Depletion attractions drive bacterial capture on both non-fouling and adhesive surfaces, enhancing cell orientation†
Wuqi Amy
Niu
,
Morgan N.
Smith
and
Maria M.
Santore
 *
*
Department of Polymer Science and Engineering University of Massachusetts, 120 Governors Drive, Amherst, MA 01003, USA. E-mail: santore@mail.pse.umass.edu; Tel: +1 413-577-1417
First published on 25th November 2022
Abstract
Depletion attractions, occurring between surfaces immersed in a polymer solution, drive bacteria adhesion to a variety of surfaces. The latter include the surfaces of non-fouling coatings such as hydrated polyethylene glycol (PEG) layers but also, as demonstrated in this work, surfaces that are bacteria-adhesive, such as that of glass. Employing a flagella free E. coli strain, we demonstrate that cell adhesion on glass is enhanced by dissolved polyethylene oxide (PEO), exhibiting a faster rate and greater numbers of captured cells compared with the slower capture of the same cells on glass from a buffer solution. After removal of depletant, any cell retention appears to be governed by the substrate, with cells immediately released from non-fouling PEG surfaces but retained on glass. A distinguishing feature of cells captured by depletion on PEG surfaces is their orientation parallel to the surface and very strong alignment with flow. This suggests that, in the moments of capture, cells are able to rotate as they adhere. By contrast on glass, captured cells are substantially more upright and less aligned by flow. On glass the free polymer exerts forces that slightly tip cells towards the surface. Free polymer also holds cells still on adhesive and non-fouling surfaces alike but, upon removal of free PEO, cells retained on glass tend to be held by one end and exhibit a Brownian type rotational rocking.
Introduction
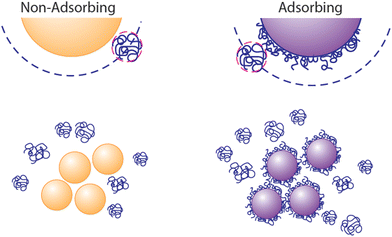
Depletion attractions, arising from the exclusion of dissolved macromolecules, micelles, or nanoparticles (the “depletant”) from the region near a fluid–solid interface,1,2 are well understood to drive aggregation of colloidal particles3–10 or colloidal deposition onto the walls of containers, templates, and flow chambers.11–15 When the particles aggregate or approach another surface, the volume available for the solvated depletant exceeds that when the colloids are dispersed, establishing the entropic origin of depletion attractions. The range of an attractive depletion potential scales as the depletant size and its strength depends on depletant concentration, with forces scaling as the osmotic pressure.7 Therefore, the potentials can be many times kT and longer range than electrostatic and van der Waals interactions.While classical descriptions of depletion employ examples, for instance polymers, that do not adsorb onto the surfaces of the particles or container walls,1,2 a lack of adsorption is not a prerequisite for the existence of depletion attractions. When ample depletant remains dissolved in solution, it exerts an osmotic attractive force between surfaces, even if they contain adsorbed polymers.16–18 The ubiquity of scenarios involving free polymer therefore explain the ubiquity of depletion interactions. Molecules in solution are effectively excluded from particles or surfaces carrying the same polymer, whether adsorbed or grafted permanently.19,20 This exclusion is pronounced in a good solvent where, for instance, polymers in free solution are repelled sterically by those on a surface. Thus, as depicted in Fig. 1, depletion forces can persist between depletant-adsorbing surfaces in the presence of excess dissolved depletant polymer. This appears to have been the case in studies employing polyethlene oxide (PEO) to generate depletion attractions between particles and a glass wall.21–23 Then, the translational entropy of the free depletant increases upon particle aggregation or deposition to a wall, producing an effective attraction. (The resulting aggregates may, however, contain adsorbed polymer depletant and experience bridging attractions in time.)
A distinguishing feature of depletion aggregation and deposition is its reversibility.7,20,24–26 Removal of the depletant from free solution erases the interparticle attraction and, as long as the particles have not drifted closer into the van der Waals minimum, they will resuspend. Likewise, if the depletant had formed an adsorbed layer on the aggregating particles, removal of the free depletant will eliminate the depletion attraction and the particles may resuspend rapidly, as long as the adsorbed chains have not entangled or bridged particles.
The mechanism of depletion is only recently being considered in the context of bacterial suspensions27–32 and biofilm formation, even though bacteria commonly live in polymer-rich environments. The most basic of these is the polysaccharides secreted by the cells themselves, which can drive cell aggregation.33 Using bacteria to degrade plastics may expose cells to polymer and nanoparticle solutions. Another example, our digestive tracts contain complex macromolecular solutions which interact with bacteria and the mucin layer of the gut wall. Common laxatives and preparations for gastro-endoscopy are based on polyethylene glycol, a few examples of the relevance of naturally occurring and synthetic polymer interactions with bacteria. Aggregation of bacteria by depletion attractions has, in one study, been shown to produce bacterial resistance by mechanisms separate from limitations on antibiotic diffusion through biofilms.34
Motivated by the possibility for negatively charged polysaccharides to produce inter-cell attractions relevant to biofilm formation, Schwarz-Linek established that the depletion phase diagram of E. coli mixed with a model anionic polymer followed expectations for depletion attractions.30,32 Secor and coworkers report that exopolysaccharide-driven depletion aggregation and co-aggregation can be species dependent.35 More recently Niu et al. established that dissolved PEO, at concentrations expected to produce substantial depletion attractions and phase separation, could not only produce reversible aggregation of E. coli cells, but also drive E. coli to adhere to a surface rendered otherwise nonadhesive to by application of a PEG coating.29 The adhesion of E. coli cells on the non-fouling PEG coating was shown to be rapidly reversed, with cells released upon removal of PEO from the free solution. The release of cells from the surface along with dissipation of aggregates upon removal of PEO was shown to be a distinguishing feature of depletion attractions experienced by bacteria, different from bridging and patch-wise attractions produced by polymers attractive towards cells.
The current work compares depletion-driven capture of E. coli cells on non-fouling PEG-coatings, a model nonadhesive biomaterial surface, to that on glass, a model adhesive surface, selected for its use in many model studies and relevance in chip assays. Here depletion is shown to occur even though PEO adsorbs to glass. Also, several of the prior studies, including that of Niu et al.,29 employed stationary phase cells, either for their rounder shape (compared to more capsular shapes of the same bacteria in log phase) or due to the need to reduce cell division and extracellular polymeric substance secretion during long settling studies. Relevant to biofilm formation, the current work examines how depletion interactions and the capture of E. coli on surfaces proceed for log phase cells.
