 Open Access Article
Open Access ArticleTwo-dimensional transition metal dichalcogenide nanomaterials for biosensing applications
Yanling
Hu†
ab,
Ying
Huang†
b,
Chaoliang
Tan
b,
Xiao
Zhang
b,
Qipeng
Lu
b,
Melinda
Sindoro
b,
Xiao
Huang
c,
Wei
Huang
ac,
Lianhui
Wang
*a and
Hua
Zhang
*b
aKey Laboratory for Organic Electronics and Information Displays (KLOEID) and Institute of Advanced Materials (IAM), National Jiangsu Synergetic Innovation Center for Advanced Materials (SICAM), Nanjing University of Posts and Telecommunications, 9 Wenyuan Road, Nanjing 210023, China. E-mail: iamlhwang@njupt.edu.cn
bCenter for Programmable Materials, School of Materials Science and Engineering, Nanyang Technological University, 50 Nanyang Avenue, Singapore 639798, Singapore. E-mail: hzhang@ntu.edu.sg
cKey Laboratory of Flexible Electronics (KLOFE) & Institute of Advanced Materials (IAM), Jiangsu National Synergistic Innovation Center for Advanced Materials (SICAM), Nanjing Tech University (NanjingTech), 30 South Puzhu Road, Nanjing 211816, China
First published on 16th November 2016
Abstract
Biosensors are powerful tools used to monitor biological and biochemical processes, ranging from clinical diagnosis to disease therapy. The huge demands for bioassays greatly promote the development of new nanomaterials as sensing platforms. Two-dimensional (2D) nanomaterials with superior properties, such as large surface areas and excellent conductivities, are excellent candidates for biosensor applications. Among them, single- or few-layered transition metal dichalcogenide (TMD) nanomaterials represent an emerging class of 2D nanomaterials with unique physical, chemical, and electronic properties. In this mini-review, we summarize the recent progress in 2D TMD nanomaterial-based biosensors for the sensitive detection of various kinds of targets, including nucleic acid, proteins, and small biomolecules, based on different sensors like optical sensors and electrochemical sensors, and bioelectronic sensors. Finally, the challenges and opportunities in this promising field are also proposed.
1. Introduction
With the increasing demand for disease diagnosis and therapies, biosensors have become powerful tools for the detection of biological analytes, such as nucleic acid, proteins, and small biomolecules. Generally, analytes extracted from biological systems are at a low concentration level, particularly for biomarkers of specific diseases.1–3 Hence, over the past few decades, great efforts have been devoted to the design and development of ultrasensitive bioassays. Besides the development of advanced instruments and detection strategies for signal amplification, the utilization of suitable materials in sensing applications is another important way to enhance the detection signal.4 It has been demonstrated that the performance of biosensors shows a close association with the intrinsic characteristics, e.g. structures, morphologies, compositions, and physicochemical properties, of the used nanomaterials.5,6 For example, nanomaterials with a large surface area, good conductivity, and excellent biological compatibility can be used as signal amplification elements in electrochemical sensors.7 Therefore, the exploration of new advanced nanomaterials is key to develop biosensors with a high sensitivity and low detection limit.Two-dimensional (2D) nanomaterials have attracted broad interest due to their unconventional properties, especially after the 2010 Nobel Prize in physics was awarded for “groundbreaking experiments regarding graphene”.8–14 Since then, a number of graphene-based biosensors have been reported.15–19 Inspired by graphene, other graphene-like 2D nanomaterials with single- and few-layer thickness came to light and have gained increasing attention in recent years, especially transition metal dichalcogenides (TMDs) which include MoS2, WS2, TaS2, TiS2, WSe2, MoSe2, etc.20–29 To date, a wide variety of synthetic methods have been developed to prepare single- and few-layered 2D TMD nanosheets, namely, mechanical cleavage, chemical vapor deposition (CVD), wet-chemical synthesis, chemical or electrochemical Li-intercalation and exfoliation, as well as liquid phase exfoliation in different solvents.30–36 Due to their ultrathin thickness and 2D structure, these TMD nanosheets present unique physical, chemical, optical, and electronic properties compared with their bulk counterparts.20–22,37,38 In addition, 2D TMD nanosheets can directly interact with biomolecules, resulting in their surface modification/decoration. This could greatly extend their range of biosensing. Taking MoS2 as an example, structural defects, such as point defects and edges, play important roles in sensing applications.39,40 On account of these attractive properties as well as large specific surface areas, 2D TMD nanosheets hold great potential in biosensing applications.41,42
This mini-review aims to provide an overview of the recent advances on 2D TMD nanosheets for biosensing applications. First, we describe the structure and properties of 2D TMD nanosheets. Then, we present the recent development of TMD nanosheet-based biosensors for the detection of different biological analytes including nucleic acid, proteins, and small molecules through various sensing approaches like optical methods, electrochemical methods, and electronic methods. Finally, based on the current studies, the opinions on the challenges and opportunities in this hot field are proposed.
2. Structure and properties of 2D TMD nanomaterials
TMDs are MX2-type compounds with layered structures, which are made by strong in-plane covalent bonding along with weak interactions between the layers, especially for group 4–7 TMDs (Fig. 1a), while some of the group 8–10 TMDs are commonly presented in non-layered structures.20 In each MX2 monolayer, a layer of transition metal atom (M) is sandwiched between two chalcogen atom layers (X = S, Se, or Te), giving 6–7 Å of thickness for each monolayer.43–45 The properties of TMD materials change along with a decrease in the number of layers. For example, the bandgap of bulk MoS2 is about 1.3 eV while that of single-layered MoS2 is 1.8–1.9 eV,46 showing an indirect-to-direct bandgap transition as the thickness decreased to a single layer. In addition, the surface areas of TMD materials greatly increase after being exfoliated into single or few layers.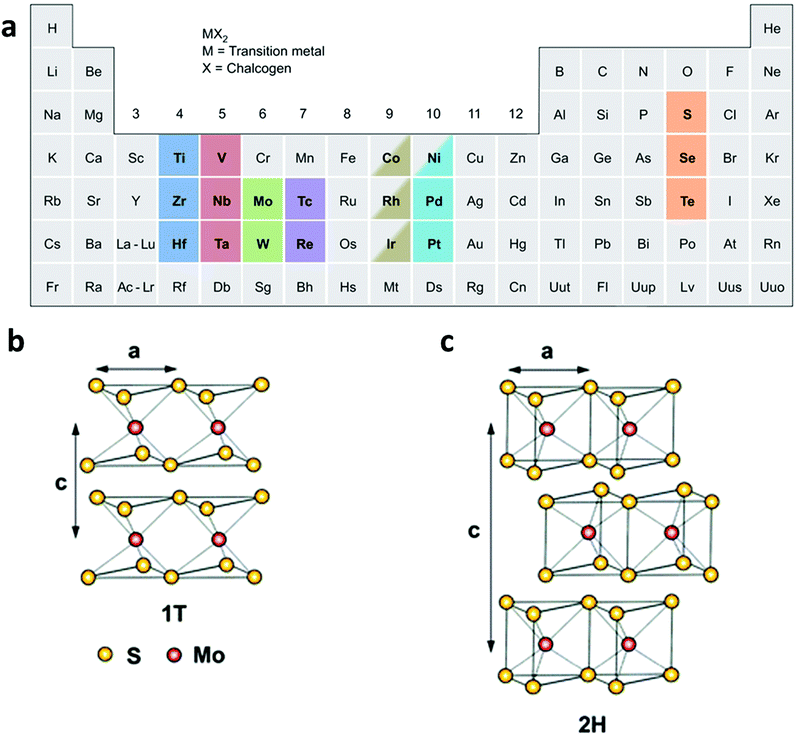 | ||
