 21546 CrossRef CAS PubMed.
- J. Guo, X. Yan, M. Xu, G. Ghimire, X. Pan and J. He, Anal. Chem., 2021, 93, 4441 CrossRef CAS PubMed.
- R. X. He, R. Liang, P. Peng and Y. Norman Zhou, J. Nanopart. Res., 2017, 19, 1 CrossRef CAS.
- C. Hu, J. Shen, J. Yan, J. Zhong, W. Qin, R. Liu, A. Aldalbahi, X. Zuo, S. Song, C. Fan and D. He, Nanoscale, 2016, 8, 2090 RSC.
- M. Zannotti, A. Rossi and R. Giovannetti, Coatings, 2020, 10, 288 CrossRef CAS.
- A. Y. F. Mahmoud, C. J. Rusin and M. T. McDermott, Analyst, 2020, 145, 1396 RSC.
- K. Li, G. Liu, S. Zhang, Y. Dai, S. Ghafoor, W. Huang, Z. Zu and Y. Lu, Nanoscale, 2019, 11, 9587 RSC.
- P. Sha, Q. Su, P. Dong, T. Wang, C. Zhu, W. Gao and X. Wu, RSC Adv., 2021, 11, 27107 RSC.
- X. Gao, Y. Yin, H. Wu, Z. Hao, J. Li, S. Wang and Y. Liu, Anal. Chem., 2021, 93, 1569 CrossRef CAS PubMed.
- Y. M. Shen, M. Y. Gao, X. Chen, A. G. Shen and J. M. Hu, Spectrochim. Acta, Part A, 2021, 252, 119566 CrossRef CAS PubMed.
- X. Li, E. Zeng, H. Di, Q. Li, J. Ji, J. Yang and D. Liu, Adv. Biosyst., 2019, 3, e1900046 CrossRef PubMed.
- C. Zhang, Z. Xu, H. Di, E. Zeng, Y. Jiang and D. Liu, Theranostics, 2020, 10, 6061 CrossRef CAS PubMed.
- W. Zhu, M. Y. Gao, Q. Zhu, B. Chi, L. W. Zeng, J. M. Hu and A. G. Shen, Nanoscale, 2020, 12, 3292 RSC.
- J. Li, J. Wang, Y. S. Grewal, C. B. Howard, L. J. Raftery, S. Mahler, Y. Wang and M. Trau, Anal. Chem., 2018, 90, 10377 CrossRef CAS PubMed.
- M. Arabi, A. Ostovan, Z. Zhang, Y. Wang, R. Mei, L. Fu, X. Wang, J. Ma and L. Chen, Biosens. Bioelectron., 2021, 174, 112825 CrossRef CAS PubMed.
- X. Wang, N. Choi, Z. Cheng, J. Ko, L. Chen and J. Choo, Anal. Chem., 2017, 89, 1163 CrossRef CAS PubMed.
- S. Ganesh, K. Venkatakrishnan and B. Tan, Nat. Commun., 2020, 11, 1135 CrossRef CAS PubMed.
- J. Plou, I. García, M. Charconnet, I. Astobiza, C. García-Astrain, C. Matricardi, A. Mihi, A. Carracedo and L. M. Liz-Marzán, Adv. Funct. Mater., 2020, 30, 1910335 CrossRef CAS.
- J. Kelly, R. Patrick, S. Patrick and S. E. J. Bell, Angew. Chem. Int. Ed., 2018, 57, 15686 CrossRef CAS PubMed.
- J. Guo, A. Sesena Rubfiaro, Y. Lai, J. Moscoso, F. Chen, Y. Liu, X. Wang and J. He, Analyst, 2020, 145, 4852 RSC.
- G. H. Yang, Q. Zhang, Y. Liang, H. Liu, L. L. Qu and H. T. Li, Colloids Surf., A, 2019, 562, 289 CrossRef CAS.
- M. N. Sanz-Ortiz, K. Sentosun, S. Bals and L. M. Liz-Marzán, ACS Nano, 2015, 9, 10489 CrossRef CAS PubMed.
- F. Yu, M. Su, L. Tian, H. Wang and H. Liu, Anal. Chem., 2018, 90, 5232 CrossRef CAS PubMed.
- L. Liu, C. J. Shangguan, J. L. Guo, K. J. Ma, S. L. Jiao, Y. Yao and J. Q. Wang, Adv. Opt. Mater., 2020, 8, 2001214 CrossRef CAS.
- X. Qin, Y. Si, D. Wang, Z. Wu, J. Li and Y. Yin, Anal. Chem., 2019, 91, 4529 CrossRef CAS PubMed.
- J. Chen, Y. Wu, C. Fu, H. Cao, X. Tan, W. Shi and Z. Wu, Biosens. Bioelectron., 2019, 143, 111619 CrossRef CAS PubMed.
