Emerging biotransduction strategies on soft interfaces for biosensing
Yuanyuan
Tian
ab,
Guoliang
Xu
a,
Kaiyu
Cai
a,
Xiao
Zhao
a,
Bo
Zhang
ab,
Lianhui
Wang
 *a and
Ting
Wang
*a and
Ting
Wang
 *a
*a
aKey Laboratory for Organic Electronics and Information Displays & Jiangsu Key Laboratory for Biosensors, Institute of Advanced Materials (IAM), National Jiangsu Synergetic Innovation Center for Advanced Materials (SICAM), Nanjing University of Posts and Telecommunications, 9 Wenyuan Road, Nanjing 210023, China. E-mail: iamtingwang@njupt.edu.cn; iamlhwang@njupt.edu.cn
bSchool of Science, Nanjing University of Posts and Telecommunications, Nanjing, 210023, China
First published on 23rd November 2022
Abstract
As a lab-on-soft biochip providing accurate and timely biomarker information, wearable biosensors can satisfy the increasing demand for intelligent e-health services, active disease diagnosis/therapy, and huge bioinformation data. As biomolecules generally could not directly produce detectable signals, biotransducers that specifically convert biomolecules to electrical or optical signals are involved, which determines the pivotal sensing performance including 3S (sensitivity, selectivity, and stability), reversibility, etc. The soft interface poses new requirements for biotransducers, especially equipment-free, facile operation, mechanical tolerance, and high sensing performance. In this review, we discussed the emerging electrochemical and optical biotransduction strategies on wearables from the aspects of the transduction mechanism, amplification strategies, biomaterial selection, and device fabrication procedures. Challenges and perspectives regarding future biotransducers for monitoring trace amounts of biomolecules with high fidelity, sensitivity, and multifunctionality are also discussed. It is expected that through fusion with functional electronics, wearable biosensors can provide possibilities to further decentralize the healthcare system and even build biomolecule-based intelligent cyber-physical systems and new modalities of cyborgs.
1. Introduction
With the prevalence of flexible and mobile electronics, wearable sensing technology has been rapidly developed due to its features including time-saving, low-cost, non-invasion, and real-time/dynamic monitoring.1,2 Soft sensors offer many opportunities to address societal concerns in hospital-centralized public healthcare systems, precision personalized medicine, artificial intelligence, environmental security, and energy management.3 It is estimated that the number of wearable devices in use will reach 1100 million in 2022,4 and wearable technology will save the global healthcare industry more than $200 billion in costs in the next 25 years and significantly reduce the clinician/patient interaction time.5 It is foreseen that wearable device-based health interventions and life-cycle health management will become the new modality of the e-healthcare system, reform our lifestyle and enrich our comprehension of the human body.As an emerging branch of micro-analytical systems, wearable sensing allows the direct sampling and measurement of biosignals, and provides feedback to the user through wireless communication. In early research activities, the wearable sensing platform mainly focused on vital parameters ranging from physical parameters (body temperature, heart rate, respiration rate, blood pressure, and humidity) to electrophysiological signals (electromyography, electroencephalogram, and electrocardiogram).6,7 Recent interest has shifted to clinically relevant chemical and biochemical indicators considering the importance of biomarkers in healthcare,8 such as analytes with high concentrations (e.g. pH, Cl−, lactate, glucose) as well as trace amounts of analytes (e.g. cortisol, dopamine, serotonin).9–11 Real-time biomarker information is expected to provide precise and dynamic information related to normal and abnormal physiological states and in situ treatment guidance in non-clinical situations.12,13
To meet the requirements of conformability, softness, and non-irritating, wearable sensors basically comprise three key components, such as a soft substrate, an active layer, and an encapsulation layer.14,15 The soft substrate is the basic construction of a device. The commonly used materials include flexible polymers (such as polyester, polyethylenimine, and polyimide), stretchable elastomers (polydimethylsiloxane (PDMS), styrene ethylene butylene styrene (SEBS), and thermoplastic polyurethanes (TPU)) and breathable films (paper, fabric).16,17 The encapsulation layer is generally employed to avoid unexpected interference and achieve good stability. The crucial part of the wearable sensor is the active layer that utilizes the functional unit through physical or biochemical interactions for perceiving the biosignals. Note that the interfacial adhesion between the active layer and the soft substrate is a long-term concern in stretchable electronics.18–20 For flexible biosensors, extra attention should also be paid to the adhesion between the biomolecules and the active layer. For physical signal monitoring, the essential feature of the active layer is stimuli-dependent electronic or ionic conductivity, such as piezoelectric or piezoresistive properties for pressure sensing and temperature-dependent ionic conductivity changes.21–23 Many reviews summarize material innovation (e.g. nanomaterials, hydrogels, liquid metals, conductive polymers, composites) and rational structural design (e.g. wave, wrinkle, island-bridge, origami, textile, and crack), which allows for conformability, high electron/hole mobility, low impedance/resistance, high throughput, and satisfactory sensing performance.24–28 For biomolecule detection, as biomolecules could not directly produce detectable signals, biosensors generally need an extra biotransducer to selectively recognize molecules and subsequently produce electrical or optical signals. Hence, it is necessary to summarize efficient biotransduction strategies for better soft biosensors.
Currently, recognition elements in biotransducers commonly include biometric receptors (such as enzymes, antibodies, and nucleic acids) and biomimetic components (such as inorganic nanomaterials and molecularly imprinted polymers (MIPs)). The recognition reaction is then coupled with a physicochemical transduction process to produce electrical or optical signals that are feasible to be quantitively or qualitatively measured.29 Hence, biotransducers determine the type and intensity of the signal output, which greatly influences the pivotal performance of biosensors such as 3S (selectivity, sensitivity, stability) performance, reversibility, and mechanical tolerance. In order to achieve high biosensing performance, traditional biotransducers rely on precise detection equipment (such as conventional optical devices), special signal amplification strategies (such as polymerase chain reaction), and multiple bioreaction steps. However, wearable modality calls for biotransducers that have equipment-free, easy operation and provide quick responses without sacrificing the biosensing performance.30 In addition to the miniaturization requirement, the concentration of biomarkers in biofluids is generally one or two magnitude lower than that of blood which calls for even higher efficient biotransducers. Moreover, a non-ideal biosensing environment with an abundance of interference species,31 mechanical deformation, disturbance of the sample flow or evaporation also poses new challenges to biotransducers.6,32–35
With this in mind, in this review, we mainly focused on efficient biotransducers that are applicable to wearables to address the challenges mentioned above. In short, we summarize currently achievable biorecognition-induced electrochemical and optical signal transduction strategies (Fig. 1), including the transduction mechanism, amplification mechanism (effective biometrics and signal enhancement), biomaterial selection, and device fabrication procedures. Specifically, electrochemical biotransduction mainly focuses on these aspects, including potentiometric sensors, amperometric sensors toward electroactive/electro-inactive species, and electrical transducer-based intelligent integration. For optical biotransduction, this system employs colorimetric and fluorescent sensors based on visible changes due to chelation or redox reactions, as well as SERS sensors with spectrum signals. Finally, the current challenges and development trends of flexible wearable biosensors are discussed regarding stability, sensitivity, and versatility.