While focusing on the relevant case of log phase cells, the current study examines behaviors important to early biofilm formation: the rates and numbers of cells captured, and their orientations in-plane and relative to the flow direction. Cell orientations are compared with previous reports for orientations of E. coli on surfaces of different chemistries.36 Cell orientation, in addition to mobility and evidence for partial adhesion is relevant to colony and biofilm growth, influencing the compactness of growing bacterial communities and their transition from a flat layer to a three dimensional structure.37–41 The cells here are alive throughout the study and in subsequent works we demonstrate their growth patterns.
Materials and methods
Depletant
For the depletant, this work employed molecular weight standard 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 g mol−1 poly(ethylene oxide) (PEO) with a polydispersity of 1.07, from Agilent Technologies.
200 g mol−1 poly(ethylene oxide) (PEO) with a polydispersity of 1.07, from Agilent Technologies.
Bacteria
This study employed E. coli that did not express flagella because the flhD gene had been knocked out. ΔflhD E. coli JW1881 were obtained from the Coli Genetic Stock Center (New Haven, CT). Prior electron microscopy studies confirmed a lack of flagella and swimming motility.42
E. coli were grown at 37 °C overnight in lysogeny broth (LB). After overnight growth, bacteria were back-diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 in LB, incubated at 37 °C for 2 hours and harvested in log phase. To remove components of the growth medium and other macromolecular constituents, bacteria cultures were washed 3 times (centrifuged at 3800 rpm for 2 min) in pH 7.4 phosphate buffered saline (PBS) (0.008 M Na2HPO4, 0.002 M KH2PO4, and 0.15 M NaCl) and resuspended in the same buffer at a concentration of approximately 1 × 108 cells per mL, as determined using D600 turbidity measurements. Cells were studied immediately, with work completed within 1 hour of preparation. Viability screening with propidium iodide (Sigma Aldrich, excitation 535 nm, emission 617 nm) confirmed cell viability before and after all experimental procedures.
50 in LB, incubated at 37 °C for 2 hours and harvested in log phase. To remove components of the growth medium and other macromolecular constituents, bacteria cultures were washed 3 times (centrifuged at 3800 rpm for 2 min) in pH 7.4 phosphate buffered saline (PBS) (0.008 M Na2HPO4, 0.002 M KH2PO4, and 0.15 M NaCl) and resuspended in the same buffer at a concentration of approximately 1 × 108 cells per mL, as determined using D600 turbidity measurements. Cells were studied immediately, with work completed within 1 hour of preparation. Viability screening with propidium iodide (Sigma Aldrich, excitation 535 nm, emission 617 nm) confirmed cell viability before and after all experimental procedures.
Drops of the E. coli suspension were imaged at 100× in phase contrast to determine cell size. Analysis via Oufti43 using the cell detection analysis tool revealed, for 350–400 cells in each suspension, an average length of 3.0 ± 0.3 μm and an average width of 0.96 ± 0.05 μm. Batch-to-batch reproducibility in cell shape is important to avoid as longer cells orient more than short ones in flow. Our cell dimensions are typical of E. coli.
Glass surfaces
Microscope slides were soaked in concentrated sulfuric acid overnight and rinsed thoroughly with deionized (DI) water before immediately sealing in a flow chamber, and then initiating flow of buffer. This process produces a silica surface on the slide.Non-bioadhesive PEG surfaces
Non-bacterial adhesive surfaces were produced in situ on acid-etched microscope slides sealed in the flow chamber. After buffer flow had been established over a slide, it was replaced by a 100 ppm buffered solution of a poly-L-lysine-PEG (PLL-PEG) graft copolymer, flowing at a wall shear rate of 22 s−1. The copolymer forms a layer adsorbed by electrostatic attractions between negative silanol groups on glass slides with cationic groups on the PLL backbone.44,45 The PEG side chains protrude from the interface to produce a solvated PEG brush which repels cells and proteins. The particular PLL-PEG architecture is key to the generation of the copolymer layer on the microscope slide and to the steric repulsions between the PEG brush and molecules or cells from solution.46 This study employed PLL of nominal molecular weight 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–30
000–30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1 (Sigma Aldrich) and PEG side chains of 5000 g mol−1 (methoxypoly(ethylene glycol)-succinimidyl valerate from Laysan Bio. Inc.) that functionalized 1/3 of the backbone PLL amines, described previously in detail.47
000 g mol−1 (Sigma Aldrich) and PEG side chains of 5000 g mol−1 (methoxypoly(ethylene glycol)-succinimidyl valerate from Laysan Bio. Inc.) that functionalized 1/3 of the backbone PLL amines, described previously in detail.47
Polymer adsorption
The adsorbed amounts of PLL-PEG or PEO depletants, and the times over which layers were established and retained were determined employing near-Brewster reflectometry, in a flow chamber of similar geometry to that of the flow microscope. While near-Brewster reflectometry has been previously summarized in detail,48 we mention here that with parallel-polarized light reflecting back to a detector from inside a glass substrate, the intensity of a laser reflected off a clean interface vanishes at the Brewster angle. However, even small amounts of adsorbed proteins and polymers, 0.01 mg m−2, can be detected based on the intensity of the weakly reflecting beam at these conditions.49 Upon adsorption of PLL-PEG to a negative silica surface, the resulting PEG brush layers were confirmed not to adhere E. coli cells or proteins, and did not desorb from the surface at or near the conditions of this study.50Bacterial capture and assessment
Bacteria captured on glass surfaces and those coated with a non-adhesive PEG brush were studied in a video flow microscope at a wall shear rate of 5 s−1. The flow chamber was oriented perpendicular to the laboratory floor so that gravity did not pull cells towards or away from the test surface. Cell capture and orientations were recorded at standard video frame rates and, as prescribed in a particular experiment, a study focused on one region of surface as the numbers of cells changed in flow, or in other parts of experiments, multiple images were recorded after the viewing position was shifted to a neighboring spot, all towards the center of the slide and near the original point of study. In this way, multiple regions of a surface were assessed for cell alignment and mobility. All experiments were run in triplicate employing bacterial cultures grown on different days. In measurements of capture kinetics, a 20× objective was employed while measurements of cell orientation employed a 40× objective.Data analysis
Images from video frames were analyzed by first subtracting a background control frame, recorded prior to bacteria introduction, to remove features and aberrations on the camera's detector array. Then, to generate cell capture and release traces (numbers of cells per time), each captured cell in the frame was located and counted by employing a self-written Python code implementing the OpenCV library. Only cells that stayed in the same position for more than 30 frames (1 s) were counted. By this method, when an aggregate attaches to the surface, the aggregated cells are not well distinguished or counted. However, as shown in the results, most (95% or more) frames contained only singlet cells. On a glass surface, when cells were captured in the presence of solvated PEO, more aggregates (making up <20% of captured cells) were observed. In this case, counts determined by the Python code were compared with manual counting to ensure accuracy.To develop images for publishable standalone figures, time-lapse averaging of video frames was employed to clearly identify when cells were immobilized as opposed to moving. Images in Fig. 3B (i), (ii), and (iii) are time-lapse images from a 5 second video.