| Fig. 1 (a) Various kinds of TMD nanomaterials, reproduced with permission from ref. 20. Copyright 2013 Nature Publishing Group. Schematic illustration of (b) the 1T-type and (c) the 2H-type structure of MoS2. Reproduced with permission from ref. 8. Copyright 2013 Royal Society of Chemistry. | ||
Of particular interest is that TMD materials exhibit various polymorphs, commonly including 1T, 2H, and 3R, which stand for trigonal, hexagonal, and rhombohedral crystal phases, respectively.47,48 However, when exfoliated into a single layer, TMD materials exhibit two polymorphs: 1T and 2H (Fig. 1b and c). It is worth noting that the phase transformation between these phases can be realized by various strategies. Taking MoS2 as an example, the 3R phase can be transformed into 2H phase upon heating.47 Moreover, the crystal structure of MoS2 can be partly engineered from 2H to 1T by intercalating alkaline metals (Li, K, or Na) in the exfoliation process.49–52 Note that the phase transformation from 2H to 1T is accompanied by a change in the density of states, resulting in the evolution of MoS2 from a semiconductor to a metal.53
These aforementioned unique properties make the single-layered TMD nanosheets good candidates for the fabrication of biosensors. For example, the increased bandgap of MoS2 nanosheets makes it a better choice for electronic devices.54 The phase transformation of MoS2 from a semiconductor to a metal can enhance its electrical conductivity which benefits the electrochemical properties.55,56 Moreover, the large surface area of single-layered TMD nanosheets enables high physical adsorption of biomolecules onto their surface. Simultaneously, 2D TMD nanosheets present an efficient fluorescence quenching ability for fluorophores, rendering them promising platforms for the construction of fluorescence-based biosensors.57,58 Our group first reported the physisorption of single-layered MoS2 nanosheets towards single-stranded DNA (ssDNA) and studied its efficient quenching ability.59 As the physisorption of aromatic and conjugated compounds on the MoS2 surface has been proposed, the ssDNA adsorption is believed to be a result of the van der Walls force between nucleobases and the basal plane of the MoS2 nanosheet.60 The principle also holds that the lower adsorption of double-stranded DNA (dsDNA), with nucleobases buried between their helical phosphate backbones, occurs on MoS2 nanosheets. Besides DNA hybridization, the adsorption of the DNA probe can also be weakened by 3D structural switching resulting from specific aptamer-target recognition.
3. 2D TMD nanomaterial-based biosensors
3.1 2D TMD nanomaterials used for optical biosensors
Optical biosensors are attracting huge attention for disease diagnosis and clinical evaluation due to their simple, fast, and direct read out properties.61–64 Fluorescence sensing and colorimetric methods are two typical sensing techniques for optical biosensors.15,65,66 Since TMD nanosheets could act as fluorescence quenchers and peroxide mimics, a series of optical biosensors based on TMD nanosheets have been developed.Fluorescent biosensors for microRNA (miRNA) detection are very important, since miRNA plays a significant role in physiological and pathological processes and can be used as a biomarker for disease diagnosis. Great efforts have been devoted to the development of biosensors by using TMD nanosheets for miRNA detection. The WS2 nanosheet was reported to show weaker affinities towards short ssDNA compared to long ssDNA.72 This property has been applied to design a fluorescent sensor for miRNA detection by Jiang et al.72 A dye-labeled ssDNA was used as the probe whose fluorescence can then be quenched by the WS2 nanosheet. In contrast, in the presence of target miRNA and duplex-specific nuclease (DSN), the formed DNA–RNA heteroduplex could be cleaved to release target miRNA to hybridize with other probe ssDNA, leading to cyclic reaction and signal amplification. This method further pushed the miRNA detection limit down to 300 fM. Obviously, the fluorophore could be replaced by other kinds of molecule beacons. Zhao et al. proposed a chemiluminescence (CL) resonance energy transfer (CRET) sensing platform for miRNA detection.26 In this case, part of the probe DNA can form a G-quadruplex-hemin-H2O2-luminol structure and give strong CL which can be quenched by the WS2 nanosheet due to the interaction between ssDNA and the G-quadruplex-hemin DNAzyme structure. In the presence of target miRNA, the formed DNA–RNA heteroduplex showed a weak affinity with the WS2 nanosheet and desorbed from the WS2 nanosheet surface, leading to restoration of the CL signal. The CL intensity and the concentration of miRNA showed a linear relationship in the range of 0.5–10 nM, yielding a detection limit of ∼180 pM.
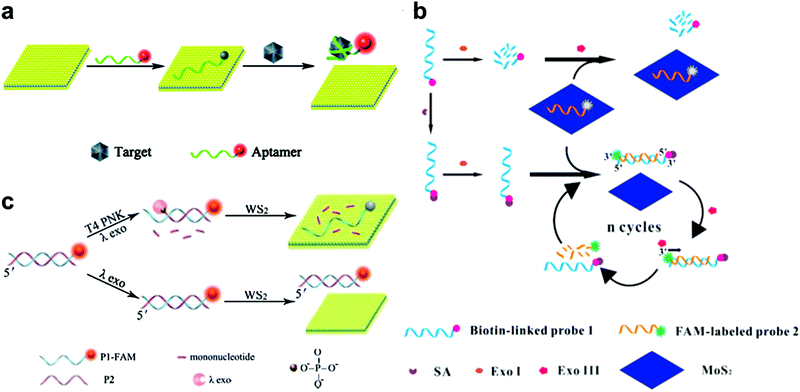 | ||
| Fig. 2 (a) Schematic illustration of the MoS2 nanosheet used as an effective sensing biosensor. Reproduced with permission from ref. 74. Copyright 2014 Royal Society of Chemistry. (b) Schematic illustration of the mechanism of a MoS2-based biosensor for the detection of SA via terminal protection of small-molecule-linked DNA and Exo III-aided DNA recycling amplification. Reproduced with permission from ref. 79. Copyright 2015 Elsevier. (c) Schematic illustration of the WS2 nanosheet-based platform for T4 PNK activity and inhibition analysis. Reproduced with permission from ref. 80. Copyright 2014 Royal Society of Chemistry. | ||
Besides protein detection using nucleic-acid aptamers, Xiang and co-workers have converted a protein assay into DNA detection by combining the terminal protection of small-molecule-linked DNA with exonuclease III (Exo III)-aided DNA recycling amplification.79 As shown in Fig. 2b, in the sensing process, a dye-labeled probe 2 could hybridize with a biotin-modified probe 1 and tether to the 3′-end of the antisense strand. Probe 1 was digested by exonuclease I (Exo I) in the absence of SA. Probe 2 was adsorbed onto the MoS2 nanosheet surface, resulting in a low fluorescence signal. While in the presence of streptavidin (SA), the SA and biotin could specifically bind together and protect probe 1 from Exo I-assisted digestion. After hybridization with probe 2, the DNA duplex formed and probe 2 could be degraded by Exo III, resulting in a cyclic amplification and the release of more probe 2 from MoS2. This strategy achieved highly sensitive SA detection with a detection limit of 0.67 ng mL−1.