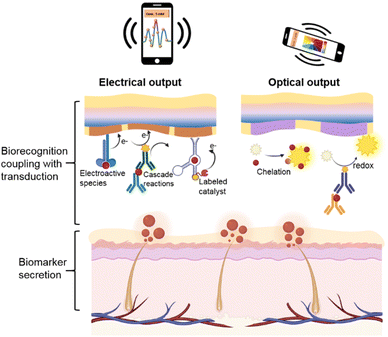 | ||
| Fig. 1 Schematic diagram of the electrical and optical output based on the biorecognition coupling with transduction in the soft biosensing system for real-time biomarker detection. | ||
2. Electrochemical biotransduction strategies
Electrochemical biotransduction is a mature technology that has been used in the past 50 years with the advantages of high sensitivity, high selectivity, cheap, fast responses, and easy operation.36 Generally speaking, when a chemical or biochemical target binds to the electrode by recognition species, electrode potential shifts (potential as the output) or electron transfer occurs at the electrode (current as the output). Due to the diversity of current amplification strategies (such as the catalytic effect), the current (amperometric) techniques are prevalent for the soft biosensing platform.25 It is noted that as electrochemical impedance generally needs extra electrochemical probes such as Fe(CN)63−/Fe(CN)64−, electrochemical impedance transduction is not widely used in soft biosensors.37 Hence, in this section, we briefly discuss the strategies devoted to transducing biomolecules to provide potential and current signals coupled with different recognition processes.2.1 Potentiometric sensors
The mechanism for potentiometric sensors is utilizing the potential of a modified work electrode versus a reference electrode under negligible current for quantitative analysis. Skin-interfaced potentiometric biosensors are widely used for biological fluid detection based on an ion-selective membrane-modified electrode. The specific binding of ions at the electrode produces a potential shift according to the Nernst equation. For example, fully integrated sensor arrays based on the printed circuit board were employed for continuous and non-invasive analysis of pH, K+, Na+, Ca2+, and Cl− in perspiration.38–41In situ detection of ions could realize the continuous monitoring of the human physiological state and the diagnosis of cystic fibrosis (Fig. 2a). In sweat analysis, iontophoresis was introduced to address insufficient sweat secretion under sedentary conditions and realize on-the-spot sweat analysis.40 Such soft sensors can be broadened to other scenarios by incorporating a flexible circuit. For example, a stretchable hybrid electronic system loaded on a dental retainer had been introduced for the real-time detection of sodium intake to manage hypertension.42 Besides the electrochemical sensing mechanism, ion-selective pH sensors with flexible charge-coupled device structures could offer higher sensitivities through multiple accumulation cycles of electron charge transfer.43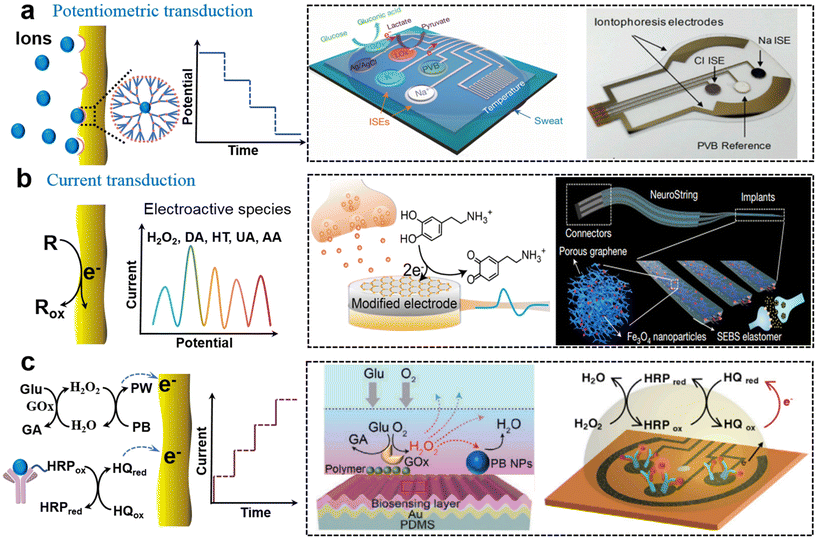 | ||
| Fig. 2 Soft biosensors with the electrical output. (a) Potentiometric sensor based on ionic chelation induced potential changes for analysis of Na+, K+, and Cl−.38,40 This figure has been reproduced from ref. 38 with permission from Spring Nature; copyright: 2016, and ref. 40 with permission from the National Academy of Science; copyright: 2017. (b) Current transduction based on direct redox reaction of electroactive molecules (DA) in the secretion on the electrode.10,46 This figure has been reproduced from ref. 10 with permission from Spring Nature; copyright: 2022, and ref. 46 with permission from Spring Nature; copyright: 2022. (c) Current transduction based on affinity process-induced redox reaction for analysis of Glu52 and cortisol.9 This figure has been reproduced from ref. 52 with permission from the WILEY-VCH Verlag GmbH & Co. KGaA; copyright: 2020, and ref. 9 with permission from Elsevier Inc.; copyright: 2020. | ||
2.2 Amperometric sensors toward electroactive species
In amperometric biosensing, electrochemical redox reactions occur at the functionalized working electrode which produces a faradaic current proportional to the concentration of the analyte. For electroactive species such as H2O2, NO, ascorbic acid (AA), uric acid (UA), dopamine (DA), and serotonin (5-HT), amperometric detection can be directly applied to the electrode based on concentration-dependent oxidation current density. The potential position of a redox peak is used to distinguish different electroactive species. The surface properties of the soft electrode including high conductivity, catalytic properties, and strong interfacial interactions toward targets are beneficial for achieving high sensitivity and selectivity. Commonly used active materials are metals, graphene oxides (GO), carbon nanotubes (CNT), conductive conjugated polymers, metal oxides, and metal–organic frameworks, which can favor electron transfer between species and electrodes.44 For example, Huang's group reported a series of stretchable electrodes for monitoring H2O2 and NO from mechanically deformed cells and tissues by utilizing metal and conjugated polymer nanowires, which allows us to understand the mechanotransduction process.34 Recently, Chen's group reported a flexible dopamine electrochemical sensor using an optimized CNT/GO structure where the conductive CNT network contributes to the high sensitivity and negatively charged GO enhances selectivity towards positively charged DA molecules with the coexistence of other interference species.10Such soft electrochemical biosensors can be further integrated to achieve multiplex metabolite sensing in vivo. For example, Gao's group reported a flexible graphene-based chemical sensor through an entirely CO2 laser-engraved method for the detection of UA and tyrosine (Tyr) and vital-sign monitoring with low concentrations.45 The establishment of such amperometric sensing systems complemented metabolic and nutritional management and dynamic health monitoring and personalized intervention. Moreover, introducing catalytic nanoparticles into stretchable conductive materials can produce stretchable functional electrodes that can work in real-time on soft tissues. Combining the graphene/iron oxide nanocomposites with elastic networks (SEBS), Bao et al. developed a stretchable, neurochemical monitoring electrode for in vivo real-time simultaneous monitoring of DA in the central nervous system and measuring 5-HT dynamics in the gastrointestinal (GI) system, respectively (Fig. 2b).46 Such electrodes could satisfy various motion modes (bending, stretching, and twisting) with soft and twisted structures, and fit the intestinal tract very well, which realizes the monitoring of the dynamics of neurotransmitters in vivo and the enteric nervous system.
2.3 Amperometric sensors toward electro-inactive species
In contrast to electroactive active substances, electrochemical biosensing of inactive species relies on a cascade process where biorecognition is coupled with physiochemical signal transduction. The biorecognition unit can be divided into enzymes (such as glucose oxidase (GOx) and lactate oxidase (LOx)), antibodies, aptamers, and bionic receptors (such as MIP). The physiochemical signal transduction generally exploited biomolecule labels such as horseradish peroxidase (HRP), and artificial mediators such as Prussian blue (PB), nanocatalyst, and hydroquinone (HQ). Soft biosensors based on different biorecognition units are discussed as follows.Soft enzyme biosensors generally involve cascade enzymatic bioreactions. Taking the glucose biosensor as an example, GOx specifically catalyzes the oxidation of glucose, producing H2O2 with the coexistence of O2. Then intermediate H2O2 is further reduced at the electrode with PB as the catalyst (Fig. 2c). For example, Gao et al. fabricated a fully integrated sweat sensor using glucose oxidase and lactate oxidase (GOx and LOx), and PB as a mediator for the selective detection of glucose and lactate, simultaneously.38 Based on a similar cascade bioreaction, a ultrathin (∼3 μm) skin-like oxidase biosensor with high sensitivity (130.4 μA mM−1) had been used for in situ intravascular blood glucose monitoring in the interstitial fluid.47 Developing an efficient catalyst is also effective to further enhance sensitivity. Gold-platinum bimetallic nanocatalysts could rapidly decompose hydrogen peroxide produced by redox reaction of glucose oxidase for amperometric responses in smart contact lenses to achieve long-term and robust glucose monitoring.48 In addition, the charge transfer mediated by nanocatalysts exhibited a fast response time and low detection limit. The glucose-sensing immobilized bimetallic nanocatalyst had high sensitivity and reversibility without hysteresis.49,50
The mechanical tolerance of cascade reaction is another challenge due to the adverse effect of deformation on an intermediate transfer pathway. Therefore, special electrode structures and designs are required to cope with the detecting strain. Recently, a stretchable electrode glucose biosensor based on enokitake mushroom-like standing gold nanowires had shown a sensitivity of 4.55 μA mM−1 cm−2 even for 30% strain.51 In addition to the standing nanostructure, Chen's group investigated a mechanically tolerant biosensor with a wavy microstructure (Fig. 2c).52 They revealed that the biosensing instability mechanism behind is the intermediate pathway that changes under mechanical deformation by simulation. Then, a wavy biosensor with an adaptive curvature was proposed to alleviate this problem. The experiment shows that when in a 50% strain state, the signal fluctuation of the wave-type bioelectrodes for detecting glucose (lactic acid) exhibited 7.0% (4.9%), which was significantly lower than that of the flat electrodes. Such epidermal biosensors via adaptive curvature engineering provided quantitative bio/chemical-transduction for the non-invasive soft healthcare platform.
