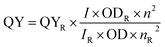Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Water stable, red emitting, carbon nanoparticles stimulate 3D cell invasion via clathrin-mediated endocytic uptake†
Udisha
Singh‡
a,
Aditya Guduru
Teja‡
b,
Shanka
Walia‡
a,
Payal
Vaswani
a,
Sameer
Dalvi
 bc and
Dhiraj
Bhatia
bc and
Dhiraj
Bhatia
 *ac
*ac
aBiological Engineering Discipline, Indian Institute of Technology Gandhinagar, Palaj, Gujarat 382355, India. E-mail: dhiraj.bhatia@iitgn.ac.in
bChemical Engineering Discipline, Indian Institute of Technology Gandhinagar, Palaj, Gujarat 382355, India
cCenter for Biomedical Engineering, Indian Institute of Technology Gandhinagar, Palaj, Gujarat 382355, India
First published on 26th January 2022
Abstract
Bright fluorescent nanoparticles with excitation and emission towards the red end of the spectrum are highly desirable in the field of bioimaging. We present here a new class of organic carbon-based nanoparticles (CNPs) with a robust quantum yield and fluorescence towards the red region of the spectrum. Using organic substrates such as para-phenylenediamine (PPDA) dispersed in diphenyl ether under reflux conditions, we achieved scalable amounts of CNPs with an average size of 27 nm. These CNPs were readily taken up by different mammalian cells, and we show that they prefer clathrin-mediated endocytosis for their cellular entry route. Not only can these CNPs be specifically taken up by cells, but they also stimulate cellular processes such as cell invasion from 3D spheroid models. This new class of CNPs, which have sizes similar to those of proteinaceous ligands, hold immense potential for their surface functionalization. These could be explored as promising bioimaging agents for biomedical imaging and intracellular drug delivery.
1 Introduction
Bioimaging of specific biomolecules, cellular processes, entire cells or tissues is one of the best non-invasive ways to understand and visualize biological activity at the cellular level.1 Recent years have witnessed the emergence of a plethora of fluorescent nanomaterials, having gained much attention in the field of bioimaging owing to their small sizes, low cost, highly tuneable fluorescent properties, and their ease of interfacing with biological systems leading to widespread applications.2,3 Many different fluorescent nanomaterials have already been used, such as rare-earth semiconductor quantum dots,4,5 organic dyes,6 and polymer dots.7However, their applications are still extremely limited due to their limited stability, toxicity, and poor water stability, thus hindering their use for biomedical and biological applications. Carbon-based nanomaterials, especially carbon dots (CDs, including carbon nanoparticles, graphene quantum dots, and carbon quantum dots), have gained much attention because of their small size, biocompatibility, low toxicity, stable fluorescent properties, and low cost of synthesis.8,9 All these carbon nanomaterials have an sp2 hybridized carbon core and surface functional groups.10
Much research had already been done in the synthesis methods, doping and functionalization, tunable emission, etc. for CNPs. Still, it is challenging to tune the emission range of CNPs towards the red region of electromagnetic spectra. The structure of CNPs (by doping, surface chemistry, and size) and their synthesis methods significantly affect their emission properties. Most of the CNPs synthesised usually show fluorescence in the blue-green region, which is not of much use in bioimaging because their excitation is in the UV region, which not only induces autofluorescence to disturb the signal of CNPs but also damages the cells and tissues.11,12 CNPs emitting in the near-infrared region could overcome all the shortcomings of lower wavelength emissive CNPs and make them applicable for bioimaging purposes. Few studies on carbon nanoparticles showing fluorescence in the near-infrared region have been reported (ESI Table 1†). For example, Hui et al. synthesised red CDs with optical emission at 627 nm.13 Xiong and co-workers synthesised red emissive CDs with optical emission at 625 nm using PPDA and urea by the hydrothermal synthesis method.14 Although a few red emissive CNPs have been obtained showing fluorescence in the near-infrared region, the products obtained are in tiny amounts, and the synthesis process can be tedious, limiting the use of these CNPs. Also, the size and shape of nanoparticles matter a lot for their use in biomedical applications.