- W. Shen, X. Lin, C. Y. Jiang, C. Y. Li, H. X. Lin, J. T. Huang, S. Wang, G. K. Liu, X. M. Yan, Q. L. Zhong and B. Ren, Angew. Chem. Int. Ed., 2015, 54, 7308 CrossRef CAS PubMed.
- M. Li, J. Y. Wang, Q. Q. Chen, L. H. Lin, P. Radjenovic, H. Zhang, S. Y. Luo, Z. Q. Tian and J. F. Li, Anal. Chem., 2019, 15025 CrossRef CAS PubMed.
- T. Zhou, D. Lu, Q. She, C. Chen, J. Chen, Z. Huang, S. Feng, R. You and Y. Lu, Sens. Actuators, B, 2021, 336, 129597 CrossRef CAS.
- F. Sun, P. Zhang, T. Bai, D. D. Galvan, H. C. Hung, N. Zhou, S. Y. Jiang and Q. M. Yu, Biosens. Bioelectron., 2015, 73, 202 CrossRef CAS PubMed.
- H. Wei, M. R. Willner, L. C. Marr and P. J. Vikesland, Analyst, 2016, 141, 5159 RSC.
- Y. Shen, L. Liang, S. Zhang, D. Huang, J. Zhang, S. Xu, C. Liang and W. Xu, Nanoscale, 2018, 10, 1622 RSC.
- Y. T. Huang, W. Liu, D. M. Wang, Z. J. Gong and M. K. Fan, Microchem. J., 2020, 154, 104565 CrossRef CAS.
- G. Zhu, L. Cheng, G. Liu and L. Zhu, Nanomaterials, 2020, 10, 1501 CrossRef CAS PubMed.
- A. Quinn, Y. H. You and M. J. McShane, Sensors, 2019, 19, 3521 CrossRef CAS PubMed.
- Y. Bi, H. Di, E. Zeng, Q. Li, W. Li, J. Yang and D. Liu, Anal. Chem., 2020, 92, 9574 CrossRef CAS PubMed.
- M. Y. Gao, Q. Chen, W. Li, A. G. Shen and J. M. Hu, Anal. Chem., 2019, 91, 13866 CrossRef CAS PubMed.
- B. Pettinger, B. Ren, G. Picardi, R. Schuster and G. Ertl, J. Raman Spectrosc., 2005, 36, 541 CrossRef CAS.
- B. R. Wood, E. Bailo, M. A. Khiavi, L. Tilley, S. Deed, T. Deckert-Gaudig, D. McNaughton and V. Deckert, Nano Lett., 2011, 11, 1868 CrossRef CAS PubMed.
- X. Gong, M. Tang, Z. Gong, Z. Qiu, D. Wang and M. Fan, Food Chem., 2019, 295, 254 CrossRef CAS PubMed.
- W. Zhu, B. Y. Wen, L. J. Jie, X. D. Tian, Z. L. Yang, P. M. Radjenovic, S. Y. Luo, Z. Q. Tian and J. F. Li, Biosens. Bioelectron., 2020, 154, 112067 CrossRef CAS PubMed.
- O. Guselnikova, P. Postnikov, R. Elashnikov, E. Miliutina, V. Svorcik and O. Lyutakov, Anal. Chim. Acta, 2019, 1068, 70 CrossRef CAS PubMed.
- D. Li, X. K. Cao, Q. M. Zhang, X. G. Ren, L. Jiang, D. W. Li, W. Deng and H. T. Liu, J. Mater. Chem. A, 2019, 7, 14108 RSC.
- Q. Z. Wang, Z. H. Xu, Y. J. Zhao, S. H. Zhang, T. Bu, C. Q. Zhang, X. Wang and L. Wang, Sens. Actuators, B, 2021, 329, 129080 CrossRef CAS.
- X. Zhou, G. Liu, H. Zhang, Y. Li and W. Cai, J. Hazard. Mater., 2019, 368, 429 CrossRef CAS PubMed.
- H. S. Lai, G. K. Li, F. G. Xu and Z. M. Zhang, J. Mater. Chem. C, 2020, 8, 2952 RSC.
- Q. Q. Li, S. S. Gong, H. Zhang, F. Z. Huang, L. N. Zhang and S. K. Li, Chem. Eng. J., 2019, 371, 26 CrossRef CAS.
- P. Jiang, Y. Hu and G. Li, Talanta, 2019, 200, 212 CrossRef CAS PubMed.
- X. Qiao, B. Su, C. Liu, Q. Song, D. Luo, G. Mo and T. Wang, Adv. Mater., 2018, 30, 1702275 CrossRef PubMed.
- S. Choi, H. J. Lee and M. Oh, Small, 2016, 12, 2425 CrossRef CAS PubMed.
- Q. Xu, W. Liu, L. Li, F. Zhou, J. Zhou and Y. Tian, Chem. Commun., 2017, 53, 1880 RSC.
| This journal is © The Royal Society of Chemistry 2021 |