Soft electrochemical immunosensors fall under an important category where antibody–antigen binding is coupled with the immobilization of biomolecule labels which are capable of producing electrical signals. The high affinity between a natural antibody and an antigen endowed biosensors with strong selectivity and sensitivity, which is very beneficial for the detection of a target with low concentration in biological fluids without extra treatment. With the competitive sensing strategy, Gao et al. showed that a wearable immune biosensor with an integrated portable system could investigate the dynamics of stress-related hormones in real-time with high sensitivity, which would promote the progress of human personalized healthcare and comprehensive mental health management.9 In this cortisol immunosensor, antibody-modified graphene was used as the electrode for competitive binding between HRP-labeled cortisol and sweat cortisol, and the current signal was generated from HQ-mediated enzymatic reduction of hydrogen peroxide (Fig. 2c). Based on the competition mechanism of sandwich configuration, they further reported a multiplexed and portable electrochemical platform on the modified graphene electrodes which aimed at the rapid and sensitive detection and quantification of COVID-19 associated proteins.53 In addition, a bioassembly strategy with a more exquisite molecular architecture had also been applied to these biosensor schemes. Compared with the labeled immunosensors, a sensitive label-free nanobody organic electrochemical transistor biosensor was introduced for the detection of single molecule COVID-19 and Middle East respiratory syndrome coronavirus antigens in 5 μL of untreated samples less than 15 min.54 The protocol of this novel biosensor was designed with a chemical self-assembled monolayer and an oriented bio-SAM architecture with specificity and programmability.
Besides the enzyme and antibody, aptamers are attractive catch probes with artificial programmability. Thanks to their good tunable affinity, aptamer-based amperometric sensors have shown good stability and selectivity in the detection of trace components in complex biological environment samples.55 Recently, wearable electrochemical aptamer biosensing (EAB) had been used to realize a precision dosage of drugs.56 In this EAB system, a redox signal reporter (methylene blue) is coupled with an aptamer which is immobilized on Au NPs. The aptamer-target binding event alters the charge transfer rate between the signal reporter and electrode surface, which can be measured via voltammetry-based approaches.56 In addition, aptamers are often used in the field-effect transistor (FET) system with high sensitivity. The FET sensor is composed of a source, drain, and gate electrode, and any slight change caused by the charged species near the gate electrode can translate into a detectable signal of the FET drain.57 Target-induced conformational changes of the aptamer-FET sensors had been reported to overcome the electrical double layer (the “Debye length” limitation) for small-molecule sensing.58,59 There have been many studies on soft amperometric biosensors for cortisol detection based on aptameric FET, including carbon nanotubes FET,60 platinum/graphene FET,57 indium oxide FETs,61 liquid-ion gated FET,62 a liquid gate graphene FET, etc.63,64 Real-time detection of cytokine in physiologically relevant environments is challenging due to a trace amount of cytokine in biofluids (hundreds fM in sweat) with the coexistence of abundant interference species.65 A flexible aptameric graphene–Nafion biosensor was utilized for cytokine storm biomarker detection in undiluted human sweat.66 The composite material not only prevented the electrode from contacting interferers in the solution but also anchored the aptamer probe, which ensured sensitivity and stability toward wearable application. However, there are still challenges in developing the FET substrate from flexibility to stretchability.
Furthermore, MIP, known as antibody mimics or artificial receptors, is capable of binding target molecules with high specificity and affinity similar to biological receptors.67 The synthesis of MIP generally involves polymerization and elution steps. Firstly, coordination compounds are synthesized using functional monomers and template molecules (usually target molecules) through covalent or non-covalent interactions, in the presence of cross-linkers and porogenic solvent. The MIP precursors were copolymerized into polymer networks in the presence of cross-linking agents and pore-forming solvents. The template is eluted from the polymer host, leaving the template-specific recognition sites. Due to the functional groups of the monomers, the exposed sites can reassociate specifically with the analyte molecule based on both physical and chemical properties.68 Meanwhile, redox-active nanoreporters (such as PB) are also often used cooperatively in this identification system. As a rapidly developing field, tailor-made chemical receptors had been employed in flexible sensor systems.69,70
For amperometric biosensing based on MIP, a sweat wearable biosensor for the human stress hormone cortisol was developed by fabricating a molecularly selective nanoporous membrane as a biorecognition layer and using an electrochemical probe.71 Subsequently, an efficient novel cortisol sensing platform was reported by Wang's group for fast, label-free, and reliable detection of natural perspiration through fingertip touch.72 In this work, the PB nanoparticles were merged in the MIP network as the redox probe to realize sensitive amperometric non-invasive stress detection. The advantage of MIP is providing versatile artificial receptors according to the sensing needs, which can greatly broaden the sensing target spectrum. Recently, a universal wearable biosensor was demonstrated for continuously monitoring all the essential amino acids and vitamins in any state using customized MIP layers, which had not been achieved by antibody-related biorecognition yet.73 Combined with PB as the electron mediator, such electrochemical biosensors also showed high sensitivity and good reversibility. In brief, MIP with unique in situ regeneration advantages acts as a supplement for nature receptors which can further advance precise metabolic profiling and personalized nutritional intervention in wearable modules.
2.4 Electrical transducer-based intelligent integration
Electrical signals can be easily integrated with functional bioelectronics to provide comprehensive healthcare data and timely feedback for multiplex precise diagnosis or intelligent interactions.74 For example, electrochemical metabolite sensing can be further integrated with vital sign monitoring.75 A soft chemical–electrophysiological hybrid biosensing system was reported to real-time monitor a biochemical (lactate) and an electrophysiological signal (electrocardiogram) with negligible cross-talk.76 Such multimodal wearable systems that fuse chemical, electrophysiological and physical sensors allow us to more comprehensively monitor human physiology.Besides multiplex biosensing, another interesting research direction is to provide intelligent feedback based on biosignals such as on-demand drug delivery and active perception for human–machine interaction. One typical example is the integration of enzyme-based amperimetric glucose sensors (GOx) and a thermoresponsive microneedle-based drug delivery system.77 This system concurrently realizes the monitoring of glucose levels and real-time treatment which brings one step closer point-of-care (POC) treatment for patients with diabetes mellitus. Another example is using neurotransmitters, a natural chemical messenger in the human brain, to build a biohybrid interaction interface. Based on the aforementioned DA electrochemical sensors, a chemically mediated artificial neuron is reported where the sensor is connected with a memristor and DA releasor. The memristor conducts the signal processing including threshold and memory function. The DA releasor provides feedback which releases DA molecules to trigger other neurons. In this way, artificial neurons realize a bidirectional communication loop with their biological counterpart (Fig. 2b).10 Such a kind of neuromorphic biosensing provides avenues for constructing a new modality of chemical brain–machine interfaces.