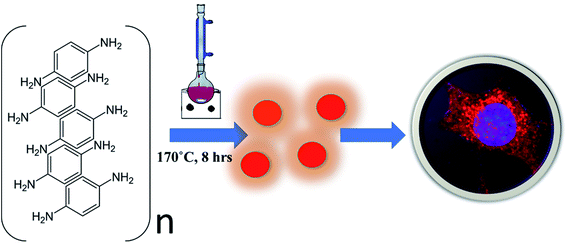
We modified the existing methods in this direction by using para-phenylenediamine (PPDA) as the precursor material to achieve the synthesis of red emissive fluorescent CNPs. The CNPs obtained by reducing para-phenylenediamine (PPDA) exhibit excitation and emission spectra in the red region of the visible spectrum. The CNPs are synthesised by a one-step reflux reaction in a round bottom flask, which is simple and cost-effective, and a large amount of product is obtained with a yield of 87.65%, which is much higher as compared to other synthesis methods. To use the CNPs for biomedical applications, it is essential to know through which pathway they enter the cells. These CNPs were seen to be readily taken up by different mammalian cells via a very specific endocytic route called clathrin-mediated endocytosis (CME). The long-wavelength emission helps in avoiding the strong absorption from biological samples and preventing enhanced emission due to autofluorescence from the cells. Hua X. et al. also synthesised red CNPs with a size of approximately 2 nm using a similar precursor material para-phenylenediamine (PPDA). However, their quantum yield in water and ethanol compared to that of the CNPs we have synthesised is lower. They used hydrothermal methods of synthesis instead of the reflux condensation reaction we have used in the current study.15 Similarly, Wu and coworkers synthesised carbon dots with the highest emission at 530 nm wavelength and used them not only to stain the nucleolus but also for drug delivery and advanced bioimaging applications. They used meta-phenylene diamine instead of para-phenylenediamine as the precursor material and used the hydrothermal method of synthesis. They successfully transferred protoporphyrin IX into the nucleolus using carbon dots (CDs). The CNPs we have synthesised do not explicitly target any organelle, but they easily enter the cell cytoplasm and can be used for drug delivery, just like the CDs synthesised by Wu and coworkers.16 Not only were the CNPs actively taken up by cells, but they also stimulated different cellular physiological processes such as cell invasion in 3D spheroid models. Overall, our results indicate that the CNPs presented here are good bioimaging materials and can lay a path for the development of advanced bioimaging tools (Scheme 1).
2 Results and discussion
2.1 Synthesis and characterization of CNPs
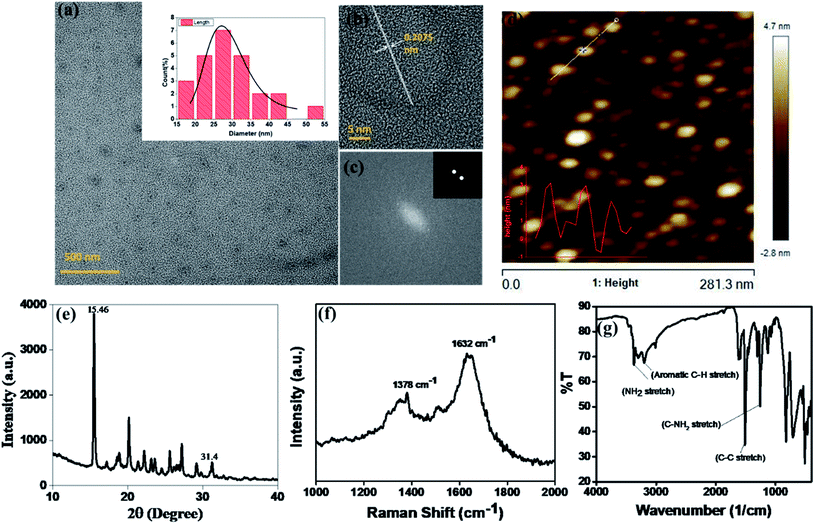
Red fluorescence emitting CNPs were synthesised through reflux reaction mediated pyrolysis of para-phenylenediamine (PPDA) in diphenyl ether for 8 h at 170 °C. The size and morphology of the CNPs were characterized using transmission electron microscopy (TEM). TEM images show that these particles are adequately dispersed and possess a spherical disk-like structure. The mean diameter of the fluorescent CNPs is 26.7 ± 3.256 nm while counting 100 nanoparticles displayed in Fig. 1a. Particle size measurement was performed with ImageJ software using a scaled image and analysed using Origin software. From the high-resolution transmission electron microscopy (HRTEM) image in Fig. 1b and c, lattice fringes with a lattice distance of 0.2075 nm, which is in good accordance with the (101) carbon planes, were obtained, suggesting that the CNPs synthesised are crystalline in nature.17 The atomic force microscopy (AFM) image reveals a topological height of 2.6 nm, indicating that fluorescent CNPs contain an average of 7 to 8 layers of graphene sheets (Fig. 1d).18To further understand the structure and composition of CNPs, we performed X-ray diffraction spectroscopy (XRD), and Raman and Fourier transform infrared (FT-IR) spectroscopies of fluorescent CNPs. The XRD profile of CNPs shows several peaks that may be due to the ordered stacking of the synthesised CNPs. The number of peaks also shows the complex mixture of crystalline components in the sample solution of CNPs.19 The peaks at 15.46° and 31.4° correspond to the precursor material PPDA, revealing some of the precursor material's chemical structure is still intact even when the CNPs are formed as shown in Fig. 1e. Raman spectroscopy was also used as a non-destructive way to understand the degree of graphitization of CNPs. We obtained two peaks