2D TMD nanosheet-based optical biosensors have also been applied to detect T4 polynucleotide kinase (PNK), which can phosphorylate DNA and plays a significant role in many cellular events. For example, Ge et al. developed a WS2 nanosheet-based optical sensing platform to analyze nucleotide kinase activity.80 In the presence of T4 PNK, dsDNA could be cleaved by λ exonuclease, leaving a FAM-labeled ssDNA. In this case, the fluorescence of the FAM-labeled ssDNA could be quenched by the WS2 nanosheet. As shown in Fig. 2c, in the absence of T4 PNK, the λ exonuclease showed low activity towards non-phosphorylated dsDNA. The remaining dsDNA showed a strong fluorescence signal because of the weak affinity towards the WS2 nanosheet. This strategy offered a detection limit of 0.01 U mL−1. Recently, Liu et al. used hairpin DNA coupled with DNA polymerase and deoxyribonucleoside triphosphates (dNTPs) for T4 PNK assay, achieving a detection limit of 0.05 U mL−1.81
Besides approaches based on the fluorescence quenching abilities of TMD nanosheets, the colorimetric method has also been used for the detection of small biomolecules due to its advantages, such as simplicity, rapidity, and visibility.82 The ability of MoS2 and WS2 nanosheets to serve as peroxidase mimics has been widely reported, and thus they can be used for the detection of H2O2 and glucose.83,84 A color change occurred in the presence of H2O2 when the MoS2 nanosheet catalyzed the reaction of the peroxidase substance such as 3,3′,5,5′-tetramethylbenzidine (TMB). The MoS2 nanosheet–TMB–H2O2 system provided a detection limit of 1.5 μM with a linear range from 5 to 100 μM for H2O2 detection. This method could be used for glucose detection because hydrogen peroxide is the main product of the GOx-catalyzed reaction. The linear range was from 5 to 150 μM with a detection limit of 1.2 μM. Furthermore, a portable test kit was developed to evaluate the glucose level in human serum samples, as a proof of concept, due to its visibility and small sample requirement.
3.2 2D TMD nanomaterials used for electrochemical biosensors
Electrochemical biosensors have been widely used as effective assay methods for clinical diagnosis, environment monitoring, and food safety in the past few decades due to their simplicity, low cost, high sensitivity, and selectivity. Recently, 2D TMD nanomaterials have attracted great attention in the construction of electrochemical biosensors for the detection of various analytes. The ability of TMD nanosheets to prompt the electron transfer rate has been demonstrated due to the exposed active sites on their edges.85 Moreover, TMD nanosheets with an atomic layer structure and a large surface area can act as substrates to support other nanomaterials, including noble metals, transition metals, carbon materials and conductive polymers, further improving their electrochemical biosensing performances. Therefore, the decoration of noble metal nanoparticles (NPs) on the surface of TMD nanosheets has been considered as an efficient way to amplify their electrochemical signals. The Au NP is the most popular one because of its excellent electrical properties and biocompatibility.11,86,87 Furthermore, the surface of Au NP-decorated TMD nanosheets can be functionalized by various molecules. Besides the noble metal NPs, the carbon materials, conductive polymers and transition metals have also been used to modify TMD nanosheets to increase the electrical conductivity and surface area, thus improving the electrochemical performance.88–91| Materials | Linear range | Detection limit | Ref. |
|---|---|---|---|
| MoS2 | 0.03–300 nM | — | 92 |
| MoS2 | 10−16–10−10 M | 1.9 × 10−17 M | 94 |
| MoS2–thionin | 0.09–1.9 ng mL−1 | — | 93 |
| MoS2-multi-walled carbon nanotube–Au nanoparticle-glucose oxidase | 10–107 fM | 0.79 fM | 95 |
| MoS2–ZnO | 10−15–10−6 M | 6.6 × 10−16 M | 96 |
| MoS2–polyaniline | 10−15–10−6 M | 2 × 10−16 M | 88 |
| MoS2–graphene | 10−16–10−13 M | 10−17 M | 89 |
| MoS2–graphene–Au NPs–horseradish peroxidase | 5.0 × 10−14–5.0 × 10−9 M | 2.2 × 10−15 M | 97 |
| WS2–graphene–Au NPs | 0.01–500 pM | 0.0023 pM | 98 |
| WS2–acetylene black | 0.001–100 pM | 0.12 fM | 99 |
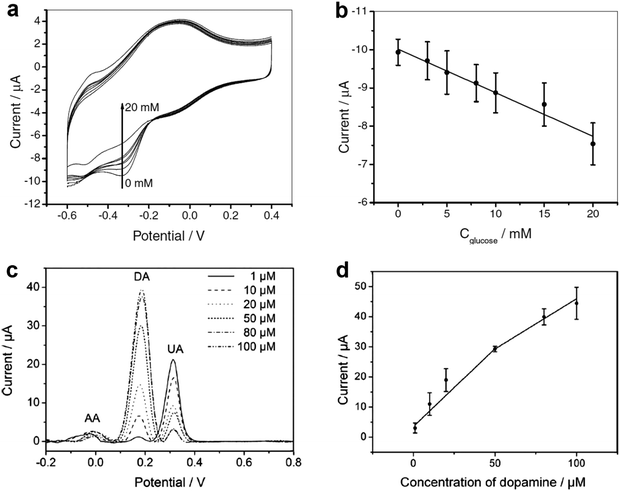 | ||
| Fig. 3 (a) Cyclic voltammograms of the GCE-APTES-chitosan-rMoS2-GOD in air-saturated 0.01 M PBS (pH = 7.4) in 0.1 M KCl solution in the presence of different concentrations of glucose (0, 3, 5, 8, 10, 15 and 20 mM). (b) The measured calibration curve corresponding to the amperometric response. (c) Differential pulse voltammograms of GCE-APTES-rMoS2 in 0.1 M PBS (pH = 7) solution containing 1 mM AA, 0.1 mM UA and DA with different concentrations (1, 10, 20, 50, 80, 100 μM). (d) The oxidation peak current of DA as a function of the concentration of DA. Reproduced with permission from ref. 105. Copyright 2012 Wiley-VCH. | ||
| Materials | Analytes | Linear range | Detection limit | Ref. |
|---|---|---|---|---|
| MoS2 | H2O2 | 5.0–100 nM | 2.5 nM | 113 |
| MoS2–graphene–horseradish peroxidase | H2O2 | 0.2 μM–1.103 mM | 0.049 μM | 114 |
| MoS2–PtW | H2O2 | 1–0.2 mM | 5 nM | 115 |
| MoS2–Cu nanoflower | H2O2/glucose | 0.04–1.88 μM/1–20 μM | 0.021 μM/0.32 μM | 116 |
| MoS2–Ni(OH)2 | Glucose | 10–1300 mM | 5.8 μM | 117 |
| MoS2–Ni NP | Glucose | 0–4 mM | 0.31 μM | 118 |
| MoS2–Au NP-glucose oxidase | Glucose | 10–300 μM | 2.8 μM | 119 |
| MoS2–PANI–Au NP | DA | 1–500 μM | 0.1 μM | 120 |
| MoS2–Au NP | DA | 0.1–200 μM | 80 nM | 121 |
| WS2–Au NP | 17β-estradiol | 1.0 × 10−11–5.0 × 10−9 M | 2.0 × 10−12 M | 122 |
| MoS2–graphene | Honokiol | 1.0 × 10−9–2.5 × 10−6 M | 6.2 × 10−10 M | 123 |
| WS2–graphene | Catechol/resorcinol/hydroquinone | 10−6–10−4 M/10−6–10−4 M/10−6–10−4 M | 2.0 × 10−7 M/1.0 × 10−7 M/1.0 × 10−7 M | 124 |
| MoS2–Ag | Tryptophan | 0.5–120 μM | 0.05 μM | 125 |
| MoS2-self-doped polyaniline | Chloramphenicol | 0.1–1000 μM | 6.5 × 10−8 M | 126 |
| MoS2–Fe3O4 | Nitrite | 1.0–2630.0 μM | 0.5 μM | 127 |
| MoS2–Au–cholesterol oxidase | Cholesterol | 0.5–48 μM | 0.26 μM | 128 |
3.3 2D TMD nanomaterials used for electronic biosensors