3. Optical biotransduction strategies
Transduction of biomolecule information to optical signals is commonly used in soft sensors where visible changes can be qualitatively evaluated by the naked eye and spectrum signals can be quantitatively detected using portable devices. Specifically, colorimetric/fluorometric reagents and surface-enhanced Raman scattering (SERS) substrates can interact with different target molecules through chelation or redox reactions, resulting in changes in color or spectrum signals. It is noted that optical biotransducers are generally combined with skin-interfaced microfluidic systems for quantitative sample collection to enhance biosensing reliability and fidelity.78,79 In this section, we focused on both the construction of epidermal microfluidics (epifluidics) system and the optical biotransduction strategies of the biosensors.3.1 Colorimetric and fluorescent sensors
The microfluidics system is critical to the epifluidics system and typically requires valves or control systems located outside the device to guild or control the solution flow. Bulky external equipment with high power consumption is incompatible with wearables. To solve this problem, soft epifludics had been developed for automatously sweat collection, which uses the sweat glands themselves as the source of pressure to drive flows instead of using external valves. Rogers's group explored a wearable microfluidic system based on the elastomer for the analysis of electrolyte balance and biomarker concentration, in which sweat flow is autonomously initiated by the sweat glands and collected by independent reservoirs, enabling directional flow in the microfluidic network through the capillary effect.11 The biosensor devices were composed of an adhesive layer with openings, sealed collection channels filled with a color-responsive dye, and an inlet and the corresponding outlet. The harvesting area on the epidermis and the rate of sweat into the reservoir and channel size parameters were key engineering aspects in the quantitative design of this system. They further explored the selection of the elastomer and skin-adhesive materials to cope with specific circumstances such as underwater80 and abnormal status calling for a timely warning.81 For fully understanding time-dependent variation in sweat composition, capillary bursting valves (CVBs) and the geometric design of microfluidic channels were also studied to quantify volumetric sweat rate and loss, and the accuracy of real-time tracking.82–86 Besides the two-dimensional (2D) microfluidic platforms, complex 3D microfluidic architectures had been developed with promise for engineered tissues and artificial organs.87Based on the well-designed wearable microfluidic devices, chelation-induced colorimetric changes of organic dyes enabled in situ quantitative analysis of pH, chloride (Fig. 3a),11,88,89 calcium, zinc, and iron.90 For example, colorimetric detection of chloride could rely on competitive binding between Hg2+ and Fe2+ in the presence of 2,4,6-tris(2-pyridyl)-s-triazine (TPTZ). Once HgCl2 was formed, the bonding of Fe2+ with TPTZ induced a blue change. Chromatism could also be successfully applied in a colorimetric barcode (twenty cross-reactive strips and each strip as a sensor) for scent fingerprint and fingerprint recognition.91 The basic bioamines lead to a splitting of the hydroxyl group of the dye (bromothymol blue) which results in a visible color change of one strip from yellow (protonated form) to blue (deprotonated form). The combination of strips forms target fingerprint information. Taking advantages of deep convolutional neural networks (DCNN) for image recognition, they obtained a rapid (within 30 s), fully integrated, and quantitative meat freshness monitoring platform. In addition to these colorimetric reagents, some conjugated polymers such as polydiacetylene (PDA) could be triggered by the deformation of the backbone structure with unique color transition. For example, a wearable sensor based on PDA/MoS2 film had been used to selectively monitor DMF vapor with a color change from blue to red.92
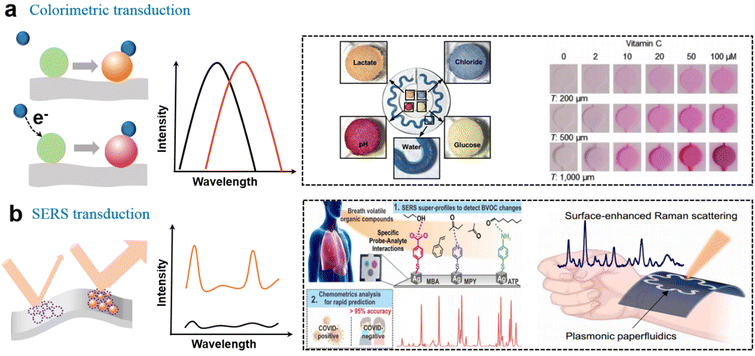 | ||
| Fig. 3 Soft biosensors with the optical output. (a) Colorimetric sensing based on the chelation reaction for the analysis of pH, Cl−, and redox reaction for analysis of glucose lactate11 and vitamin C.90 This figure has been reproduced from ref. 11 with permission from the American Association for the Advancement of Science; copyright: 2016, and ref. 90 with permission from the WILEY-VCH Verlag GmbH & Co. KGaA; copyright: 2022. (b) Overview of the hand-held SERS-based platform for the identification of COVID-19 within 5 min,96 and a SERS–based microfluidics system for continuous sweat analysis.97 This figure has been reproduced from ref. 96 with permission from the American Chemical Society; copyright: 2022, and ref. 97 with permission from the American Association for the Advancement of Science; copyright: 2022. | ||
Moreover, the redox reaction between the analyte and the colorimetric substrate is another effective colorimetric method. For example, vitamin C reduced ferric ion (Fe3+) to ferrous ion (Fe2+) utilizing a ferric reducing/antioxidant and ascorbic acid system, resulting in the naked-eye visible change (Fig. 3a).90 Utilizing enzymatic cascade redox reactions, researchers also developed colorimetric metabolite sensors. Typically, glucose could be oxidized by GOx to produce hydrogen peroxide, which oxidized iodide to yield a change in color from yellow to brown.11,88 Lactate could be reduced by lactate dehydrogenase (LDH) in the presence of a coenzyme (NAD or NADP), which further induces the reduction of chromogenic reagent to a yellow product (formazan dyes).
In addition to the aforementioned visible light colorimetric detection, fluorescence-based sweat analysis could serve as a supplementary approach.93 The fluorometric sensing modalities integrated with an available microfluidic system were employed for the in situ measurement of chloride, sodium and zinc in sweat via smartphone-based imaging. Probes that react with specific ions were prefilled in the microfluidic micro reservoir, and the fluorescence intensity changes generated by the reaction of the probes were measured for quantitative analysis. Moreover, fluorometric sensing can also be combined with colorimetric and electrochemical analyses for tracking metabolites in sweat.94 For example, in order to investigate potential relevance in physical and mental stresses, multimodal lateral flow competition immunoassays are reported which allow cortisol detection, fluorescence-based enzymatic assays for glucose and vitamin C, and measurement of galvanic skin responses. Moreover, an epidermal UV colorimetric sensor based on microfluidic systems had also been used for the accurate measurement of UV exposure levels and skin temperature.95
3.2 SERS sensors
It is well known that SERS technology has been widely used in biosensors with high sensitivity, non-invasion, label-free and providing abundant fingerprint information.98 Due to the local electromagnetic enhancement effect, Raman signals can be amplified by several orders of magnitude from the plasmonic coupling of hot spots, thus realizing SERS sensing for single-molecule detection.99 As the fundamental sensing component, the SERS active substrate with hot spots had been developed from rigid to flexible.100 Recently, flexible SERS sensors had attracted numerous attention due to their tunable SERS substrate, sampling strategy, and in situ detection for POC diagnostics.101 With the unique advantages of low cost, rapidity, mass production, and integrability, SESR technology as a powerful tool has been pushed into the ranks of next-generation wearable sensors for POC diagnosis and home-care medicine.The tunability of the SERS substrate facilitates the development of versatile flexible SERS sensors. The soft sensing platforms were developed for pH analysis based on the SERS-active substrate which was prepared by electrospinning of TPU nanofibers coated with Au.102 In 1 μL of sweat, the SERS pH sensor showed a good resolution, repeatability and reversibility by utilizing two pH-responsive molecules in the pH range of 5.5–7.0. A SERS patch sensor was utilized for label-free drug detection in sweat. The patch consisted of a silk fibroin protein layer decorated with plasmonic silver nanowires (Ag NWs) to enhance the SERS signal.103 With a portable Raman spectrometer, Raman measurements could be performed in situ on the skin surface. Since each molecule has a unique SERS fingerprint spectrum, SERS offers a wide range of target with high specificity.