at 1378 cm−1 and 1632 cm−1 representing the D band and G band, respectively (Fig. 1f). The D band constitutes the vibrations of disordered graphite or glassy carbon. The G band represents the in-plane displacement of carbon atoms in a two-dimensional hexagonal lattice. Also, the value of ID/IG is being used to evaluate the degree of graphitization.21 The intensity of the G band is higher than the peak of the D band with a ID/IG value of 0.844 (Fig. 1f), implying that more sp2-hybridized carbon is present compared to sp3 hybridized carbon. The FTIR spectrum gives a qualitative idea about the functional groups present on the surface of CNPs. CNPs mainly contain C, O and N elements forming chemical bonds –NH2 (3364 cm−1), C–C (1510 cm−1) and C–NH2 (1257 cm−1) as shown in Fig. 1g. The absorption peak obtained at 3364 cm−1 confirms the presence of an amine group on the surface of FCNPs, also proving the nitrogen doping of CNPs. The C–H stretching occurs around 3180 cm, ensuring that the CDs contain the aromatic ring of carbon.
2.2 Optical properties of CNPs reveal stable, pH-responsive, red-emitting fluorescence
To understand the optical properties of CNPs, UV-visible absorption and fluorescence emission and excitation spectra were measured. We obtain three absorbance peaks in the UV-vis spectra at 241 nm, 306 nm, and 415 nm (Fig. 2a). The absorbance peak at 241 nm is assigned to the π–π* transition from the aromatic carbon structure, the shoulder peak at 306 nm is assigned to the π–n* transition from functional groups with lone pair electrons and the absorbance peak at the longer wavelength 415 nm is assigned to the energy level transition from the angstrom-sized conjugated π-structure present in CNPs.22 The fluorescence emission spectra of fluorescent CNPs were measured as a function of different excitation wavelengths. The fluorescence (FL) spectra of CNPs show a maximum peak at 622 nm with an excitation wavelength of 480 nm. As shown in (Fig. 2b), the FL spectra of CNPs don't shift with respect to the change in the excitation wavelength of 400 to 520 nm range, indicating that fluorescent CNPs have distinct excitation independent FL behaviour, which is very seldom observed. This also shows that the CNPs exhibit a quite homogeneous size distribution, composition and surface state. The sp2 hybridized carbon network in CNPs can also be one of the reasons for independent excitation fluorescence. The CNPs show a brownish-red colour under white light and a dark red colour under UV light illumination in aqueous solution, as shown in the inset of Fig. 2a.The fluorescence quantum yield of optimal CNPs is 5.86% in Milli-Q water and 87% in ethanol by referring to the standard rhodamine B (QY = 33%). This also shows that the synthesised CNPs are more dispersed in organic solvents and show a higher QY and fluorescence. The CNPs synthesised were redispersed in different solvents: water, hexane, acetone, acetonitrile, dimethyl sulfoxide (DMSO), N,N-dimethyl formamide (DMF), ethyl acetate, isopropyl alcohol (IPA), ethanol and methanol. All ten solvents have different polarities, and their emission spectra also shift with a change in solvent, as shown in ESI Fig. S2a.† The colour of the CNPs changes in different solvents showing that the fluorescence of CNPs not only depends on functional groups present on their surface but also the type of the dispersant used.20
For cellular stability applications, pH-dependent studies of CNPs were done by monitoring changes in their fluorescence signal as a function of pH. Different pH solutions in Milli-Q water were prepared (3, 5, 7 & 12) and CNPs (0.5 mg mL−1) were dispersed in it, and emission spectra were recorded as shown in Fig. 2d. The fluorescent CNPs show an increase in fluorescence with an increase in pH, hence showing great potential in pH sensing. The lowest fluorescence intensity was obtained at pH 3 and the highest intensity at pH 12. The percentage increase in intensity from pH 3 to pH 7 is 38%, whereas the increase in fluorescence intensity from pH 7 to pH 12 is approximately 31%. The pH sensitivity of CNPs may be due to protonation and deprotonation of oxygen-containing groups on the surface.23 Also, there is a significant colorimetric change of CNPs dispersed in different pH solutions.