As mentioned in Part 2, the single-layered MoS2 nanosheet has an intrinsic bandgap of 1.8–1.9 eV, making it an ideal candidate for FET sensors.129 In a typical FET sensing mode, the adsorption of target molecules on the surface of the semiconducting channel affects the response signal. Recently, Lee et al. successfully fabricated a few-layered MoS2-based bio-FET for DNA detection (Fig. 4a).130 The probe DNA molecules were physically adsorbed on the MoS2 channel. When the hybridization between the probe DNA and the target DNA occurred, the signal of the threshold voltage shifted in the negative direction and the drain current increased. This FET sensor could achieve a detection limit of 10 fM and a high dynamic range of 106 with a high sensitivity of 17 mV dec−1. In addition, DNA hybridization can also be detected with a detection limit of ∼aM through the FET sensors fabricated using MoS2/graphene heterostructures.131 Moreover, 2D TMD nanosheet-based FET biosensors can also be used for protein detection (Fig. 4b). In a recent study, biotin and SA were chosen as the receptor and target molecules, respectively.132 Biotin was used to functionalize the channel of the FET biosensor. When the target molecules were introduced in the FET biosensor, the current of the device decreased due to the negative charge of the SA and the strong bonding between SA and biotin. A high sensitivity of 196 was achieved at a concentration of 100 fM. Furthermore, 2D TMD nanosheet-based FET biosensors can also be used for the detection of other proteins such as PSA.133 Moreover, the pH sensor based on MoS2 was also realized, which is 74-fold higher in sensitivity compared to that of the graphene-based FET sensor.132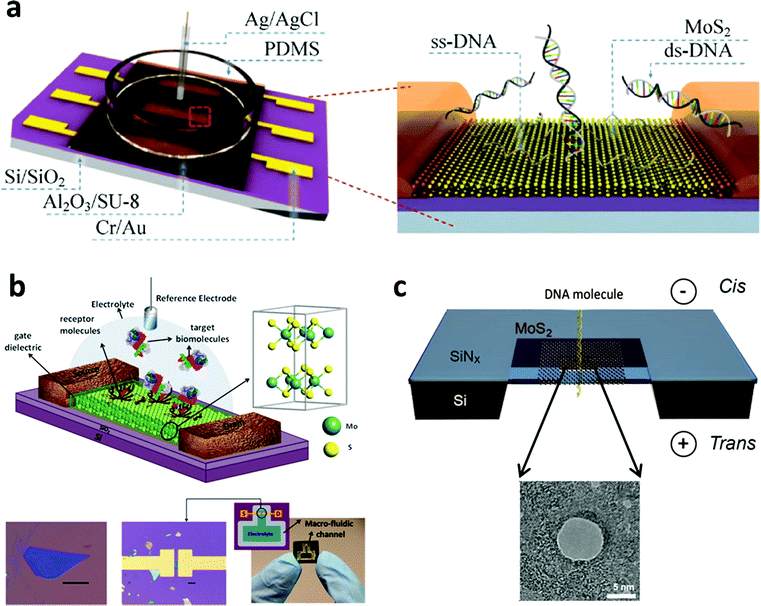 | ||
| Fig. 4 (a) Schematic illustration of MoS2-based FET. Reproduced with permission from ref. 130. Copyright 2015 Springer. (b) Schematic diagram of the MoS2-based FET biosensor. Reproduced with permission from ref. 132. Copyright 2014 American Chemical Society. (c) Schematic illustration of a MoS2 nanopore membrane for DNA translocation. Reproduced with permission from ref. 139. Copyright 2014 American Chemical Society. | ||
Importantly, TMD nanosheets have been demonstrated to present appealing properties for the fabrication of thin nanopores for DNA translocation with a high sensitivity. Nanopore sequencing has been a promising method in DNA sequencing technologies over the past two decades.134 During the sensing process, negatively charged ssDNA or dsDNA is electrophoretically driven to pass through the nanopore, resulting in a temporary blockade with different degrees according to the four DNA nucleotides.135 By observing and analyzing the current change, DNA sequencing can be realized. The first use of nanopore sequencing was based on a biological pore.136 However, biopores are unstable, often too thick to perform sequencing with a single-base resolution, and a critical conducting environment is required.137,138 As a result, the solid-state nanopore with sub-nanometer thickness becomes more important and attractive. As a typical 2D TMD nanomaterial, the MoS2 nanosheet has been used as a promising material to fabricate nanopores for DNA sequencing.139–141 As a typical example, a single- or few-layered MoS2 nanosheet was suspended on a 20 nm thick SiNx membrane which was pre-etched to form a square-shaped opening (Fig. 4c). The MoS2 nanosheet was drilled using a transmission electron microscope to fabricate nanopores ranging between 2 and 20 nm. Compared with graphene, MoS2 has an intrinsic bandgap and shows a higher mechanical stability.142 Moreover, unlike graphene143,144 and boron nitride,145 the MoS2 nanopore showed less interaction with DNA molecules due to the presence of hydrophilic Mo sites in the MoS2 membrane.146 As a result, no surface treatments are required to avoid DNA sticking to the surface of the MoS2 nanopore. This MoS2 nanopore technique improved the signal-to-noise ratio and showed a promising application in DNA sequencing at a single-base level. The simulation result was also consistent with the experimental result.146 Therefore, TMD nanosheets hold great promise in the detection of DNA sequencing in the single-base resolution.
4. Conclusion and prospects
Taking advantages of their unique physical and chemical properties, such as a distinctive layered structure, high surface area, and variation of chemical compositions, 2D TMD nanosheets can be used for a great variety of applications, especially the diverse sensing platforms for biological detection. In this mini-review, we introduced the state-of-the-art biosensors based on 2D TMD nanomaterials, including optical, electrochemical, and electronic biosensors for detecting various targets, such as nucleic acid, proteins, and small molecules. We believe that this research field will continuously grow due to the splendid merits of TMD nanomaterials and their synergistic compatibility with other nanomaterials and advanced nanotechnologies.Although great efforts have been made, there are still lots of challenges in the exploration of 2D TMD nanosheets for biosensing applications. First of all, simple and efficient synthetic methods still need to be established for the preparation of 2D TMD nanomaterials. In spite of the currently available approaches, a method, which can control the number of layers, size, defects, and morphology of the synthesized 2D TMD nanosheets with low cost and good reproducibility, still needs to be developed since all the aforementioned parameters greatly affect the sensing performance. Second, most studies focused on MoS2 and WS2 for biosensing applications, while other TMD nanosheets, e.g., MoSe2, WSe2 and ReSe2, have been ignored even though they might show potential to serve as sensing platforms. In the future, more efforts should be devoted to study other kinds of 2D TMD nanosheets. Third, the mechanism of electron transfer between 2D TMD nanosheets and biomaterials should be investigated completely. Better understanding of the interaction between TMD nanosheets and various substances can offer more opportunities to explore sensing applications beyond the current target libraries and to improve the sensitivity beyond the current detection limit. Additionally, the stability, biocompatibility, and long-term toxicity of 2D TMD nanosheets need to be studied in depth. Due to the complex physiological environment, conducting their in vivo biosensing application is still a challenge. Combining the advancement of recently developed technologies, we believe that 2D TMD nanosheets hold great promise for single molecule detection. Moreover, inspired by the 2D TMD nanomaterials, many other kinds of 2D nanomaterials have been synthesized, such as graphitic carbon nitride (g-C3N4), graphdiyne, 2D polymer and 2D metal–organic frameworks (MOFs).147–151 Our group has made an attempt to use 2D MOF nanosheets as sensing platforms for the detection of DNA and H2O2.149,152 Based on the aforementioned description, it is anticipated that beyond the 2D TMD nanomaterials, more 2D nanomaterial-based biosensors will be fabricated in the future, which might have applications in diagnosis, therapy and other biological studies.