With the rising interest in soft SERS sensing, the fabrication and amplification of the hotspot strategy on soft substrates have become the focus of the sensing system in recent years. An enhancement mechanism was verified by an electrically modulated SERS substrate based on asymmetric Au/ZnO nanorods, and the SERS signal was significantly increased by 6.7 times.104 In addition to composite materials, the enhancement factor (EF) of a stretchable gold nanomesh was estimated to be 108 in a flexible and adhesive SERS sensor system.105 An Ag nanocube array, as another widely used plasmonic nanomaterial, could generate strong electromagnetic enhancement from sharp edges with an EF of 1.4 × 1010. With the SERS-based sensor chip, COVID-19 was identified under 5 min by a hand-held breathalyzer with >95% sensitivity and specificity (Fig. 3b).96 This means that SERS sensors have taken a substantial step in the practical application of the next generation of POC diagnostic tools. Cui et al. reported an ultra-sensitive SERS sensor that employed omnidirectional plasmonic nanovoid array to produce a plenty of hot spots with an ultralow DOL of 10−16 M (R6G).106 The soft armored SERS sensor exhibited high specificity and sensitivity for the H2S gas sensing and the 1 pM of DA in sweat. Combined with a flexible electronic system, a wearable plasmonic SERS sensor with ordered silver nanocubes was reported to track the real-time change of drug concentration in the human body with high specificity and stability.107 Moreover, recent results confirmed that the plasmonic SERS sensor could also be successfully combined with the microfluidic system for continuous sweat analysis (Fig. 3b).97 The plasmonic nanosensors integrated with Au nanorods were employed to monitor UA, and the paper microfluidics device was used to quantify sweat. The combination of the multi-modal SERS sensing platform is promising to continuously measure other small volumes of biological liquids, which enables more comprehensive disease diagnosis and health monitoring.
4. Challenge and perspectives
Soft biosensors provide dynamic biomolecule information with potential application to reform the hospital-centralized healthcare system and enrich the cyber-physical system. This review has summarized the emerging biotransduction strategies with electrical and optical output for soft biosensors. Despite many achievements, there are still many challenges to be addressed in terms of sensitivity, reliability, antifouling ability, and multifunctionality.For soft electrochemical biosensors, it is paramountcy to improve the accuracy and long-term sensing performance in complex body fluid environments and tissues. Biosensors in wearable application scenarios face serious biological fouling issues due to the close contact with untreated biological fluids. Burgeoning antifouling strategies including hydrophobic coating, microstructure design, and self-cleaning properties can be employed on the soft biosensing interfaces to address this issue. Moreover, as the current output relies on the electron transfer process, considerable efforts are still needed to develop efficient mediators to accelerate electron transfer or even push toward a direct electrochemical strategy for higher electron transfer efficiency.13 To further enhance the sensitivity by magnitude, it is highly desired to develop other biotransducers such as charge-coupled mechanisms and chemomechanical transduction where the signal output is not limited to traditional electrochemical reactions. The amplified signals are easier to be transmitted and integrated without extra complex data procedures.
For optical transducers, the main challenge is the trade-off between the spectrometer miniaturization and sensing performance in terms of accuracy, linear range and repeatability. Typically, optical biosensors show a relatively narrow linear detection range due to inaccessibility to lab-based instruments. It is urgent to develop a miniaturized, intelligent, and inexpensive spectrometer system. As the SERS signal is highly dependent on substrates, it is necessary to further consider both the substrate/active nanostructures, substrate/tissue surface interfacial adhesion, and the effect of mechanical deformation. Moreover, the development of novel composite nanomaterials for colorimetric sensing or SERS active substances and the combination of multimodal detection methods can provide new capabilities and distinct features, which are also important directions in the future.
Last but not least is that the deep fusion of the biosensor with functional electronics is still in its infancy. The recent decade has witnessed the development of a cyber-physical system which has significantly reformed our lifestyle. Such a cyber-physical system relies on a deep fusion of sensors with electronic processors and algorithms. It is expected that wearable biosensors that are capable of providing huge bioinformation data will greatly enrich the cyber-physical system. Recently, the fusion of neurotransmitter-responsive sensors with neuromorphic devices such as resistive switching memristors and transistors has been reported which aims to simulate the interneuron communication ability.10,108 Biohybrid interactions with live neurons are successfully demonstrated using neurotransmitters as the communication messengers. Such research studies provide preliminary demonstration of future biomolecule-based intelligent cyber-physical systems, and even new modality of cyborg. The integral role of machine learning algorithms can bolster the performance of nanosensors in detecting diseases.109 Combining machine learning algorithms with state-of-the-art technologies to create integrated nanoscale sensing platforms with high sensitivity and adaptability is almost necessary, and machine learning-empowered nanosensors are expected to be key to avoiding/minimizing a future disease outbreak. This research direction is promising and multidisciplinary. We envision much more breakthrough achievements to be made with continuous efforts of scientists in chemistry, biology, materials science, electronics, medicine, and computer science.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge the financial support from the National Key Research and Development Program of China (2017YFA0205302), the Key Program of the National Natural Science Foundation of China (62235008), the Natural Science Foundation of Jiangsu Province-Major Project (BK20212012), the Natural Science Foundation for Young Scholars (62201286), the National Key R&D Program of China under Grant (2021YFB3601200), the Natural Science Foundation for Young Scholars of Jiangsu Province (BK20210596, BK20220400), the Program of Jiangsu Specially-Appointed Professor, the General Project of the Natural Science Research of Jiangsu Province Higher Education Institutions (Grant No. 22KJB510036, 21KJB150021), and the Science Foundation of Nanjing University of Post and Telecommunications (NUPTSF, NY221004, NY221027, NY221028).References
- J. Kim, A. S. Campbell, B. E. de Avila and J. Wang, Nat. Biotechnol., 2019, 37, 389–406 CrossRef CAS PubMed.
- J. M. Buriak, L. M. Liz-Marzán, W. J. Parak and X. Chen, ACS Nano, 2022, 16, 1681–1684 CrossRef CAS.
- H. Zhao, R. Su, L. Teng, Q. Tian, F. Han, H. Li, Z. Cao, R. Xie, G. Li, X. Liu and Z. Liu, Nanoscale, 2022, 14, 1653–1669 RSC.
- S. O. Ajakwe, C. I. Nwakanma, D. S. Kim and J. M. Lee, IEEE Access, 2022, 10, 49956–49974 Search PubMed.
- V. Vijayan, J. P. Connolly, J. Condell, N. McKelvey and P. Gardiner, Sensors, 2021, 21, 5589 CrossRef PubMed.
- S. P. Sreenilayam, I. U. Ahad, V. Nicolosi, V. Acinas Garzon and D. Brabazon, Mater. Today, 2020, 32, 147–177 CrossRef.
- Kenry, J. C. Yeo and C. T. Lim, Microsyst. Nanoeng., 2016, 2, 16043 CrossRef CAS PubMed.
- J. Kim, A. S. Campbell, B. E. F. de Avila and J. Wang, Nat. Biotechnol., 2019, 37, 389–406 CrossRef CAS PubMed.
- R. M. Torrente-Rodriguez, J. Tu, Y. Yang, J. Min, M. Wang, Y. Song, Y. Yu, C. Xu, C. Ye, W. W. IsHak and W. Gao, Matter, 2020, 2, 921–937 CrossRef PubMed.
- T. Wang, M. Wang, J. Wang, L. Yang, X. Ren, G. Song, S. Chen, Y. Yuan, R. Liu, L. Pan, Z. Li, W. R. Leow, Y. Luo, S. Ji, Z. Cui, K. He, F. Zhang, F. Lv, Y. Tian, K. Cai, B. Yang, J. Niu, H. Zou, S. Liu, G. Xu, X. Fan, B. Hu, X. J. Loh, L. Wang and X. Chen, Nat. Electron., 2022, 5(9), 622 CrossRef.
- A. Koh, D. Kang, Y. Xue, S. Lee, R. M. Pielak, J. Kim, T. Hwang, S. Min, A. Banks, P. Bastien, M. C. Manco, L. Wang, K. R. Ammann, K. I. Jang, P. Won, S. Han, R. Ghaffari, U. Paik, M. J. Slepian, G. Balooch, Y. Huang and J. A. Rogers, Sci. Transl. Med., 2016, 8, 366ra165 Search PubMed.
- J. R. Sempionatto, I. Jeerapan, S. Krishnan and J. Wang, Anal. Chem., 2020, 92, 378–396 CrossRef CAS PubMed.
- J. Tu, R. M. Torrente-Rodríguez, M. Wang and W. Gao, Adv. Funct. Mater., 2019, 30, 1906713 CrossRef.
- W. Gao, H. Ota, D. Kiriya, K. Takei and A. Javey, Acc. Chem. Res., 2019, 52, 523–533 CrossRef CAS PubMed.