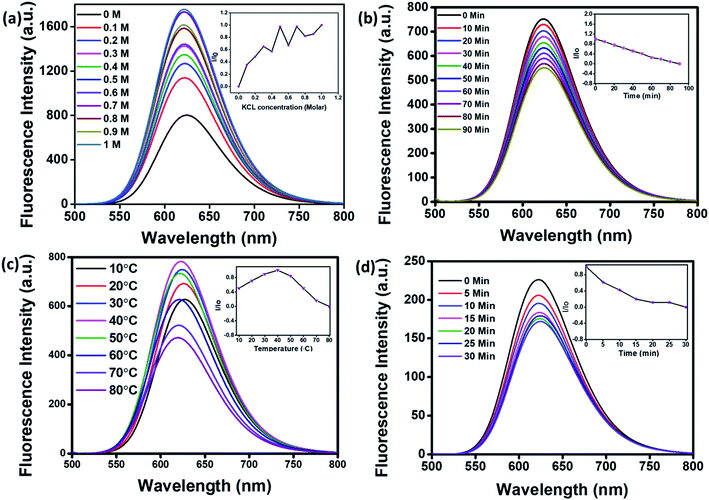
For application in cellular bioimaging, the stability of fluorescent CNPs in different ionic concentrations is very crucial. To check the fluorescence stability of CNPs in a highly ionic environment, we dispersed CNPs (0.5 mg mL−1) in different concentrations of potassium chloride (KCl) salt (0 to 1 M) solution. Our results show a significant increase in the fluorescence of CNPs with an increase in the concentration of KCl (Fig. 3a). We also analysed the photostability of CNPs under constant visible light at 480 nm. The CNPs were irradiated with uninterrupted excitation wavelength light for 90 min, and readings were taken at an interval of every 10 min. The resultant fluorescence intensity (I/Io) stipulates an insignificant decrease in fluorescence with time, indicating the photostability of synthesized CNPs (Fig. 3b).