Acknowledgements
This work was supported by MOE under AcRF Tier 2 (ARC 26/13, No. MOE2013-T2-1-034; ARC 19/15, No. MOE2014-T2-2-093; MOE2015-T2-2-057) and AcRF Tier 1 (RG5/13), NTU under a Start-Up Grant (M4081296.070.500000) and an iFood Research Grant (M4081458.070.500000), and the Singapore Millennium Foundation in Singapore. It was also supported by the Joint Research Fund for Overseas Chinese, Hong Kong and Macao Scholars (Grant No. 51528201), the National Natural Science Foundation of China (Grant No. GZ213054, 51322202) and the Natural Science Foundation of Jiangsu Province in China (Grant No. BK20130927). H. X. thanks the financial support from the National Natural Science Foundation of China (Grant No. GZ213054, 51322202) and the Natural Science Foundation of Jiangsu Province in China (Grant No. BK20130927).References
- D. Sidransky, Nat. Rev. Cancer, 2002, 2, 210–219 Search PubMed.
- G. Sozzi, D. Conte, M. Leon, R. Ciricione, L. Roz, C. Ratcliffe, E. Roz, N. Cirenei, M. Bellomi, G. Pelosi, M. A. Pierotti and U. Pastorino, J. Clin. Oncol., 2003, 21, 3902–3908 Search PubMed.
- M. Peifer, L. Fernandez-Cuesta, M. L. Sos, J. George, D. Seidel, L. H. Kasper, D. Plenker, F. Leenders, R. Sun, T. Zander, R. Menon, M. Koker, I. Dahmen, C. Muller, V. Di Cerbo, H. U. Schildhaus, J. Altmuller, I. Baessmann, C. Becker, B. de Wilde, J. Vandesompele, D. Bohm, S. Ansen, F. Gabler, I. Wilkening, S. Heynck, J. M. Heuckmann, X. Lu, S. L. Carter, K. Cibulskis, S. Banerji, G. Getz, K. S. Park, D. Rauh, C. Grutter, M. Fischer, L. Pasqualucci, G. Wright, Z. Wainer, P. Russell, I. Petersen, Y. Chen, E. Stoelben, C. Ludwig, P. Schnabel, H. Hoffmann, T. Muley, M. Brockmann, W. Engel-Riedel, L. A. Muscarella, V. M. Fazio, H. Groen, W. Timens, H. Sietsma, E. Thunnissen, E. Smit, D. A. Heideman, P. J. Snijders, F. Cappuzzo, C. Ligorio, S. Damiani, J. Field, S. Solberg, O. T. Brustugun, M. Lund-Iversen, J. Sanger, J. H. Clement, A. Soltermann, H. Moch, W. Weder, B. Solomon, J. C. Soria, P. Validire, B. Besse, E. Brambilla, C. Brambilla, S. Lantuejoul, P. Lorimier, P. M. Schneider, M. Hallek, W. Pao, M. Meyerson, J. Sage, J. Shendure, R. Schneider, R. Buttner, J. Wolf, P. Nurnberg, S. Perner, L. C. Heukamp, P. K. Brindle, S. Haas and R. K. Thomas, Nat. Genet., 2012, 44, 1104–1110 Search PubMed.
- M. Holzinger, A. Le Goff and S. Cosnier, Front. Chem., 2014, 2, 63 Search PubMed.
- J. Lei and H. Ju, Chem. Soc. Rev., 2012, 41, 2122–2134 Search PubMed.
- X. Bin, E. H. Sargent and S. O. Kelley, Anal. Chem., 2010, 82, 5928–5931 Search PubMed.
- L. Wu, E. H. Xiong, X. Zhang, X. H. Zhang and J. H. Chen, Nano Today, 2014, 9, 197–211 Search PubMed.
- X. Huang, Z. Zeng and H. Zhang, Chem. Soc. Rev., 2013, 42, 1934–1946 Search PubMed.
- M. J. Allen, V. C. Tung and R. B. Kaner, Chem. Rev., 2009, 110, 132–145 Search PubMed.
- D. Chen, L. Tang and J. Li, Chem. Soc. Rev., 2010, 39, 3157–3180 Search PubMed.
- A. Chen and S. Chatterjee, Chem. Soc. Rev., 2013, 42, 5425–5438 Search PubMed.
- A. K. Geim and K. S. Novoselov, Nat. Mater., 2007, 6, 183–191 Search PubMed.
- Y. Liu, X. Dong and P. Chen, Chem. Soc. Rev., 2012, 41, 2283–2307 Search PubMed.
- K. S. Novoselov, A. K. Geim, S. V. Morozov, D. Jiang, Y. Zhang, S. V. Dubonos, I. V. Grigorieva and A. A. Firsov, Science, 2004, 306, 666–669 Search PubMed.
- S. J. He, B. Song, D. Li, C. F. Zhu, W. P. Qi, Y. Q. Wen, L. H. Wang, S. P. Song, H. P. Fang and C. H. Fan, Adv. Funct. Mater., 2010, 20, 453–459 CrossRef CAS.
- P. J. Huang and J. Liu, Small, 2012, 8, 977–983 Search PubMed.
- J. Li, Y. Huang, D. Wang, B. Song, Z. Li, S. Song, L. Wang, B. Jiang, X. Zhao, J. Yan, R. Liu, D. He and C. Fan, Chem. Commun., 2013, 49, 3125–3127 Search PubMed.
- D. Bitounis, H. Ali-Boucetta, B. H. Hong, D. H. Min and K. Kostarelos, Adv. Mater., 2013, 25, 2258–2268 Search PubMed.
- J. Liu, C. Wang, Y. Jiang, Y. Hu, J. Li, S. Yang, Y. Li, R. Yang, W. Tan and C. Z. Huang, Anal. Chem., 2013, 85, 1424–1430 CrossRef CAS PubMed.
- M. Chhowalla, H. S. Shin, G. Eda, L. J. Li, K. P. Loh and H. Zhang, Nat. Chem., 2013, 5, 263–275 Search PubMed.
- V. Nicolosi, M. Chhowalla, M. G. Kanatzidis, M. S. Strano and J. N. Coleman, Science, 2013, 340, 1226419 Search PubMed.
- Y. Sun, S. Gao and Y. Xie, Chem. Soc. Rev., 2014, 43, 530–546 Search PubMed.
- X. Cao, C. Tan, X. Zhang, W. Zhao and H. Zhang, Adv. Mater., 2016, 28, 6167–6196 Search PubMed.
- C. Tan and H. Zhang, Nat. Commun., 2015, 6, 7873 Search PubMed.
- C. Tan and H. Zhang, J. Am. Chem. Soc., 2015, 137, 12162–12174 Search PubMed.
- J. Zhao, X. Jin, M. Vdovenko, L. Zhang, I. Y. Sakharov and S. Zhao, Chem. Commun., 2015, 51, 11092–11095 Search PubMed.
- C. Tan, Z. Liu, W. Huang and H. Zhang, Chem. Soc. Rev., 2015, 44, 2615–2628 Search PubMed.
- C. Tan and H. Zhang, Chem. Soc. Rev., 2015, 44, 2713–2731 Search PubMed.
- X. Zhang, Z. Lai, C. Tan and H. Zhang, Angew. Chem., Int. Ed., 2016, 55, 8816–8838 Search PubMed.