- Y. Ling, T. An, L. W. Yap, B. Zhu, S. Gong and W. Cheng, Adv. Mater., 2020, 32, e1904664 CrossRef PubMed.
- H. X. Liu, L. Wang, G. M. Lin and Y. H. Feng, Biomater. Sci., 2022, 10, 614–632 RSC.
- F. Yi, H. Y. Ren, J. Y. Shan, X. Sun, D. Wei and Z. F. Liu, Chem. Soc. Rev., 2018, 47, 3152–3188 RSC.
- Z. Liu, X. Wang, D. Qi, C. Xu, J. Yu, Y. Liu, Y. Jiang, B. Liedberg and X. Chen, Adv. Mater., 2017, 29, 1603382 CrossRef PubMed.
- Z. Liu, H. Wang, P. Huang, J. Huang, Y. Zhang, Y. Wang, M. Yu, S. Chen, D. Qi, T. Wang, Y. Jiang, G. Chen, G. Hu, W. Li, J. Yu, Y. Luo, X. J. Loh, B. Liedberg, G. Li and X. Chen, Adv. Mater., 2019, 31, e1901360 CrossRef PubMed.
- D. Qi, K. Zhang, G. Tian, B. Jiang and Y. Huang, Adv. Mater., 2021, 33, e2003155 CrossRef PubMed.
- S. Shrivastava, T. Q. Trung and N. E. Lee, Chem. Soc. Rev., 2020, 49, 1812–1866 RSC.
- Y. J. Ma, Y. C. Zhang, S. S. Cai, Z. Y. Han, X. Liu, F. L. Wang, Y. Cao, Z. H. Wang, H. F. Li, Y. H. Chen and X. Feng, Adv. Mater., 2020, 32, 1902062 CrossRef CAS PubMed.
- X. Zhao, G. Chen, Y. Zhou, A. Nashalian, J. Xu, T. Tat, Y. Song, A. Libanori, S. Xu, S. Li and J. Chen, ACS Nano, 2022, 16, 6013–6022 CrossRef CAS PubMed.
- C. Wang, C. Wang, Z. Huang and S. Xu, Adv. Mater., 2018, 30, e1801368 CrossRef PubMed.
- T. R. Ray, J. Choi, A. J. Bandodkar, S. Krishnan, P. Gutruf, L. Tian, R. Ghaffari and J. A. Rogers, Chem. Rev., 2019, 119, 5461–5533 CrossRef CAS PubMed.
- C. M. Tringides and D. J. Mooney, Adv. Mater., 2022, 34, e2107207 CrossRef PubMed.
- K. A. Deo, M. K. Jaiswal, S. Abasi, G. Lokhande, S. Bhunia, T. U. Nguyen, M. Namkoong, K. Darvesh, A. Guiseppi-Elie, L. Tian and A. K. Gaharwar, ACS Nano, 2022, 16, 8798–8811 CrossRef CAS PubMed.
- T. Tat, G. Chen, X. Zhao, Y. Zhou, J. Xu and J. Chen, ACS Nano, 2022, 16, 13301–13313 CrossRef CAS PubMed.
- B. K. Walther, C. Z. Dinu, D. M. Guldi, V. G. Sergeyev, S. E. Creager, J. P. Cooke and A. Guiseppi-Elie, Mater. Today, 2020, 39, 23–46 CrossRef CAS.
- T. Wang, M. Wang, L. Yang, Z. Li, X. J. Loh and X. Chen, Adv. Mater., 2020, 32, e1905522 CrossRef PubMed.
- J. Liu, F. Li, Y. Wang, L. Pan, P. Lin, B. Zhang, Y. Zheng, Y. Xu, H. Liao, G. Ko, F. Fei, C. Xu, Y. Du, K. Shin, D. Kim, S. S. Jang, H. J. Chung, H. Tian, Q. Wang, W. Guo, J. M. Nam, Z. Chen, T. Hyeon and D. Ling, Nat. Nanotechnol., 2020, 15, 321–330 CrossRef CAS PubMed.
- Y. Song, D. Mukasa, H. Zhang and W. Gao, Acc. Mater. Res., 2021, 2, 184–197 CrossRef CAS.
- H. Teymourian, A. Barfidokht and J. Wang, Chem. Soc. Rev., 2020, 49, 7671–7709 RSC.
- Y.-L. Liu and W.-H. Huang, Angew. Chem., Int. Ed., 2021, 60, 2757–2767 CrossRef CAS PubMed.
- J. Yu, P. Cai, X. Zhang, T. Zhao, L. Liang, S. Zhang, H. Liu and X. Chen, ACS Nano, 2021, 15, 7618–7627 CrossRef CAS PubMed.
- D. W. Kimmel, G. LeBlanc, M. E. Meschievitz and D. E. Cliffel, Anal. Chem., 2012, 84, 685–707 CrossRef CAS PubMed.
- A. Ganguly, K. C. Lin, S. Muthukumar and S. Prasad, ACS Sens., 2021, 6, 63–72 CrossRef CAS PubMed.
- W. Gao, S. Emaminejad, H. Y. Y. Nyein, S. Challa, K. Chen, A. Peck, H. M. Fahad, H. Ota, H. Shiraki, D. Kiriya, D. H. Lien, G. A. Brooks, R. W. Davis and A. Javey, Nature, 2016, 529, 509–514 CrossRef CAS PubMed.
- H. Y. Nyein, W. Gao, Z. Shahpar, S. Emaminejad, S. Challa, K. Chen, H. M. Fahad, L. C. Tai, H. Ota, R. W. Davis and A. Javey, ACS Nano, 2016, 10, 7216–7224 CrossRef CAS PubMed.
- S. Emaminejad, W. Gao, E. Wu, Z. A. Davies, H. Y. Y. Nyein, S. Challa, S. P. Ryan, H. M. Fahad, K. Chen, Z. Shahpar, S. Talebi, C. Milla, A. Javey and R. W. Davis, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 4625–4630 CrossRef CAS PubMed.
- Q. Zhai, L. W. Yap, R. Wang, S. Gong, Z. Guo, Y. Liu, Q. Lyu, J. Wang, G. P. Simon and W. Cheng, Anal. Chem., 2020, 92, 4647–4655 CrossRef CAS PubMed.
- Y. Lee, C. Howe, S. Mishra, D. S. Lee, M. Mahmood, M. Piper, Y. Kim, K. Tieu, H. S. Byun, J. P. Coffey, M. Shayan, Y. Chun, R. M. Costanzo and W. H. Yeo, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 5377–5382 CrossRef CAS PubMed.
- S. Nakata, M. Shiomi, Y. Fujita, T. Arie, S. Akita and K. Takei, Nat. Electron., 2018, 1, 596–603 CrossRef.
- D. Xu, L. Wu, H. Yao and L. Zhao, Small, 2022, 37, e2203400 CrossRef PubMed.
- Y. Yang, Y. Song, X. Bo, J. Min, O. S. Pak, L. Zhu, M. Wang, J. Tu, A. Kogan, H. Zhang, T. K. Hsiai, Z. Li and W. Gao, Nat. Biotechnol., 2020, 38, 217–224 CrossRef CAS PubMed.
- J. Li, Y. Liu, L. Yuan, B. Zhang, E. S. Bishop, K. Wang, J. Tang, Y.-Q. Zheng, W. Xu, S. Niu, L. Beker, T. L. Li, G. Chen, M. Diyaolu, A.-L. Thomas, V. Mottini, J. B. H. Tok, J. C. Y. Dunn, B. Cui, S. P. Paşca, Y. Cui, A. Habtezion, X. Chen and Z. Bao, Nature, 2022, 606, 94–101 CrossRef CAS PubMed.
- Y. H. Chen, S. Y. Lu, S. S. Zhang, Y. Li, Z. Qu, Y. Chen, B. W. Lu, X. Y. Wang and X. Feng, Sci. Adv., 2017, 3, e1701629 CrossRef PubMed.
- S. K. Kim, G. H. Lee, C. Jeon, H. H. Han, S. J. Kim, J. W. Mok, C. K. Joo, S. Shin, J. Y. Sim, D. Myung, Z. Bao and S. K. Hahn, Adv. Mater., 2022, 34, e2110536 CrossRef PubMed.