The vertical orientation of each cell was classified by human eye based on its shape (round-like was standing; rod-like with large aspect ratio was considered tipped; others were considered leaning) as defined in the Results section. When a cell was identified as tipped or leaning, its shape was fit to an ellipse, using a self-written Python code, to determine its major axis. The cell angle was found by calculating the angle between the major axis and the flow horizontal flow direction. 15–20 frames for each run were chosen for orientation analysis. These frames were in adjacent fields both upstream and downstream of the original field of view, where the movie of the capture process was recorded.
Results
Cell capture in the presence of free polymer
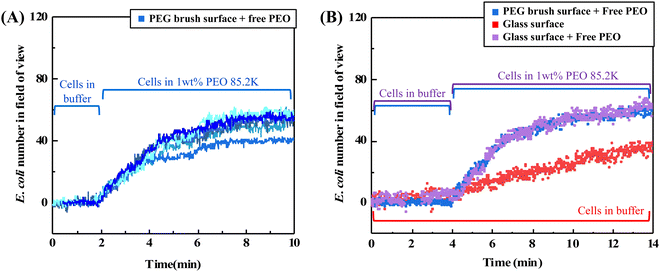
Fig. 2A shows how log-phase E. coli cells flowing over a non-adhesive surface can be made to adhere by adding dissolved 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 g mol−1 PEO (1 wt%) to the bacterial suspension. In each of five runs employing bacterial cells grown on different days, cells were initially flowed over an adhesion-resistant PEG brush coating for 20 minutes and did not adhere. Only the last 2 minutes of this step are included in Fig. 2A. Then, when the PEO homopolymer depletant is introduced into the bulk cell suspension, cells are captured on the adhesion-resistant surface with excellent reproducibility.
200 g mol−1 PEO (1 wt%) to the bacterial suspension. In each of five runs employing bacterial cells grown on different days, cells were initially flowed over an adhesion-resistant PEG brush coating for 20 minutes and did not adhere. Only the last 2 minutes of this step are included in Fig. 2A. Then, when the PEO homopolymer depletant is introduced into the bulk cell suspension, cells are captured on the adhesion-resistant surface with excellent reproducibility.
The cell suspension initially contains individual bacterial cells but, after the depletant is added, cells aggregate on the time scale of minutes in free solution. This was established for stationary phase E. coli29,32 and, in the ESI,† for the log-phase cells in this work. Aggregation reduces the numbers of singlet cells in suspension while capture is occurring. However, the adhered cells, those counted in Fig. 2A, are found to be mostly singlets. (The predominance of singlet adhered cells is evident in the micrographs later in the paper.)
Even though aggregates and singlet cells both exist in the suspension, hydrodynamic forces give rise to a preference for the capture of singlet cells because the shear force experienced by a cell or aggregate, resisting its capture, scales with the square of its size.51 The flow of aggregates through the chamber during capture of singlets is shown in the ESI.† The different hydrodynamic forces on singlets and aggregates, and the progressive reduction of singlets in the suspension from depletion aggregation explain why the initially rapid depletion-driven cell capture rates in Fig. 2A slow with time. These features, rapid initial cell capture and leveling off of the capture rate as a result of cell aggregation in the bulk suspension, reported here for log phase E. coli cells, were previously reported for stationary phase E. coli cells exposed to PEO depletants.29
In addition to producing cell adhesion on surfaces that otherwise would not capture cells, free polymer can enhance bacterial capture on surfaces that are moderately adhesive towards bacteria, such as acid-etched glass. Fig. 2B compares E. coli accumulation from buffer on acid etched glass (red data) to that from a suspension to which PEO depletant has been added (purple data). Without depletant, on the lower red curve, cells accumulate at a modest rate. Such slow capture is suggestive of an electrostatic barrier between the cell and a negative glass surface.36,52,53 When PEO is added to the free solution, cell capture occurs more rapidly, approaching the rates seen for depletion-limited capture of E. coli on a nonadhesive PEG surface. Here one of the blue curves from Fig. 2A has been replotted in Fig. 2B for comparison. It is seen that once depletant is added, the greater cell capture amounts and kinetics are dominated by depletion attractions rather than physico-chemical interactions between the cells and the glass.
Reversibility of cell capture
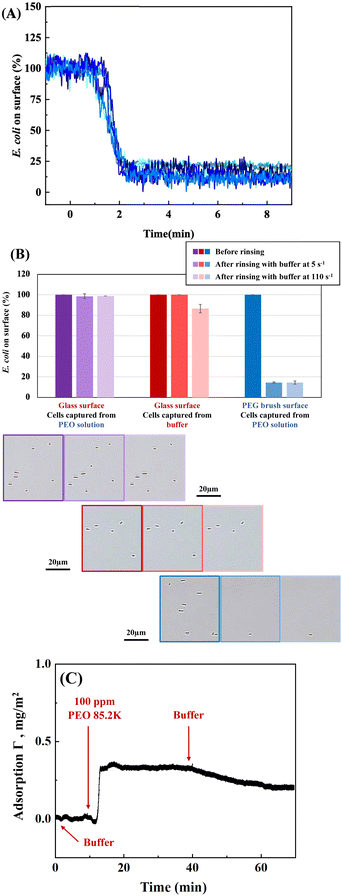
A key feature of depletion attractions is that they vanish upon removal of the depletant from solution. Consistent with this, Fig. 3A shows that cells captured on the nonadhesive PEG-coated surface are mostly released when the flowing PEO solution is replaced by buffer. The small fraction of cells remaining is reproducible for 5 runs conducted on different days, and cells are not removed by increasing the wall shear rate to 110 s−1. These E. coli are apparently retained by physico-chemical interactions as a result of flaws in the nonadhesive PEG brush54 or specific interactions with PEG. Alternately, it may be the case that the PEG brush surfaces are just at the cusp (considering variations in brush architecture) of inadequately shielding the substrate. Then osmotic pressure from free PEO slightly may dehydrate the brush or compresses cells against it to establish adhesion of some cells which are retained in the after PEO removal. In isolated instances when E. coli aggregates had been captured on the surface, when free PEO was replaced by buffer, the aggregates both dispersed and desorbed, consistent with the reversibility of depletion forces.29In contrast to the near complete release of cells from the non-adhesive PEG brush surfaces upon removal of free PEO, cells were almost entirely retained on glass at the original flow rate or after the wall shear rate was increased from 5 to 110 s−1. This is summarized in Fig. 3B, for depletion–enhanced cell capture on glass followed by replacement of the PEO depletant by flowing buffer. The results are compared with two controls: cells captured directly from buffer on glass and cells captured by PEO depletion on nonadhesive PEG surfaces, followed by removal of PEO depletant. A slight decrease in the numbers of retained cells, initially adsorbed from only buffer on glass (red data), is seen when the wall shear rate is increased from 5 to 110 s−1. Fig. 3B establishes that the presence of free polymer during cell capture strengthens physico-chemical adhesion on glass, at least within the first 30 minutes of cell capture, and this persists after removal of the free PEO.