- Y. H. Lee, X. Q. Zhang, W. J. Zhang, M. T. Chang, C. T. Lin, K. D. Chang, Y. C. Yu, J. T. W. Wang, C. S. Chang, L. J. Li and T. W. Lin, Adv. Mater., 2012, 24, 2320–2325 Search PubMed.
- K. Chang and W. X. Chen, ACS Nano, 2011, 5, 4720–4728 Search PubMed.
- Z. Yin, H. Li, H. Li, L. Jiang, Y. Shi, Y. Sun, G. Lu, Q. Zhang, X. Chen and H. Zhang, ACS Nano, 2012, 6, 74–80 Search PubMed.
- P. Joensen, R. Frindt and S. R. Morrison, Mater. Res. Bull., 1986, 21, 457–461 Search PubMed.
- K. G. Zhou, N. N. Mao, H. X. Wang, Y. Peng and H. L. Zhang, Angew. Chem., Int. Ed., 2011, 50, 10839–10842 Search PubMed.
- J. N. Coleman, M. Lotya, A. O'Neill, S. D. Bergin, P. J. King, U. Khan, K. Young, A. Gaucher, S. De, R. J. Smith, I. V. Shvets, S. K. Arora, G. Stanton, H. Y. Kim, K. Lee, G. T. Kim, G. S. Duesberg, T. Hallam, J. J. Boland, J. J. Wang, J. F. Donegan, J. C. Grunlan, G. Moriarty, A. Shmeliov, R. J. Nicholls, J. M. Perkins, E. M. Grieveson, K. Theuwissen, D. W. McComb, P. D. Nellist and V. Nicolosi, Science, 2011, 331, 568–571 Search PubMed.
- Z. Y. Zeng, Z. Y. Yin, X. Huang, H. Li, Q. Y. He, G. Lu, F. Boey and H. Zhang, Angew. Chem., Int. Ed., 2011, 50, 11093–11097 Search PubMed.
- A. H. Loo, A. Bonanni, Z. Sofer and M. Pumera, Electrochem. Commun., 2015, 50, 39–42 Search PubMed.
- Q. Lu, Y. Yu, Q. Ma, B. Chen and H. Zhang, Adv. Mater., 2016, 28, 1917–1933 Search PubMed.
- R. Lv, J. A. Robinson, R. E. Schaak, D. Sun, Y. Sun, T. E. Mallouk and M. Terrones, Acc. Chem. Res., 2015, 48, 56–64 Search PubMed.
- Y. H. Kim, K. Y. Kim, Y. R. Choi, Y.-S. Shim, J.-M. Jeon, J.-H. Lee, S. Y. Kim, S. Han and H. W. Jang, J. Mater. Chem. A, 2016, 4, 6070–6076 Search PubMed.
- X. Huang, Z. Zeng, S. Bao, M. Wang, X. Qi, Z. Fan and H. Zhang, Nat. Commun., 2013, 4, 1444 Search PubMed.
- Y. Chen, C. Tan, H. Zhang and L. Wang, Chem. Soc. Rev., 2015, 44, 2681–2701 Search PubMed.
- N. J. Doran, Physica B+C, 1980, 99, 227–237 Search PubMed.
- A. D. Yoffe, Solid State Ionics, 1990, 39, 1–7 Search PubMed.
- Q. H. Wang, K. Kalantar-Zadeh, A. Kis, J. N. Coleman and M. S. Strano, Nat. Nanotechnol., 2012, 7, 699–712 Search PubMed.
- K. F. Mak, C. Lee, J. Hone, J. Shan and T. F. Heinz, Phys. Rev. Lett., 2010, 105, 136805 Search PubMed.
- A. N. Enyashin, L. Yadgarov, L. Houben, I. Popov, M. Weidenbach, R. Tenne, M. Bar-Sadan and G. Seifert, J. Phys. Chem. C, 2011, 115, 24586–24591 Search PubMed.
- D. J. Late, C. S. Rout, D. Chakravarty and S. Ratha, Can. Chem. Trans., 2015, 3, 118–157 Search PubMed.
- M. A. Py and R. R. Haering, Can. J. Phys., 1983, 61, 76–84 Search PubMed.
- C. Tan, W. Zhao, A. Chaturvedi, Z. Fei, Z. Zeng, J. Chen, Y. Huang, P. Ercius, Z. Luo, X. Qi, B. Chen, Z. Lai, B. Li, X. Zhang, J. Yang, Y. Zong, C. Jin, H. Zheng, C. Kloc and H. Zhang, Small, 2016, 12, 1866–1874 Search PubMed.
- H. Wang, Z. Lu, D. Kong, J. Sun, T. M. Hymel and Y. Cui, ACS Nano, 2014, 8, 4940–4947 Search PubMed.
- X. F. Wang, X. Shen, Z. X. Wang, R. C. Yu and L. Q. Chen, ACS Nano, 2014, 8, 11394–11400 Search PubMed.
- F. Wypych and R. Schollhorn, J. Chem. Soc., Chem. Commun., 1992, 1386–1388 Search PubMed.
- H. Li, Z. Yin, Q. He, H. Li, X. Huang, G. Lu, D. W. Fam, A. I. Tok, Q. Zhang and H. Zhang, Small, 2012, 8, 63–67 Search PubMed.
- G. Eda, T. Fujita, H. Yamaguchi, D. Voiry, M. Chen and M. Chhowalla, ACS Nano, 2012, 6, 7311–7317 Search PubMed.
- M. Acerce, D. Voiry and M. Chhowalla, Nat. Nanotechnol., 2015, 10, 313–318 Search PubMed.
- Y. Yang, T. Liu, L. Cheng, G. Song, Z. Liu and M. Chen, ACS Appl. Mater. Interfaces, 2015, 7, 7526–7533 Search PubMed.
- K. Mao, Z. Wu, Y. Chen, X. Zhou, A. Shen and J. Hu, Talanta, 2015, 132, 658–663 Search PubMed.
- C. Zhu, Z. Zeng, H. Li, F. Li, C. Fan and H. Zhang, J. Am. Chem. Soc., 2013, 135, 5998–6001 Search PubMed.
- P. G. Moses, J. J. Mortensen, B. I. Lundqvist and J. K. Norskov, J. Chem. Phys., 2009, 130, 104709 Search PubMed.
- M. Bucci, Nat. Chem. Biol., 2014, 10, 240 Search PubMed.
- X. P. He and H. Tian, Small, 2016, 12, 144–160 Search PubMed.
- D. Chimene, D. L. Alge and A. K. Gaharwar, Adv. Mater., 2015, 27, 7261–7284 Search PubMed.
- G. Yang, C. Zhu, D. Du, J. Zhu and Y. Lin, Nanoscale, 2015, 7, 14217–14231 RSC.
- X. Liu, F. Wang, R. Aizen, O. Yehezkeli and I. Willner, J. Am. Chem. Soc., 2013, 135, 11832–11839 Search PubMed.
- P. M. Sabale, J. T. George and S. G. Srivatsan, Nanoscale, 2014, 6, 10460–10469 Search PubMed.
- Y. Zhang, B. Zheng, C. Zhu, X. Zhang, C. Tan, H. Li, B. Chen, J. Yang, J. Chen, Y. Huang, L. Wang and H. Zhang, Adv. Mater., 2015, 27, 935–939 Search PubMed.
- C. Tan, P. Yu, Y. Hu, J. Chen, Y. Huang, Y. Cai, Z. Luo, B. Li, Q. Lu, L. Wang, Z. Liu and H. Zhang, J. Am. Chem. Soc., 2015, 137, 10430–10436 Search PubMed.