- D. Keum, S. K. Kim, J. Koo, G. H. Lee, C. Jeon, J. W. Mok, B. H. Mun, K. J. Lee, E. Kamrani, C. K. Joo, S. Shin, J. Y. Sim, D. Myung, S. H. Yun, Z. N. Bao and S. K. Hahn, Sci. Adv., 2020, 6, eaba3252 CrossRef CAS PubMed.
- G. H. Lee, H. Moon, H. Kim, G. H. Lee, W. Kwon, S. Yoo, D. Myung, S. H. Yun, Z. Bao and S. K. Hahn, Nat. Rev. Mater., 2020, 5, 149–165 CrossRef PubMed.
- Q. Zhai, S. Gong, Y. Wang, Q. Lyu, Y. Liu, Y. Ling, J. Wang, G. P. Simon and W. Cheng, ACS Appl. Mater. Interfaces, 2019, 11, 9724–9729 CrossRef CAS PubMed.
- T. Wang, Q. L. Lei, M. Wang, G. Deng, L. Yang, X. Liu, C. Li, Q. Wang, Z. Liu, J. Wang, Z. Cui, K. G. Utama, R. Ni and X. Chen, Adv. Mater., 2020, 32, e2000991 CrossRef PubMed.
- R. M. Torrente-Rodriguez, H. Lukas, J. Tu, J. Min, Y. Yang, C. Xu, H. B. Rossiter and W. Gao, Matter, 2020, 3, 1981–1998 CrossRef PubMed.
- K. Guo, S. Wustoni, A. Koklu, E. Diaz-Galicia, M. Moser, A. Hama, A. A. Alqahtani, A. N. Ahmad, F. S. Alhamlan, M. Shuaib, A. Pain, I. McCulloch, S. T. Arold, R. Grunberg and S. Inal, Nat. Biomed. Eng., 2021, 5, 666–677 CrossRef CAS PubMed.
- S. Dalirirad and A. J. Steckl, Sens. Actuators, B, 2019, 283, 79–86 CrossRef CAS.
- S. Lin, X. Cheng, J. Zhu, B. Wang, D. Jelinek, Y. Zhao, T. Y. Wu, A. Horrillo, J. Tan, J. Yeung, W. Yan, S. Forman, H. A. Coller, C. Milla and S. Emaminejad, Sci. Adv., 2022, 8, eabq4539 CrossRef CAS PubMed.
- S. Sheibani, L. Capua, S. Kamaei, S. S. A. Akbari, J. Zhang, H. Guerin and A. M. Ionescu, Commun. Mater., 2021, 2, 10 CrossRef CAS PubMed.
- N. Nakatsuka, K. A. Yang, J. M. Abendroth, K. M. Cheung, X. Xu, H. Yang, C. Zhao, B. Zhu, Y. S. Rim, Y. Yang, P. S. Weiss, M. N. Stojanovic and A. M. Andrews, Science, 2018, 362, 319–324 CrossRef CAS PubMed.
- B. Zhu, On-Skin Chemical Sensors, in Handbook of Biochips, ed. M. Sawan, Springer, New York, NY, 2022, pp. 129–141 Search PubMed.
- B. J. Sanghavi, J. A. Moore, J. L. Chavez, J. A. Hagen, N. Kelley-Loughnane, C. F. Chou and N. S. Swami, Biosens. Bioelectron., 2016, 78, 244–252 CrossRef CAS PubMed.
- B. Wang, C. Z. Zhao, Z. Q. Wang, K. A. Yang, X. B. Cheng, W. F. Liu, W. Z. Yu, S. Y. Lin, Y. C. Zhao, K. M. Cheung, H. S. Lin, H. Hojaiji, P. S. Weiss, M. N. Stojanovic, A. J. Tomiyama, A. M. Andrews and S. Emaminejad, Sci. Adv., 2022, 8, eabk0967 CrossRef CAS PubMed.
- J. E. An, K. H. Kim, S. J. Park, S. E. Seo, J. Kim, S. Ha, J. Bae and O. S. Kwon, ACS Sens., 2022, 7, 99–108 CrossRef CAS PubMed.
- N. K. Singh, S. Chung, M. Sveiven and D. A. Hall, ACS Omega, 2021, 6, 27888–27897 CrossRef CAS PubMed.
- Z. Wang, Z. Hao, S. Yu, C. G. De Moraes, L. H. Suh, X. Zhao and Q. Lin, Adv. Funct. Mater., 2019, 29, 1905202 CrossRef CAS PubMed.
- T. Kaya, G. C. Liu, J. Ho, K. Yelamarthi, K. Miller, J. Edwards and A. Stannard, Electroanalysis, 2019, 31, 411–421 CrossRef CAS.
- Z. Wang, Z. Hao, X. Wang, C. Huang, Q. Lin, X. Zhao and Y. Pan, Adv. Funct. Mater., 2020, 31, 2005958 CrossRef.
- K. Haupt, P. X. M. Rangel and B. T. S. Bui, Chem. Rev., 2020, 120, 9554–9582 CrossRef CAS PubMed.
- A. Koklu, D. Ohayon, S. Wustoni, V. Druet, A. Saleh and S. Inal, Chem. Rev., 2022, 122, 4581–4635 CrossRef CAS PubMed.
- S. M. Mugo and J. Alberkant, Anal. Bioanal. Chem., 2020, 412, 1825–1833 CrossRef CAS PubMed.
- B. T. S. Bui, T. Auroy and K. Haupt, Angew. Chem., Int. Ed., 2022, 61, e202106493 Search PubMed.
- O. Parlak, S. T. Keene, A. Marais, V. F. Curto and A. Salleo, Sci. Adv., 2018, 4, eaar2904 CrossRef CAS PubMed.
- W. Tang, L. Yin, J. R. Sempionatto, J. M. Moon, H. Teymourian and J. Wang, Adv. Mater., 2021, 33, e2008465 CrossRef PubMed.
- M. Wang, Y. Yang, J. Min, Y. Song, J. Tu, D. Mukasa, C. Ye, C. Xu, N. Heflin, J. S. McCune, T. K. Hsiai, Z. Li and W. Gao, Nat. Biomed. Eng., 2022, 6(11), 1225–1235 CrossRef CAS PubMed.
- S. Premachandran, R. Haldavnekar, S. Das, K. Venkatakrishnan and B. Tan, ACS Nano, 2022, 16(11), 17948–17964 CrossRef CAS PubMed.
- J. Wang, C. Wang, P. Cai, Y. Luo, Z. Cui, X. J. Loh and X. Chen, ACS Nano, 2021, 15, 18671–18678 CrossRef CAS PubMed.
- S. Imani, A. J. Bandodkar, A. M. Mohan, R. Kumar, S. Yu, J. Wang and P. P. Mercier, Nat. Commun., 2016, 7, 11650 CrossRef CAS PubMed.
- H. Lee, C. Song, Y. S. Hong, M. S. Kim, H. R. Cho, T. Kang, K. Shin, S. H. Choi, T. Hyeon and D. H. Kim, Sci. Adv., 2017, 3, e1601314 CrossRef PubMed.
- L. Shang, Y. Cheng and Y. Zhao, Chem. Rev., 2017, 117, 7964–8040 CrossRef CAS PubMed.
- J. Choi, R. Ghaffari, L. B. Baker and J. A. Rogers, Sci. Adv., 2018, 4, eaar3921 CrossRef PubMed.
- J. T. Reeder, J. Choi, Y. G. Xue, P. Gutruf, J. Hanson, M. Liu, T. Ray, A. J. Bandodkar, R. Avila, W. Xia, S. Krishnan, S. Xu, K. Barnes, M. Pahnke, R. Ghaffari, Y. Huang and J. A. Rogers, Sci. Adv., 2019, 5, eaau6356 CrossRef PubMed.
- J. T. Reeder, Y. Xue, D. Franklin, Y. Deng, J. Choi, O. Prado, R. Kim, C. Liu, J. Hanson, J. Ciraldo, A. J. Bandodkar, S. Krishnan, A. Johnson, E. Patnaude, R. Avila, Y. Huang and J. A. Rogers, Nat. Commun., 2019, 10, 5513 CrossRef CAS PubMed.