The depletion–enhanced capture of E. coli on glass occurs despite the fact that PEO likely adsorbs on glass, established in the literature48,55–57 and shown for our particular PEO in Fig. 3C. In near-Brewster reflectometry, scattering and refractive index effects from the concentrated 1 wt% PEO solutions mask the adsorbed layer; therefore reflectometry was conducted with PEO solutions of 100 ppm. PEO adsorption on glass is found to be fast (transport limited) and, upon rinsing, PEO is retained on the surface for long times. However, prior studies demonstrated that within minutes of adsorption, even high molecular weight PEO chains can be displaced by challenger species,56,58 one explanation for bacterial capture. PEO displacement might occur due to the weak hydrogen bonding nature of PEO adsorption and, there may also be the opportunity for cell adhesion in the presence of some retained PEO chains. These observations confirm that adsorption of a depletant does not prevent the development of depletion attractions. Depletion attractions require polymer free in solution but persist in the presence of polymer adsorption. Further when PEO adsorption occurs concurrently with bacterial cell capture, cells can access the underlying surface and remain adhered after the depletant is removed. Hence depletion has an enhancement effect on cell adhesion already taking place through physico-chemical routes.
Cell orientation
An important factor in the developing morphology of biofilms is the orientation of the initially adhered cells.Fig. 4 compares the orientations of cells captured via depletion attractions on a non-adhesive surface to the orientations of cells captured on glass with or without free PEO in solution. The micrograph of Fig. 4A first establishes metrics of cell orientation showing, for example, a field of E. coli cells captured on a non-adhesive PEG-coated surface in the presence of a depletant. All the adhered cells in the field of view are singlets as was often the case. The schematic defines metrics for cell orientation, adhered by one end and standing vertically; adhered by one end and leaning over a bit; or appearing tipped almost flat to the surface (where it is not possible to see if only one end or the entire side of the bacterium is in contact). We estimate that standing cells are vertical to the surface to within 15°, that leaning cells are 15°–70° to the surface normal, and tipped cells are 70°–90° (flat) to the surface.36 In-plane cell orientation, which could be measured for cells that were leaning or tipped, is reported with respect to the flow direction.
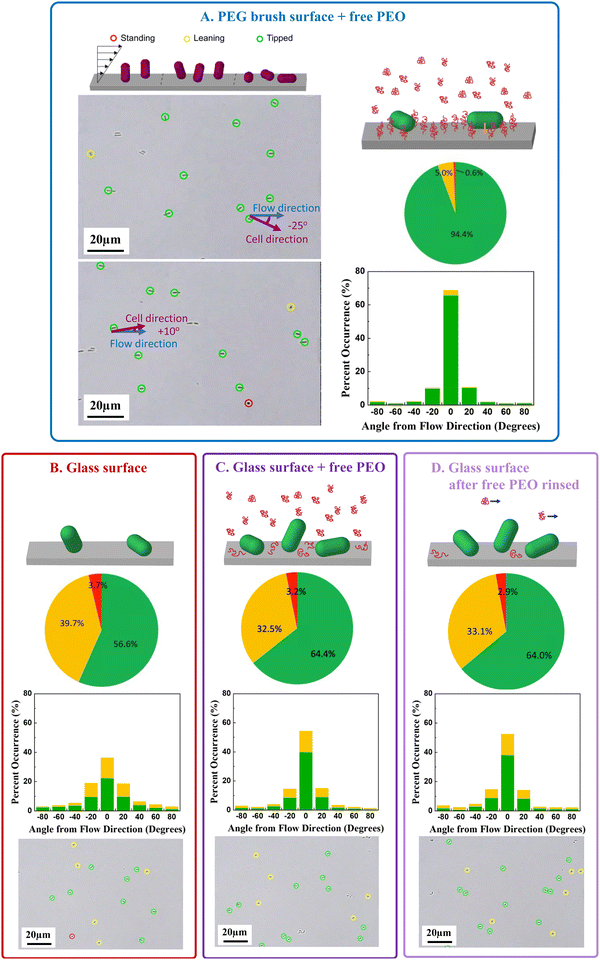 | ||
| Fig. 4 (A) Definition of standing/leaning/tipped cells and typical micrographs showing examples of each, along with cell alignment angles. Each panel in (B–D) includes a schematic, an example micrograph, a pie chart summarizing standing/leaning/tipped data from 15–20 different surface regions for 3 separate runs on separate days, and histograms for flow alignment of same cells. Four conditions are compared: (A) PEO-depletion driven capture on a PEG brush surface (B) adhesive cell capture on glass from buffer (C) cell capture on glass enhanced by PEO depletant and (D) PEO enhanced cell capture on glass after removal of PEO depletant. For all data, there is flow at a wall shear rate of 5 s−1. Color coding of frames matches Fig. 2 and 3. | ||
Fig. 4A–D summarize the cell orientation and reveal a dramatic influence of physico-chemical attractions versus depletion forces on cell orientation. Most pronounced is the strong in-plane orientation (pie chart) and flow alignment of cells (histogram) captured on the non-adhesive surfaces by depletion forces. Here the large majority of cells (94+%) leaned mostly flat to the surface and aligned with the flow, giving a remarkable appearance to the surface in Fig. 4A. This behavior contrasts previous reports of E. coli orientations on surfaces to which they adhered more nearly vertically by physico-chemical interactions on hydrophobic, cationic, and anionic glass surfaces.36 In Fig. 4B, E. coli adsorbed to glass from buffer, only 56% of the cells are tipped almost flat to the surface (pie chart) and their alignment in the shear direction (histograms) is less pronounced than cells held by depletion on nonadhesive surfaces in Fig. 4A. The data in Fig. 4B quantitatively reproduce our prior study on glass,36 though they were conducted by different personnel years apart.