We further studied the thermal stability of CNPs using a fluorescence spectrophotometer and changed the temperature from 10 °C to 80 °C. With increase in temperature, initially, the fluorescence intensity increased from 10 °C to 40 °C followed by a decrease from 50 °C to 80 °C (Fig. 3c). The increase in fluorescence intensity from 10 °C to 40 °C is 20%, and the reduction in fluorescence intensity from 50 °C to 80 °C is 40%, indicating that the fluorescence intensity of CNPs is temperature-dependent, and the best fluorescence is obtained at physiological temperatures of cells and tissues. The CNPs were also kept at 80 °C for 30 minutes to check their thermal stability. There was an insignificant decrease in the fluorescence intensity of CNPs with time at 80 °C (Fig. 3d). The reduction in fluorescence intensity with increase in temperature from 40 °C to 80 °C can be explained by thermally activated non-radiative channels. At high temperatures, the non-radiative channels are activated, leading to excited electrons not effectively releasing photons. This is why we see a reduction in PL intensity at high temperatures.24
2.3 Cellular uptake of CNPs follows clathrin-mediated endocytosis
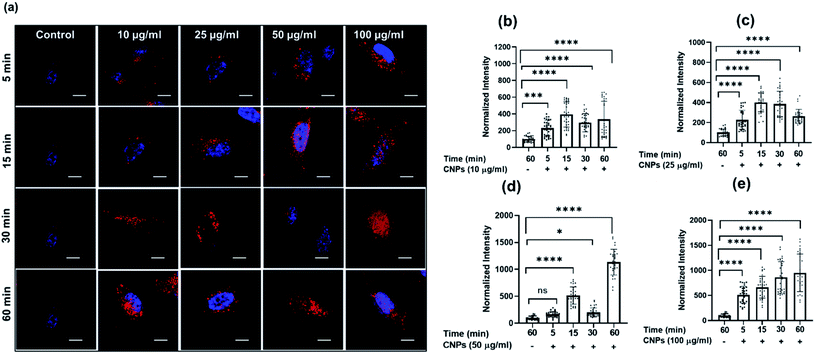
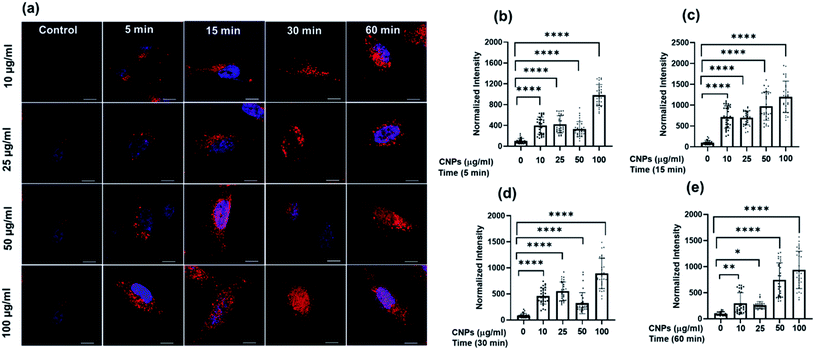
To check the suitability of the developed CNPs as molecular imaging probes toward biological systems and also to monitor the effect of the same on normal and cancer cells, microculture tetrazolium (MTT) assay was performed with mesenchymal triple-negative breast cancer cells (SUM159A) and mouse embryonic fibroblast (MEF) cells (ESI Fig. S5a and b).† The experiments were carried out using different concentrations, namely 10, 20, 30, 50, 100, 200, and 500 μg mL−1 of CNPs. After 24 h of incubation with CNPs, SUM159A showed ∼80% cell viability up to 50 μg mL−1 concentration, decreased to ∼62% up to 200 μg mL−1 and finally 52% at 500 μg mL−1 of CNP concentration (ESI Fig. SI5a†). In the case of MEF, the CNPs showed cell viabilities of 70% and 50% at a concentration of 10–20 μg mL−1 and 30–100 μg mL−1, respectively. The cell viability percentage significantly dropped, with ∼37% and 16% cell viabilities at 200 and 500 μg mL−1 concentrations of CNPs. The cytocompatibility studies with SUM159A suggested that CNPs show high toxicity at a concentration of 100 μg mL−1 even after 24 h, whereas the same limit for MEF was found to be 20 μg mL−1. This also suggests that different cells have different capacities to interact with CNPs, thus affecting their cytotoxicity.To explore the cellular uptake properties of CNPs, confocal microscopy studies were done to analyse the time and concentration-dependent cellular internalization of CNPs using SUM159A and MEF cells (Fig. 4 and SI4i,† respectively). Cells, when incubated with CNPs of different concentrations at 37 °C and analysed by using a confocal microscope, revealed the successful internalization of CNPs. Further, the confocal images and quantification data suggested that CNPs showed a strong intracellular fluorescence response regarding time and concentration. Concentration-dependent studies showed that the fluorescence intensity was increased ∼1–2 times from 10 to 100 μg mL−1 concentration of CNPs. The cell morphology was retained unchanged at lower concentrations. However, when cells were exposed to a higher concentration of 100 μg mL−1 CNPs, the fluorescence signal was observed from the nucleus, and cells appeared to be in stress (Fig. 4a and ESI Fig. SI6i).† Further, to evaluate the time dependent uptake of CNPs, the same data from Fig. 4 were plotted with respect to different time intervals. The time-dependent studies showed an increase in fluorescence signal up to 60 min with enhanced fluorescence intensity (Fig. 5 and ESI Fig. SI6ii).† Confocal microscopy studies showed that the synthesized CNPs could be used for bioimaging applications due to their stable and robust fluorescence emission in the red channel.