- A. H. Loo, A. Bonanni and M. Pumera, Analyst, 2016, 141, 4654–4658 Search PubMed.
- Y. Huang, Y. Shi, H. Y. Yang and Y. Ai, Nanoscale, 2015, 7, 2245–2249 Search PubMed.
- S. Wang, Y. Zhang, Y. Ning and G. J. Zhang, Analyst, 2015, 140, 434–439 Search PubMed.
- Q. Xi, D. M. Zhou, Y. Y. Kan, J. Ge, Z. K. Wu, R. Q. Yu and J. H. Jiang, Anal. Chem., 2014, 86, 1361–1365 Search PubMed.
- D. H. Bunka and P. G. Stockley, Nat. Rev. Microbiol., 2006, 4, 588–596 Search PubMed.
- J. Ge, E.-C. Ou, R.-Q. Yu and X. Chu, J. Mater. Chem. B, 2014, 2, 625–628 Search PubMed.
- C. H. Lu, H. H. Yang, C. L. Zhu, X. Chen and G. N. Chen, Angew. Chem., Int. Ed., 2009, 48, 4785–4787 Search PubMed.
- Y. Yuan, R. Li and Z. Liu, Anal. Chem., 2014, 86, 3610–3615 Search PubMed.
- R. M. Kong, L. Ding, Z. Wang, J. You and F. Qu, Anal. Bioanal. Chem., 2015, 407, 369–377 Search PubMed.
- X. Yin, J. Cai, H. Feng, Z. Wu, J. Zou and Q. Cai, New J. Chem., 2015, 39, 1892–1898 Search PubMed.
- X. Xiang, J. Shi, F. Huang, M. Zheng, Q. Deng and J. Xu, Biosens. Bioelectron., 2015, 74, 227–232 Search PubMed.
- J. Ge, L. J. Tang, Q. Xi, X. P. Li, R. Q. Yu, J. H. Jiang and X. Chu, Nanoscale, 2014, 6, 6866–6872 Search PubMed.
- X. Liu, J. Ge, X. Wang, Z. Wu, G. Shen and R. Yu, Anal. Methods, 2014, 6, 7212–7217 Search PubMed.
- S. Cai, Q. Han, C. Qi, Z. Lian, X. Jia, R. Yang and C. Wang, Nanoscale, 2016, 8, 3685–3693 Search PubMed.
- T. Lin, L. Zhong, Z. Song, L. Guo, H. Wu, Q. Guo, Y. Chen, F. Fu and G. Chen, Biosens. Bioelectron., 2014, 62, 302–307 Search PubMed.
- T. Lin, L. Zhong, L. Guo, F. Fu and G. Chen, Nanoscale, 2014, 6, 11856–11862 Search PubMed.
- S. M. Ahmed and H. Gerischer, Electrochim. Acta, 1979, 24, 705–711 Search PubMed.
- K. Saha, S. S. Agasti, C. Kim, X. Li and V. M. Rotello, Chem. Rev., 2012, 112, 2739–2779 Search PubMed.
- J. Ju and W. Chen, Anal. Chem., 2015, 87, 1903–1910 Search PubMed.
- T. Yang, L. Meng, H. Chen, S. Luo, W. Li and K. Jiao, Adv. Mater. Interfaces, 2016, 3, 1500700 Search PubMed.
- Y. Chu, B. Cai, Y. Ma, M. Zhao, Z. Ye and J. Huang, RSC Adv., 2016, 6, 22673–22678 Search PubMed.
- Q. Wang, Y. Zhang, W. Ye and C. Wang, J. Solid State Electrochem., 2015, 20, 133–142 Search PubMed.
- Z. Xing, X. Yang, A. M. Asiri and X. Sun, ACS Appl. Mater. Interfaces, 2016, 8, 14521–14526 Search PubMed.
- A. H. Loo, A. Bonanni, A. Ambrosi and M. Pumera, Nanoscale, 2014, 6, 11971–11975 Search PubMed.
- T. Wang, R. Zhu, J. Zhuo, Z. Zhu, Y. Shao and M. Li, Anal. Chem., 2014, 86, 12064–12069 Search PubMed.
- X. Wang, F. Nan, J. Zhao, T. Yang, T. Ge and K. Jiao, Biosens. Bioelectron., 2015, 64, 386–391 Search PubMed.
- K. J. Huang, Y. J. Liu, H. B. Wang, Y. Y. Wang and Y. M. Liu, Biosens. Bioelectron., 2014, 55, 195–202 Search PubMed.
- T. Yang, M. Chen, Q. Kong, X. Luo and K. Jiao, Biosens. Bioelectron., 2016 DOI:10.1016/j.bios.2016.03.025.
- X. Cao, Microchim. Acta, 2014, 181, 1133–1141 Search PubMed.
- K.-J. Huang, Y.-J. Liu, H.-B. Wang, T. Gan, Y.-M. Liu and L.-L. Wang, Sens. Actuators, B, 2014, 191, 828–836 Search PubMed.
- H. L. Shuai, K. J. Huang and Y. X. Chen, J. Mater. Chem. B, 2016, 4, 1186–1196 Search PubMed.
- X. Wang, C. Chu, L. Shen, W. Deng, M. Yan, S. Ge, J. Yu and X. Song, Sens. Actuators, B, 2015, 206, 30–36 Search PubMed.
- S. Su, M. Zou, H. Zhao, C. Yuan, Y. Xu, C. Zhang, L. Wang, C. Fan and L. Wang, Nanoscale, 2015, 7, 19129–19135 Search PubMed.
- P. Jing, H. Yi, S. Xue, Y. Chai, R. Yuan and W. Xu, Anal. Chim. Acta, 2015, 853, 234–241 Search PubMed.
- K.-J. Huang, Y.-J. Liu, J.-T. Cao and H.-B. Wang, RSC Adv., 2014, 4, 36742–36748 Search PubMed.
- S. Su, H. Sun, W. Cao, J. Chao, H. Peng, X. Zuo, L. Yuwen, C. Fan and L. Wang, ACS Appl. Mater. Interfaces, 2016, 8, 6826–6833 Search PubMed.
- S. X. Wu, Z. Y. Zeng, Q. Y. He, Z. J. Wang, S. J. Wang, Y. P. Du, Z. Y. Yin, X. P. Sun, W. Chen and H. Zhang, Small, 2012, 8, 2264–2270 Search PubMed.
- G. X. Wang, W. J. Bao, J. Wang, Q. Q. Lu and X. H. Xia, Electrochem. Commun., 2013, 35, 146–148 Search PubMed.
- S. Su, J. Chao, D. Pan, L. H. Wang and C. H. Fan, Electroanalysis, 2015, 27, 1062–1072 Search PubMed.
- P. Bollella, G. Fusco, C. Tortolini, G. Sanzo, G. Favero, L. Gorton and R. Antiochia, Biosens. Bioelectron., 2016 DOI:10.1016/j.bios.2016.03.068.
- H. F. Sun, J. Chao, X. L. Zuo, S. Su, X. F. Liu, L. H. Yuwen, C. H. Fan and L. H. Wang, RSC Adv., 2014, 4, 27625–27629 Search PubMed.
- K. J. Huang, Y. J. Liu, Y. M. Liu and L. L. Wang, J. Hazard. Mater., 2014, 276, 207–215 Search PubMed.
- K.-J. Huang, L. Wang, J. Li and Y.-M. Liu, Sens. Actuators, B, 2013, 178, 671–677 Search PubMed.
- T. Yang, R. Yang, H. Chen, F. Nan, T. Ge and K. Jiao, ACS Appl. Mater. Interfaces, 2015, 7, 2867–2872 CAS.
- T. Wang, H. Zhu, J. Zhuo, Z. Zhu, P. Papakonstantinou, G. Lubarsky, J. Lin and M. Li, Anal. Chem., 2013, 85, 10289–10295 Search PubMed.
- H. Song, Y. Ni and S. Kokot, Biosens. Bioelectron., 2014, 56, 137–143 Search PubMed.
- L. Zhu, Y. Zhang, P. Xu, W. Wen, X. Li and J. Xu, Biosens. Bioelectron., 2016, 80, 601–606 Search PubMed.
- X. Lin, Y. Ni and S. Kokot, Biosens. Bioelectron., 2016, 79, 685–692 Search PubMed.
- S. S. Ji, Z. Yang, C. Zhang, Y. E. Miao, W. W. Tjiu, J. S. Pan and T. X. Liu, Microchim. Acta, 2013, 180, 1127–1134 Search PubMed.
- J. W. Huang, Y. Q. He, J. Jin, Y. R. Li, Z. P. Dong and R. Li, Electrochim. Acta, 2014, 136, 41–46 CrossRef CAS.
- S. Su, H. F. Sun, F. Xu, L. H. Yuwen, C. H. Fan and L. H. Wang, Microchim. Acta, 2014, 181, 1497–1503 Search PubMed.
- K. J. Huang, J. Z. Zhang, Y. J. Liu and L. L. Wang, Sens. Actuators, B, 2014, 194, 303–310 Search PubMed.
- S. Su, H. F. Sun, F. Xu, L. H. Yuwen and L. H. Wang, Electroanalysis, 2013, 25, 2523–2529 Search PubMed.
- K. J. Huang, Y. J. Liu, J. Z. Zhang and Y. M. Liu, Anal. Methods, 2014, 6, 8011–8017 Search PubMed.
- X. J. Zhao, X. H. Xia, S. J. Yu and C. M. Wang, Anal. Methods, 2014, 6, 9375–9382 Search PubMed.
- K. J. Huang, L. Wang, Y. J. Liu, T. Gan, Y. M. Liu, L. L. Wang and Y. Fan, Electrochim. Acta, 2013, 107, 379–387 Search PubMed.
- X. Xia, Z. Zheng, Y. Zhang, X. Zhao and C. Wang, Sens. Actuators, B, 2014, 192, 42–50 Search PubMed.
- R. Yang, J. Zhao, M. Chen, T. Yang, S. Luo and K. Jiao, Talanta, 2015, 131, 619–623 Search PubMed.
- Y. Zhang, P. Chen, F. Wen, B. Yuan and H. Wang, J. Electroanal. Chem., 2016, 761, 14–20 Search PubMed.
- X. Lin, Y. Ni and S. Kokot, Sens. Actuators, B, 2016, 233, 100–106 Search PubMed.
- B. Radisavljevic, A. Radenovic, J. Brivio, V. Giacometti and A. Kis, Nat. Nanotechnol., 2011, 6, 147–150 Search PubMed.
- D. W. Lee, J. Lee, I. Y. Sohn, B. Y. Kim, Y. M. Son, H. Bark, J. Jung, M. Choi, T. H. Kim, C. Lee and N. E. Lee, Nano Res., 2015, 8, 2340–2350 Search PubMed.
- P. T. Loan, W. Zhang, C. T. Lin, K. H. Wei, L. J. Li and C. H. Chen, Adv. Mater., 2014, 26, 4838–4844 Search PubMed.
- D. Sarkar, W. Liu, X. Xie, A. C. Anselmo, S. Mitragotri and K. Banerjee, ACS Nano, 2014, 8, 3992–4003 Search PubMed.
- L. Wang, Y. Wang, J. I. Wong, T. Palacios, J. Kong and H. Y. Yang, Small, 2014, 10, 1101–1105 Search PubMed.
- L. J. Steinbock and A. Radenovic, Nanotechnology, 2015, 26, 074003 Search PubMed.
- M. Fyta, J. Phys.: Condens. Matter, 2015, 27, 273101 Search PubMed.
- J. J. Kasianowicz, E. Brandin, D. Branton and D. W. Deamer, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 13770–13773 Search PubMed.
- D. Branton, D. W. Deamer, A. Marziali, H. Bayley, S. A. Benner, T. Butler, M. Di Ventra, S. Garaj, A. Hibbs, X. Huang, S. B. Jovanovich, P. S. Krstic, S. Lindsay, X. S. Ling, C. H. Mastrangelo, A. Meller, J. S. Oliver, Y. V. Pershin, J. M. Ramsey, R. Riehn, G. V. Soni, V. Tabard-Cossa, M. Wanunu, M. Wiggin and J. A. Schloss, Nat. Biotechnol., 2008, 26, 1146–1153 Search PubMed.
- S. M. Iqbal, D. Akin and R. Bashir, Nat. Nanotechnol., 2007, 2, 243–248 Search PubMed.
- K. Liu, J. Feng, A. Kis and A. Radenovic, ACS Nano, 2014, 8, 2504–2511 Search PubMed.
- J. Feng, K. Liu, M. Graf, M. Lihter, R. D. Bulushev, D. Dumcenco, D. T. Alexander, D. Krasnozhon, T. Vuletic, A. Kis and A. Radenovic, Nano Lett., 2015, 15, 3431–3438 Search PubMed.
- J. Feng, K. Liu, R. D. Bulushev, S. Khlybov, D. Dumcenco, A. Kis and A. Radenovic, Nat. Nanotechnol., 2015, 10, 1070–1076 Search PubMed.
- H. Arjmandi-Tash, L. A. Belyaeva and G. F. Schneider, Chem. Soc. Rev., 2016, 45, 476–493 RSC.
- C. Sathe, X. Q. Zou, J. P. Leburton and K. Schulten, ACS Nano, 2011, 5, 8842–8851 CrossRef CAS PubMed.
- Z. S. Siwy and M. Davenport, Nat. Nanotechnol., 2010, 5, 697–698 Search PubMed.
- Z. Zhou, Y. Hu, H. Wang, Z. Xu, W. Wang, X. Bai, X. Shan and X. Lu, Sci. Rep., 2013, 3, 3287 Search PubMed.
- A. B. Farimani, K. Min and N. R. Aluru, ACS Nano, 2014, 8, 7914–7922 Search PubMed.
- C. Wang, P. Yu, S. Guo, L. Mao, H. Liu and Y. Li, Chem. Commun., 2016, 52, 5629–5632 Search PubMed.
- T. Rodenas, I. Luz, G. Prieto, B. Seoane, H. Miro, A. Corma, F. Kapteijn, F. X. Llabres i Xamena and J. Gascon, Nat. Mater., 2015, 14, 48–55 Search PubMed.
- M. Zhao, Y. Wang, Q. Ma, Y. Huang, X. Zhang, J. Ping, Z. Zhang, Q. Lu, Y. Yu, H. Xu, Y. Zhao and H. Zhang, Adv. Mater., 2015, 27, 7372–7378 Search PubMed.
- T. Y. Ma, Y. Tang, S. Dai and S. Z. Qiao, Small, 2014, 10, 2382–2389 Search PubMed.
- P. Kissel, D. J. Murray, W. J. Wulftange, V. J. Catalano and B. T. King, Nat. Chem., 2014, 6, 774–778 Search PubMed.
- Y. Wang, M. Zhao, J. Ping, B. Chen, X. Cao, Y. Huang, C. Tan, Q. Ma, S. Wu, Y. Yu, Q. Lu, J. Chen, W. Zhao, Y. Ying and H. Zhang, Adv. Mater., 2016, 28, 4149–4155 Search PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © the Partner Organisations 2017 |