When cells adhere on glass in the presence of 1 wt% dissolved PEO (Fig. 4C), the combined physico-chemical and depletion forces produce a cell population in which slightly greater numbers of cells are tipped flat to the surface (pie chart) compared with direct adhesion onto glass (Fig. 4B). With both depletion and physico-chemical attraction on glass (Fig. 4C), however, the cells are tipped far less flat to the surface (pie charts) compared with the case when depletants trapped cells on a non-adhesive substrate (Fig. 4A). The combination of depletion and physico-chemical interactions (Fig. 4C) also allows cells to align more with the flow (histograms) compared with direct adhesion on glass without depletion, but less than depletion-driven capture on non-adhesive surfaces (Fig. 4A). Fig. 4D shows that when cell capture on glass is enhanced by PEO in free solution, the cell alignment in the plane of the surface and in the flow direction is mostly retained after the PEO solution is replaced by buffer after 30 minutes. The greater fraction of cells tipped towards the surface in the presence of depletant is likely a result of the depletion attraction of the cell to the surface; however, since cells achieve a greater range of orientations on rapidly and strongly adherent cationic surfaces,36 the current findings suggest that cells turn over on brushy surfaces in the moments their adhesion to the surface is established, facilitating greater alignment in the flow direction than on physio-chemically adhesive surfaces. It follows that the orientations of cells adhered by depletion attractions are more aligned than their free orientations in flowing solution.
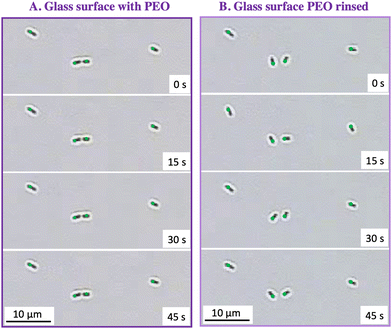
Surface mobility of captured cells
When the PEO solution is replaced by buffer, the depletion attraction is eliminated at the chamber wall and, if the surface is non-adhesive, cells escape as shown in Fig. 3A. However on glass, physico-chemical interactions hold cells on the surface after removal of depletant, summarized in Fig. 3B. We observed that cells which were mostly immobile and were flat to glass surfaces in the presence of depletant, tipped slightly up and rocked in place, adhering by only one end after removal of depletant. This was particularly obvious if the flow was stopped after removal of depletant. This rocking on glass was Brownian rather than active, since cells lacked flagella.42Table 1 summarizes the percentages of cells that rocked or wiggled, averaging data for three capture runs on separate days, examining two fields in each run, and tallying behaviors of 30–80 cells in each run.| Fraction of wiggling or rocking cells | |
|---|---|
| PEG brush surface + PEO depletant | 3% ± 1% |
| Glass surface + PEO depletant | 18% ± 3% |
| Glass surface + PBS | 56% ± 1% |
| Glass surface + PEO depletant, then replace PEO with PBS | 67 ± 3% |
The results in Table 1 were compiled by manually observing video footage of each surface region after the conditions of interest were achieved and then the pump was shut off. This information is made clear, as shown in the examples Fig. 5, for a glass surface in (A) with PEO depletant and later (B) after replacement of PEO by buffer.
The ability of cells to wiggle on glass in the absence of PEO suggests that the physico-chemical adhesion between the cell envelope and glass either is very weak or it occurs over a very small contact area. This may be, in part, a consequence of the short range character of physico-chemical adhesion to glass, contrasted with the longer range PEO-induced depletion attractions that drive cells to tip down a surface and hold them there. Further, the in-place mobility of cells adhered to glass appears to be a consequence of interactions with the glass itself, evidenced by the behavior of cells captured on glass in the absence of PEO. Thus, PEO adsorption during cell capture, which might be expected to block adsorption sites on the glass and weaken cell adhesion, seems not to be the cause of cell rocking on glass after PEO rinsing.
Discussion
Adhesive interactions and kinetics
The features of E. coli capture on glass in the presence of free PEO are consistent with the expected concentration of polymer needed to produce substantial depletion attractions and with our understanding of the additivity of interfacial potentials. The reversibility of depletion attractions reveals the presence or lack of other interactions: on fundamentally nonadhesive surfaces such as PEG brushes, cells are released upon removal of depletant, but on adhesive surfaces such as glass, the underlying cell-glass interactions remain, allowing cells to rock or wiggle in place. Relevant to biofilm formation, the presence of depletant can enhance cell capture, quickly elevating the numbers of captured cells which would be otherwise slow to accumulate on a surface such as glass, for instance as a result of electrostatic barriers to adhesion.When PEO acts as a depletant, its concurrent adsorption to glass appears not to hinder cell capture. The reason may be because PEO chains residing on a surface for a short time are easily displaced by challenging species, especially those higher in molecular weight.56,58 Indeed, the fact that PEO homopolymer adsorption alone provides poor protection against cell and protein adhesion is the reason why more sophisticated approaches have been pursued to anchor PEG to surfaces, enabling retention of a non-adhesive coating. Our observations further underline the fact that it is the presence of free polymer in solution which gives rise to depletion attractions at an interface. With polymer remaining in solution, depletion forces can cause cell adhesion to a bare surface or to a surface containing some adsorbed depletant molecules. Thus, adsorption of the depletant is not contrary to depletion-attractions, as long as some free polymer remains.
Another interesting fact borne out in this study is that strong cell adhesion to a surface does not necessarily ensure a flat cell configuration. On cationic surfaces that strongly adhere oppositely charged objects, both E. coli cells36 and negative rod-shaped silica microparticles59 are captured and trapped in mostly end-adhered configurations, unable to rotate down and lie flat on the surface. In contrast the flat (or nearly so) adhered cell configurations produced by depletion attractions suggest cell mobility in the instants of capture, so that cells may turn flat upon initial surface contact. Once flat to the surface, however, depletion attractions can be sufficient to hold cells still, depending on the depletant concentration which dictates the strength of the depletion attraction. The adhesion of E. coli on glass and other negative surfaces is interesting, exhibiting a preference for end-on adsorption36,60 and the observed rotational wiggling of nonmotile cells,61 or aggressive circling of trapped flagella-containing cells also including Pseudomonas.62–64
Potential impact on colony and biofilm growth
Recent work has suggested the orientation of adhered cells has a large impact on the structure of growing microcolonies and biofilms.37–40 When founder cells, those captured initially from solution, are flat to the surface, early cell division produces daughter cells which also lie flat to the surface. The resulting microcolony can consist of a monolayer containing hundreds of cells. In such a monolayer, all the cells have relatively good transport and are directly wetted by solution. At some point, cell division causes cells towards the center of the colony to buckle upwards, so that further division produces at least one cellular overlayer, producing 3D structures in the biofilm. One might imagine that end-adsorbed cells never form much of a monolayer with daughter cells potentially escaping the interface, or adsorbing nearby, especially in the presence of depletant. Thus the depletion interactions, especially in combination with surfaces that hold cells by physio-chemical interactions, hold potential to produce colonies and biofilms of markedly different structures.Conclusions
This work established how depletion attractions from PEO in free solution can combine with other surface forces giving diverse cell capture behavior on different surfaces. Free PEO in solution drives the capture of log phase E. coli cells on PEG-coated surfaces that otherwise would not capture these cells. This behavior was compared to capture of the same cells on cell-adhesive glass surfaces from buffer and PEO solutions. Depletion attractions increased the rate of cell capture and the numbers of cells captured above what they would otherwise be on glass. This enhanced cell capture occurred because the attractive depletion potential can combine with other attractive interactions and dominate kinetics. The result is particularly pronounced when the other potentials are shorter in range or contain repulsive barriers.Removal of free PEO from solution eliminates depletion forces, leaving the physico-chemical interactions of E. coli with the underlying surface, either a mostly non-adhesive PEG coating or adhesive glass. In the former case, cells are mostly released from the surface, but on glass, cells are retained by physico-chemical interactions at one end of the cell and their in-place mobility increases. This work further demonstrated how the adsorption of the depletant does not prevent the development of depletion forces, which only require sufficient concentrations of free polymer (or other depletant species) in solution. Depletion forces, for the case of 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 g mol−1 PEO at 1 wt% in solution, are sufficient to hold cells still on the surface and flat to the nonadhesive surfaces. On glass, in the presence of PEO, cells are held still, but their physico-chemical adhesion by one end becomes evident upon removal of the depletion interaction.
200 g mol−1 PEO at 1 wt% in solution, are sufficient to hold cells still on the surface and flat to the nonadhesive surfaces. On glass, in the presence of PEO, cells are held still, but their physico-chemical adhesion by one end becomes evident upon removal of the depletion interaction.
These findings may have impact on our understanding of biofilm formation under conditions that give rise to different cell-surface interactions, especially considering the prevalence of polymers in bacteria-rich environments and the non-specific character of depletion and electrostatic interactions, which act across different bacterial types and also colloidal particles.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by NSF1848065. M. N. Smith acknowledges partial support from an NIH traineeship, the Biotechnology Program Fellowship, T32 GM135096. We are grateful to support from the Siegrist Lab: R. Gordon and S. Rivera for growing bacteria and measuring their dimensions and to S. Siegrist for helpful discussions. We also thank Z. Xu for sharing his computer codes to analyzing cell counts.References
- S. Asakura and F. Oosawa, On Interactions between 2 Bodies Immersed in a Solution of Macromolecules, J. Chem. Phys., 1954, 22, 1255–1256 CrossRef CAS.
- S. Asakura and F. Oosawa, Interaction between Particles Suspended in Solutions of Macromolecules, J. Polym. Sci., 1958, 33, 183–192 CrossRef CAS.
- J. F. Joanny, L. Leibler and P. G. DeGennes, Effects of Polymer Solutions on Colloid Stability, J. Polym. Sci., Polym. Phys., 1979, 17, 1073–1084 CrossRef CAS.
- M. Dijkstra, R. van Roij and R. Evans, Phase Diagram of Highly Asymmetric Binary Hard-Sphere Mixtures, Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top., 1999, 59, 5744–5771 CrossRef CAS PubMed.
- J. K. Armstrong, R. B. Wenby, H. J. Meiselman and T. C. Fisher, The Hydrodynamic Radii of Macromolecules and Their Effect on Red Blood Cell Aggregation, Biophys. J., 2004, 87, 4259–4270 CrossRef CAS PubMed.
- J. Bergenholtz, W. C. K. Poon and M. Fuchs, Gelation in Model Colloid-Polymer Mixtures, Langmuir, 2003, 19, 4493–4503 CrossRef CAS.
- W. C. K. Poon, The Physics of a Model Colloid-Polymer Mixture, J. Phys.: Condens. Matter, 2002, 14, R859–R880 CrossRef CAS.
- W. C. K. Poon, A. D. Pirie and P. N. Pusey, Gelation in Colloid-Polymer Mixtures, Faraday Discuss., 1995, 101, 65–76 RSC.
- I. Szilagyi, G. Trefalt, A. Tiraferri, P. Maroni and M. Borkovec, Polyelectrolyte Adsorption, Interparticle Forces, and Colloidal Aggregation, Soft Matter, 2014, 10, 2479–2502 RSC.
- M. M. Santore, W. B. Russel and R. K. Prudhomme, A 2-Component Model for the Phase Behavior of Dispersions Containing Associative Polymer, Macromolecules, 1989, 22, 1317–1325 CrossRef CAS.
- K. Binder, P. Virnau and A. Statt, Perspective: The Asakura Oosawa Model: A Colloid Prototype for Bulk and Interfacial Phase Behavior, J. Chem. Phys., 2014, 141, 140901 CrossRef PubMed.
- J. M. Brader, M. Dijkstra and R. Evans, Inhomogeneous Model Colloid-Polymer Mixtures: Adsorption at a Hard Wall, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2001, 63, 041405 CrossRef CAS PubMed.
- A. A. Louis, P. G. Bolhuis, E. J. Meijer and J. P. Hansen, Density Profiles and Surface Tension of Polymers near Colloidal Surfaces, J. Chem. Phys., 2002, 116, 10547–10556 CrossRef CAS.
- K. Sandomirski, E. Allahyarov, H. Lowen and S. U. Egelhaaf, Heterogeneous Crystallization of Hard-Sphere Colloids near a Wall, Soft Matter, 2011, 7, 8050–8055 RSC.
- S. X. Ji and J. Y. Walz, Depletion Forces and Flocculation with Surfactants, Polymers and Particles - Synergistic Effects, Curr. Opin. Colloid Interface Sci., 2015, 20, 39–45 CrossRef CAS.
- M. M. Santore, W. B. Russel and R. K. Prudhomme, Experimental and Theoretical Study of Phase Transitions Indcued in Colloidal Dipsersions by Associative Polymers, Faraday Discuss., 1990, 90, 323–333 RSC.
- B. Vincent, J. Clarke and K. G. Barnett, The Flocculation of Nonaqueous, Sterically-Stabilized Latex Dispersions in the Presence of Free Polymer, Colloids Surf., 1986, 17, 51–65 CrossRef CAS.
- B. Vincent, J. Edwards, S. Emmett and A. Jones, Depletion Flocculation in Dispersions of Sterically-Stabilized Particles (Soft Spheres), Colloids Surf., 1986, 18, 261–281 CrossRef CAS.
- J. Nam and M. M. Santore, Depletion Versus Deflection: How Membrane Bending Can Influence Adhesion, Phys. Rev. Lett., 2011, 107, 078101 CrossRef PubMed.
- N. J. Smith and P. A. Williams, Depletion Flocculation of Polystyrene Latices by Water-Soluble Polymers, J. Chem. Soc., Faraday Trans., 1995, 91, 1483–1489 RSC.
- Y. N. Ohshima, H. Sakagami, K. Okumoto, A. Tokoyoda, T. Igarashi, K. B. Shintaku, S. Toride, H. Sekino, K. Kabuto and I. Nishio, Direct Measurement of Infinitesimal Depletion Force in a Colloid-Polymer Mixture by Laser Radiation Pressure, Phys. Rev. Lett., 1997, 78, 3963–3966 CrossRef CAS.
- D. Rudhardt, C. Bechinger and P. Leiderer, Direct Measurement of Depletion Potentials in Mixtures of Colloids and Nonionic Polymers, Phys. Rev. Lett., 1998, 81, 1330–1333 CrossRef CAS.
- D. Rudhardt, C. Bechinger and P. Leiderer, Repulsive Depletion Interactions in Colloid-Polymer Mixtures, J. Phys.: Condens. Matter, 1999, 11, 10073–10078 CrossRef CAS.
- J. Bibette, F. L. Calderon and P. Poulin, Emulsions: Basic Principles, Rep. Prog. Phys., 1999, 62, 969–1033 CrossRef CAS.
- M. W. Rampling, H. J. Meiselman, B. Neu and O. K. Baskurt, Influence of Cell-Specific Factors on Red Blood Cell Aggregation, Biorheology, 2004, 41, 91–112 CAS.
- J. Surh, E. A. Decker and D. J. McClements, Influence of Ph and Pectin Type on Properties and Stability of Sodium-Caseinate Stabilized Oil-in-Water Emulsions, Food Hydrocolloids, 2006, 20, 607–618 CrossRef CAS.
- M. K. Porter, A. P. Steinberg and R. F. Ismagilov, Interplay of Motility and Polymer-Driven Depletion Forces in the Initial Stages of Bacterial Aggregation, Soft Matter, 2019, 15, 7071–7079 RSC.
- K. E. Eboigbodin, J. R. A. Newton, A. F. Routh and C. A. Biggs, Role of Nonadsorbing Polymers in Bacterial Aggregation, Langmuir, 2005, 21, 12315–12319 CrossRef CAS PubMed.
- W. A. Niu, S. L. Rivera, M. S. Siegrist and M. M. Santore, Depletion Forces Drive Reversible Capture of Live Bacteria on Non-Adhesive Surfaces, Soft Matter, 2021, 17, 8185–8194 RSC.
- J. Schwarz-Linek, G. Dorken, A. Winkler, L. G. Wilson, N. T. Pham, C. E. French, T. Schilling and W. C. K. Poon, Polymer-Induced Phase Separation in Suspensions of Bacteria, EPL, 2010, 89, 68003 CrossRef.
- J. Schwarz-Linek, C. Valeriani, A. Cacciuto, M. E. Cates, D. Marenduzzo, A. N. Morozov and W. C. K. Poon, Phase Separation and Rotor Self-Assembly in Active Particle Suspensions, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 4052–4057 CrossRef CAS PubMed.
- J. Schwarz-Linek, A. Winkler, L. G. Wilson, N. T. Pham, T. Schilling and W. C. K. Poon, Polymer-Induced Phase Separation in Escherichia Coli Suspensions, Soft Matter, 2010, 6, 4540–4549 RSC.
- G. Dorken, G. P. Ferguson, C. E. French and W. C. K. Poon, Aggregation by Depletion Attraction in Cultures of Bacteria Producing Exopolysaccharide, J. R. Soc., Interface, 2012, 9, 3490–3502 CrossRef PubMed.
- P. R. Secor, L. A. Michaels, A. Ratjen, L. K. Jennings and P. K. Singh, Entropically Driven Aggregation of Bacteria by Host Polymers Promotes Antibiotic Tolerance in Pseudomonas Aeruginosa, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 10780–10785 CrossRef CAS PubMed.
- P. R. Secor, L. A. Michaels, D. C. Bublitz, L. K. Jennings and P. K. Singh, The Depletion Mechanism Actuates Bacterial Aggregation by Exopolysaccharides and Determines Species Distribution & Composition in Bacterial Aggregates, Front. Cell. Infect. Microbiol., 2022, 12, 869736 CrossRef CAS PubMed.
- Z. Xu, W. A. Niu, S. L. Rivera, M. T. Tuominen, M. S. Siegrist and M. M. Santore, Surface Chemistry Guides the Orientations of Adhering E. Coli Cells Captured from Flow, Langmuir, 2021, 37, 7720–7729 CrossRef CAS PubMed.
- D. Dell’Arciprete, M. L. Blow, A. T. Brown, F. D. C. Farrell, J. S. Lintuvuori, A. F. McVey, D. Marenduzzo and W. C. K. Poon, A Growing Bacterial Colony in Two Dimensions as an Active Nematic, Nat. Commun., 2018, 9, 4190 CrossRef PubMed.
- M. C. Duvernoy, T. Mora, M. Ardre, V. Croquette, D. Bensimon, C. Quilliet, J. M. Ghigo, M. Balland, C. Beloin, S. Lecuyer and N. Desprat, Asymmetric Adhesion of Rod-Shaped Bacteria Controls Microcolony Morphogenesis, Nat. Commun., 2018, 9, 1120 CrossRef PubMed.
- C. Y. Fei, S. Mao, J. Yan, R. Alert, H. A. Stone, B. L. Bassler, N. S. Wingreen and A. Kosmrlj, Nonuniform Growth and Surface Friction Determine Bacterial Biofilm Morphology on Soft Substrates, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 7622–7632 CrossRef CAS PubMed.
- D. Volfson, S. Cookson, J. Hasty and L. S. Tsimring, Biomechanical Ordering of Dense Cell Populations, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 15346–15351 CrossRef PubMed.
- P. Nghe, S. Boulineau, S. Gude, P. Recouvreux, J. S. van Zon and S. J. Tans, Microfabricated Polyacrylamide Devices for the Controlled Culture of Growing Cells and Developing Organisms, PLoS One, 2013, 8, e75537 CrossRef CAS PubMed.
- M. K. Shave and M. M. Santore, Motility Increases the Numbers and Durations of Cell-Surface Engagements for Escherichia Coli Flowing near Poly(Ethylene Glycol)-Functionalized Surfaces, ACS Appl. Mater. Interfaces, 2022, 14, 34342–34353 CrossRef CAS PubMed.
- https://Https://Onlinelibrary.Wiley.Com/Doi/Pdf/10.1111/Mmi.13264 .
- G. L. Kenausis, J. Voros, D. L. Elbert, N. P. Huang, R. Hofer, L. Ruiz-Taylor, M. Textor, J. A. Hubbell and N. D. Spencer, Poly(L-Lysine)-G-Poly(Ethylene Glycol) Layers on Metal Oxide Surfaces: Attachment Mechanism and Effects of Polymer Architecture on Resistance to Protein Adsorption, J. Phys. Chem. B, 2000, 104, 3298–3309 CrossRef CAS.
- N. P. Huang, R. Michel, J. Voros, M. Textor, R. Hofer, A. Rossi, D. L. Elbert, J. A. Hubbell and N. D. Spencer, Poly(L-Lysine)-G-Poly(Ethylene Glycol) Layers on Metal Oxide Surfaces: Surface-Analytical Characterization and Resistance to Serum and Fibrinogen Adsorption, Langmuir, 2001, 17, 489–498 CrossRef CAS.
- J. L. Dalsin, L. J. Lin, S. Tosatti, J. Voros, M. Textor and P. B. Messersmith, Protein Resistance of Titanium Oxide Surfaces Modified by Biologically Inspired Mpeg-Dopa, Langmuir, 2005, 21, 640–646 CrossRef CAS PubMed.
- S. Gon, M. Bendersky, J. L. Ross and M. M. Santore, Manipulating Protein Adsorption Using a Patchy Protein-Resistant Brush, Langmuir, 2010, 26, 12147–12154 CrossRef CAS PubMed.
- Z. G. Fu and M. M. Santore, Poly(Ethylene Oxide) Adsorption onto Chemically Etched Silicates by Brewster Angle Reflectivity, Colloids Surf., A, 1998, 135, 63–75 CrossRef CAS.
- A. Toscano and M. M. Santore, Fibrinogen Adsorption on Three Silica-Based Surfaces: Conformation and Kinetics, Langmuir, 2006, 22, 2588–2597 CrossRef CAS PubMed.
- B. Fang, S. Gon, K. Nusslein and M. M. Santore, Surfaces for Competitive Selective Bacterial Capture from Protein Solutions, ACS Appl. Mater. Interfaces, 2015, 7, 10275–10282 CrossRef CAS PubMed.
- A. J. Goldman, R. G. Cox and H. Brenner, Slow Viscous Motion of a Sphere Parallel to a Plane Wall. 2. Couette Flow, Chem. Eng. Sci., 1967, 22, 653–660 CrossRef CAS.
- N. P. Boks, W. Norde, H. C. van der Mei and H. J. Busscher, Forces Involved in Bacterial Adhesion to Hydrophilic and Hydrophobic Surfaces, Microbiol. Sgm, 2008, 154, 3122–3133 CrossRef CAS PubMed.
- B. K. Li and B. E. Logan, Bacterial Adhesion to Glass and Metal-Oxide Surfaces, Colloids Surf., B, 2004, 36, 81–90 CrossRef CAS PubMed.
- S. Gon, K. N. Kumar, K. Nusslein and M. M. Santore, How Bacteria Adhere to Brushy Peg Surfaces: Clinging to Flaws and Compressing the Brush, Macromolecules, 2012, 45, 8373–8381 CrossRef CAS PubMed.
- Z. L. Fu and M. Santore, Effect of Layer Age and Interfacial Relaxations on the Self-Exchange Kinetics of Poly(Ethylene Oxide) Adsorbed on Silica, Macromolecules, 1999, 32, 1939–1948 CrossRef CAS.
- E. Mubarekyan and M. M. Santore, Influence of Molecular Weight and Layer Age on Self-Exchange Kinetics for Saturated Layers of Peo in a Good Solvent, Macromolecules, 2001, 34, 4978–4986 CrossRef CAS.
- R. J. Owen, J. C. Crocker, R. Verma and A. G. Yodh, Measurement of Long-Range Steric Repulsions between Microspheres Due to an Adsorbed Polymer, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2001, 64, 011401 CrossRef CAS PubMed.
- E. Mubarekyan and M. M. Santore, Energy Barrier to Self-Exchange between Peo Adsorbed on Silica and in Solution, Macromolecules, 2001, 34, 7504–7513 CrossRef CAS.
- M. K. Shave, A. Balciunaite, Z. Xu and M. M. Santore, Rapid Electrostatic Capture of Rod-Shaped Particles on Planar Surfaces: Standing up to Shear, Langmuir, 2019, 35, 13070–13077 CrossRef CAS PubMed.
- J. F. Jones, J. D. Feick, D. Imoudu, N. Chukwumah, M. Vigeant and D. Velegol, Oriented Adhesion of Escherichia Coli to Polystyrene Particles, Appl. Environ. Microbiol., 2003, 69, 6515–6519 CrossRef CAS PubMed.
- T. Vissers, N. Koumakis, M. Hermes, A. T. Brown, J. Schwarz-Linek, A. Dawson and W. C. K. Poon, Dynamical Analysis of Bacteria in Microscopy Movies, PLoS One, 2019, 14, e0217823 CrossRef CAS PubMed.
- J. C. Conrad, M. L. Gibiansky, F. Jin, V. D. Gordon, D. A. Motto, M. A. Mathewson, W. G. Stopka, D. C. Zelasko, J. D. Shrout and G. C. L. Wong, Flagella and Pili-Mediated near-Surface Single-Cell Motility Mechanisms in P. Aeruginosa, Biophys. J., 2011, 100, 1608–1616 CrossRef CAS PubMed.
- S. Sharma, Y. A. Jaimes-Lizcano, R. B. McLay, P. C. Cirino and J. C. Conrad, Subnanometric Roughness Affects the Deposition and Mobile Adhesion of Escherichia Coli on Silanized Glass Surfaces, Langmuir, 2016, 32, 5422–5433 CrossRef CAS PubMed.
- K. C. Marshall, R. Stout and R. Mitchell, Mechanism of Initial Events in Sorption of Marine Bacteria to Surfaces, J. Gen. Microbiol., 1971, 68, 337–348 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Supporting information includes documentation of depletion aggregation in bulk solution and flow of aggregates through the flow chamber. See DOI: https://doi.org/10.1039/d2sm01248k |
| This journal is © The Royal Society of Chemistry 2022 |