- A. J. Bandodkar, J. Choi, S. P. Lee, W. J. Jeang, P. Agyare, P. Gutruf, S. Wang, R. A. Sponenburg, J. T. Reeder, S. Schon, T. R. Ray, S. Chen, S. Mehta, S. Ruiz and J. A. Rogers, Adv. Mater., 2019, 31, e1902109 CrossRef PubMed.
- J. Choi, D. Kang, S. Han, S. B. Kim and J. A. Rogers, Adv. Healthc. Mater., 2017, 6, 1601355 CrossRef PubMed.
- S. B. Kim, K. Lee, M. S. Raj, B. Lee, J. T. Reeder, J. Koo, A. Hourlier-Fargette, A. J. Bandodkar, S. M. Won, Y. Sekine, J. Choi, Y. Zhang, J. Yoon, B. H. Kim, Y. Yun, S. Lee, J. Shin, J. Kim, R. Ghaffari and J. A. Rogers, Small, 2018, 14, e1802876 CrossRef PubMed.
- J. Choi, S. Chen, Y. Deng, Y. Xue, J. T. Reeder, D. Franklin, Y. S. Oh, J. B. Model, A. J. Aranyosi, S. P. Lee, R. Ghaffari, Y. Huang and J. A. Rogers, Adv. Healthcare Mater., 2021, 10, e2000722 CrossRef PubMed.
- S. Liu, D. S. Yang, S. Wang, H. Luan, Y. Sekine, J. B. Model, A. J. Aranyosi, R. Ghaffari and J. A. Rogers, EcoMat, 2022 DOI:10.1002/eom2.12270.
- H. Luan, Q. Zhang, T.-L. Liu, X. Wang, S. Zhao, H. Wang, S. Yao, Y. Xue, J. W. Kwak, W. Bai, Y. Xu, M. Han, K. Li, Z. Li, X. Ni, J. Ye, D. Choi, Q. Yang, J.-H. Kim, S. Li, S. Chen, C. Wu, D. Lu, J.-K. Chang, Z. Xie, Y. Huang and J. A. Rogers, Sci. Adv., 2021, 7, e1601314 Search PubMed.
- J. Choi, A. J. Bandodkar, J. T. Reeder, T. R. Ray, A. Turnquist, S. B. Kim, N. Nyberg, A. Hourlier-Fargette, J. B. Model, A. J. Aranyosi, S. Xu, R. Ghaffari and J. A. Rogers, ACS Sens., 2019, 4, 379–388 CrossRef CAS PubMed.
- A. J. Bandodkar, P. Gutruf, J. Choi, K. Lee, Y. Sekine, J. T. Reeder, W. J. Jeang, A. J. Aranyosi, S. P. Lee, J. B. Model, R. Ghaffari, C.-J. Su, J. P. Leshock, T. Ray, A. Verrillo, K. Thomas, V. Krishnamurthi, S. Han, J. Kim, S. Krishnan, T. Hang and J. A. Rogers, Sci. Adv., 2019, 5, eaav3294 CrossRef PubMed.
- J. Kim, Y. Wu, H. Luan, D. S. Yang, D. Cho, S. S. Kwak, S. Liu, H. Ryu, R. Ghaffari and J. A. Rogers, Adv. Sci., 2022, 9, e2103331 CrossRef PubMed.
- L. Guo, T. Wang, Z. Wu, J. Wang, M. Wang, Z. Cui, S. Ji, J. Cai, C. Xu and X. Chen, Adv. Mater., 2020, 32, e2004805 CrossRef PubMed.
- T. Wang, Y. Guo, P. Wan, X. Sun, H. Zhang, Z. Yu and X. Chen, Nanoscale, 2017, 9, 869–874 RSC.
- Y. Sekine, S. B. Kim, Y. Zhang, A. J. Bandodkar, S. Xu, J. Choi, M. Irie, T. R. Ray, P. Kohli, N. Kozai, T. Sugita, Y. Wu, K. Lee, K. T. Lee, R. Ghaffari and J. A. Rogers, Lab Chip, 2018, 18, 2178–2186 RSC.
- S. Kim, B. Lee, J. Reeder, S. H. Seo, S. U. Lee, A. Hourlier-Fargette, J. Shin, Y. Sekine, H. Jeong, Y. S. Oh, A. J. Aranyosi, S. P. Lee, J. B. Model, G. Lee, M. H. Seo, S. S. Kwak, S. Jo, G. Park, S. Han, I. Park, H. I. Jung, R. Ghaffari, J. Koo, P. V. Braun and J. A. Rogers, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 27906–27915 CrossRef CAS PubMed.
- H. Araki, J. Kim, S. Zhang, A. Banks, K. E. Crawford, X. Sheng, P. Gutruf, Y. Shi, R. M. Pielak and J. A. Rogers, Adv. Funct. Mater., 2017, 27, 1604465 CrossRef.
- S. X. Leong, Y. X. Leong, E. X. Tan, H. Y. F. Sim, C. S. L. Koh, Y. H. Lee, C. Chong, L. S. Ng, J. R. T. Chen, D. W. C. Pang, L. B. T. Nguyen, S. K. Boong, X. Han, Y. C. Kao, Y. H. Chua, G. C. Phan-Quang, I. Y. Phang, H. K. Lee, M. Y. Abdad, N. S. Tan and X. Y. Ling, ACS Nano, 2022, 16, 2629–2639 CrossRef CAS PubMed.
- U. Mogera, H. Guo, M. Namkoong, M. S. Rahman, T. Nguyen and L. Tian, Sci. Adv., 2022, 8, eabn1736 CrossRef CAS PubMed.
- Y. Tian, Z. Shuai, J. Shen, L. Zhang, S. Chen, C. Song, B. Zhao, Q. Fan and L. Wang, Small, 2018, 14, e1800669 CrossRef PubMed.
- Y. Y. Tian, L. Zhang and L. H. Wang, Biotechnol. J., 2020, 15, 9 Search PubMed.
- L. Song, J. Chen, B. B. Xu and Y. Huang, ACS Nano, 2021, 15, 18822–18847 CrossRef CAS PubMed.
- K. Xu, R. Zhou, K. Takei and M. Hong, Adv. Sci., 2019, 6, 1900925 CrossRef PubMed.
- M. Chung, W. H. Skinner, C. Robert, C. J. Campbell, R. M. Rossi, V. Koutsos and N. Radacsi, ACS Appl. Mater. Interfaces, 2021, 13, 51504–51518 CrossRef CAS PubMed.
- E. H. Koh, W. C. Lee, Y. J. Choi, J. I. Moon, J. Jang, S. G. Park, J. Choo, D. H. Kim and H. S. Jung, ACS Appl. Mater. Interfaces, 2021, 13, 3024–3032 CrossRef CAS PubMed.
- J. Xu, H. He, X. Jian, K. Qu, J. Xu, C. Li, Z. Gao and Y. Y. Song, Anal. Chem., 2021, 93, 9286–9295 CrossRef CAS PubMed.
- L. Liu, P. M. Pancorbo, T. H. Xiao, S. Noguchi, M. Marumi, H. Segawa, S. Karhadkar, J. G. De Pablo, K. Hiramatsu, Y. Kitahama, T. Itoh, J. Qu, K. Takei and K. Goda, Adv. Opt. Mater., 2022, 10, 2200054 CrossRef CAS.
- K. Zhu, K. Yang, Y. Zhang, Z. Yang, Z. Qian, N. Li, L. Li, G. Jiang, T. Wang, S. Zong, L. Wu, Z. Wang and Y. Cui, Small, 2022, 18, e2201508 CrossRef PubMed.
- Y. Wang, C. Zhao, J. Wang, X. Luo, L. Xie, S. Zhan, J. Kim, X. Wang, X. Liu and Y. Ying, Sci. Adv., 2021, 7, eabe4553 CrossRef CAS PubMed.
- Y. X. Leong, E. X. Tan, S. X. Leong, C. S. L. Koh, L. B. T. Nguyen, J. R. T. Chen, K. Xia and X. Y. Ling, ACS Nano, 2022, 16, 13279–13293 CrossRef CAS PubMed.
- S. T. Keene, C. Lubrano, S. Kazemzadeh, A. Melianas, Y. Tuchman, G. Polino, P. Scognamiglio, L. Cina, A. Salleo, Y. van de Burgt and F. Santoro, Nat. Mater., 2020, 19, 969–973 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |