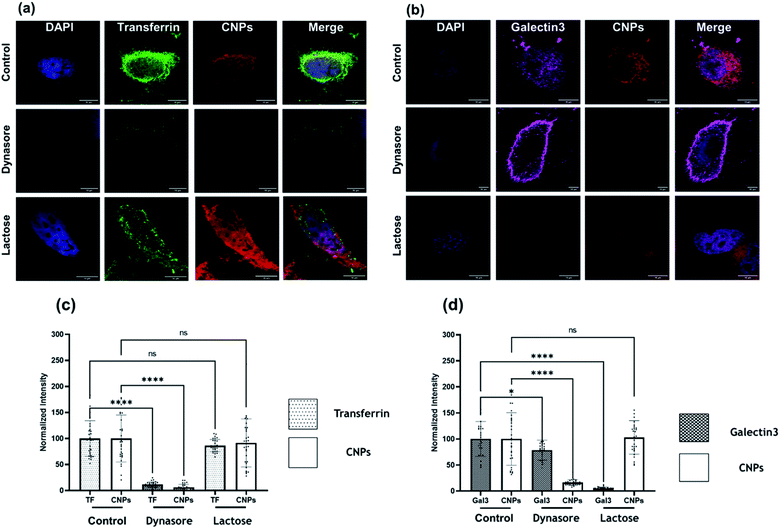
Clathrin-mediated endocytosis (CME) and clathrin-independent endocytosis (CIE) are the most critical pathways considered for NP uptake.25 We thus investigated the plausible endocytosis pathway responsible for the cellular uptake of CNPs by using cellular cargo markers viz., transferrin (Tf), a well-known target of CME, whereas galectin3 (Gal3) for marking CIE was included as a reference. Specific inhibitors, namely dynasore to block CME and lactose for CIE inhibition, were used to validate their effect on CNP uptake. When cells were pulsed with these markers and CNPs in the presence of these inhibitors, we observed that Tf and CNP uptake were significantly reduced by dynasore treatment in SUM-159A cells (Fig. 6a and b). At the same time, lactose inhibition resulted in a non-significant change in fluorescence intensity in the case of Tf and CNPs, while the intensity of Gal3 uptake was significantly decreased.
We also examined the cellular uptake of CNPs via the CIE pathway using Gal3 as a reference marker. Lactose is known to compete with extracellular Gal3 for binding to specific β-galactosides on the plasma membrane receptors and, therefore, used to inhibit the uptake of extracellular Gal3.26 It was observed that the uptake of Gal3 was significantly decreased when inhibited by lactose (CIE blocker), whereas the CNP uptake was marginally affected in cells (Fig. 6c and d). However, upon inhibition via dynasore, the uptake of Gal3 was only marginally reduced. On the contrary to this, CNP uptake was decreased significantly upon treatment with dynasore, indicating the strong involvement of dynamin which plays the role of scission of the clathrin-coated vesicles formed on the plasma membrane. These results confirmed our previous observation that CNP uptake was CME dependent.
2.4 CNP uptake stimulates cellular invasion in 3D spheroid models
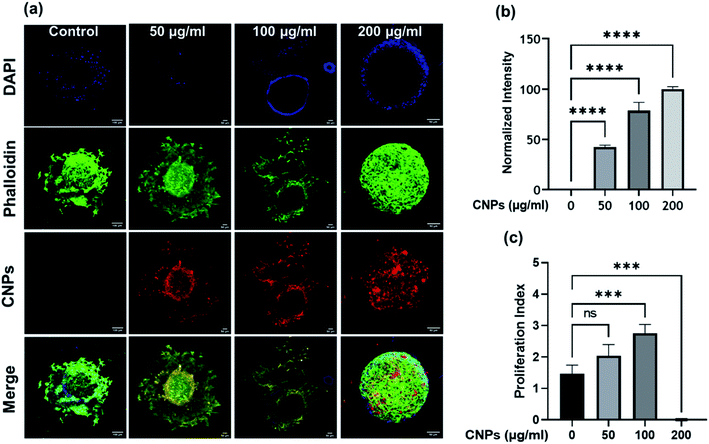
Three-dimensional (3D) spheroid-based cancer models are well explored to study tumour progression, cell activity, and the effect of pharmacokinetic drugs. Three-dimensional spheroids provide better insight into the disease state and proliferation, invasion, or migration of tumour cells. Therefore, we further explored the cellular uptake of CNPs by 3D spheroids and its effect on cell invasion in a 3D matrix. Spheroids were made from triple-negative breast cancer MDA-MB-231 cells using the hanging drop method. A T25 flask having approximately 85% cellular confluency was used for preparation of spheroids. We use 1 mL trypsin for trypsinization and add 4 mL DMEM complete media. Media containing the cells were placed on a Petri dish lid in the form of droplets and put upside down, covering the Petri dish, which was filled with around 30 mL PBS. PBS gives a moist environment for spheroid growth. The excellent quality spheroids are collected from the hanging drop by softly dispersing in 50 μL of collagen-complete media (at 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 vol ratio). These spheroids were suspended in collagen with or without CNPs. The spheroids were allowed to grow in the matrix for 24 h at 37 °C, after which they were fixed using paraformaldehyde (4% PFA), stained for nucleus using DAPI and actin-cytoskeleton using Phalloidin green.27 Spheroids treated with three different concentrations of CNPs viz., 50, 100, and 200 μg mL−1 were incubated for 24 h at 37 °C (Fig. 7a and b). After staining and fixing, the spheroids were mounted on glass slides using Mowiol as a mounting medium. The prepared glass slides were then used for analysis on a Leica DMI8 Confocal microscope. 10× magnification is used for imaging of spheroids treated with different concentrations of CNPs. Images were in the form of z-stacks. Three lasers of 405 nm, 488 nm and 561 nm were used as excitation sources for confocal microscope imaging. Cells, while migrating in 3D, had actively endocytosed the CNPs from the collagen matrix, probably via the CME pathway. To study the effect of CNPs on the invasion capacity of cells in 3D spheroid models, the invasion index was plotted. The cell invasion index of every 3D tumour cell was calculated using the formula:
1 vol ratio). These spheroids were suspended in collagen with or without CNPs. The spheroids were allowed to grow in the matrix for 24 h at 37 °C, after which they were fixed using paraformaldehyde (4% PFA), stained for nucleus using DAPI and actin-cytoskeleton using Phalloidin green.27 Spheroids treated with three different concentrations of CNPs viz., 50, 100, and 200 μg mL−1 were incubated for 24 h at 37 °C (Fig. 7a and b). After staining and fixing, the spheroids were mounted on glass slides using Mowiol as a mounting medium. The prepared glass slides were then used for analysis on a Leica DMI8 Confocal microscope. 10× magnification is used for imaging of spheroids treated with different concentrations of CNPs. Images were in the form of z-stacks. Three lasers of 405 nm, 488 nm and 561 nm were used as excitation sources for confocal microscope imaging. Cells, while migrating in 3D, had actively endocytosed the CNPs from the collagen matrix, probably via the CME pathway. To study the effect of CNPs on the invasion capacity of cells in 3D spheroid models, the invasion index was plotted. The cell invasion index of every 3D tumour cell was calculated using the formula:| Cell invasion index = distance migrated by the cells from the surface of spheroids/diameter of core spheroid |
The migration distances were calculated using ImageJ software. To study the effect of CNPs on the invasion capacity of cells in 3D spheroid models, the invasion index was plotted, which showed that 3D tumour models treated with 100 μg mL−1 of CNPs had a cell invasion index of approximately 3, which was higher than the cell invasion index obtained for the tumour model for the 50 μg mL−1 and control samples (Fig. 7c). In the 200 μg mL−1 sample, there was no proliferation observed, indicating an inhibitory effect of CNPs on cell invasion at higher concentrations.
3 Conclusions
We designed near-infrared red emissive CNPs by the reflux reaction method of heating using para-phenylenediamine (PPDA) as the precursor material and diphenyl ether as the solvent. Owing to red emissive fluorescence and good biocompatibility, the CNPs have great potential in the field of bioimaging.28–32 Different characterization techniques were used to properly analyse the structure, morphology, and optical properties of CNPs. The QY of CNPs is ∼6% in water and 87% in ethanol, showing that the fluorescence of CNPs also depends on the type of solvent used. The fluorescence intensity of CNPs also depends on the pH of the dispersing medium (water), which increases with an increase in pH. Ionic stability, photostability, and thermal stability were also checked. In cell viability studies, we observed that CNPs did not show toxicity up to 100 μg mL−1 even after 24 h in SUM159A cells. Concentration-dependent and time-dependent studies on cellular uptake of CNPs were done and observed that most CNPs were taken up into the cells through clathrin-mediated endocytosis. The same effect was observed not only in 2D but also in the 3D spheroid model, and it was observed that CNPs could trigger the invasion of MDA-MB-231 cells in a collagen matrix. The present work provides not only a promising fluorescent nanomaterial for bioimaging but also opens other platforms for modulating the surface properties of nanoparticles along with the capacity to bioconjugate to different biomolecules, which will enable widespread biological and biomedical applications of CNPs in the future.4 Materials and methods
4.1 Materials
Para-phenylenediamine (PPDA), Moviol, Hoechst, dynasore, galectin3 (Gal3) (alexa 647), transferrin (Tf)-A488, and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were obtained from Sigma-Aldrich. Diphenyl ether (99%) was obtained from Avra, and n-hexane of HPLC grade, acetone (>99.5%), N,N-dimethyl formamide (>99.5%), ethanol(>99.5%), methanol (>99.8%), acetone, acetonitrile, isopropyl alcohol (IPA) and lactose were purchased from Merck. Paraformaldehyde, dimethyl sulfoxide (DMSO), rhodamine B, and cell culture dishes for adherent cells (treated surface) were procured from Himedia. Ham's Nutrient Mixture F12 (HAMs F12), Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), penicillin–streptomycin, trypsin–EDTA (0.25%), and collagen1 rat tail were purchased from Gibco. All the chemicals were of analytical grade and no further purification was required.4.2 Synthesis of fluorescent CNPs
Highly fluorescent red light-emitting CNPs were synthesised using the pyrolysis method by a reflux reaction.19 We have modified the experimental conditions to obtain the CNPs of desired size and properties. Para-phenylenediamine (PPDA) is used as the precursor material and dissolved in diphenyl ether. The reaction takes place for eight hours at 170 °C. After the heating process, the product was allowed to cool down to room temperature. The CNPs synthesised were precipitated out using hexane by the solvent displacement method. The product obtained after heating was added dropwise to 100 mL of hexane solution for the separation of CNPs. The precipitated CNPs were centrifuged and washed three times using hexane and then kept under normal atmospheric conditions to dry up.4.3 Various analytical methods used for the characterization of CNPs
The shape and size of the fluorescent CNPs were analysed using an FEI Titan Themis transmission electron microscope (TEM) (60–300 kV). An atomic force microscope (AFM) was used to obtain both two-dimensional (2D) and three-dimensional (3D) images of CNPs. The X-ray diffraction (XRD) technique was used to characterize the crystalline nature of CNPs and for phase identification. The patterns from an X-ray diffractometer (Brüker-D8 Discover) were recorded with a speed of 0.2° min−1 from 10° to 60° with Cu Kα radiation. Raman spectroscopy was performed using a Kaiser Raman spectrometer with a 785 nm YAG laser. The fluorescence spectra were obtained using an FP-8300 Jasco spectrofluorometer (Japan) in the excitation range of 400 to 520 nm. Rhodamine B was used as a standard reference dye for fluorescence measurement, with a relative quantum yield of 5.86% in water and 87% in ethanol. UV-vis absorbance spectra of CNPs were obtained by using a Spectrocord-210 Plus Analytikjena (Germany). Fourier transform infrared spectroscopy (FT-IR) spectra of CNPs were recorded by using an FTIR spectrometer from Spectrum 2, PerkinElmer in the ATR mode. Scanning was done in the range of 400 cm−1 to 4000 cm−1. The cell imaging for the fixed samples was performed under 63× resolution using a confocal laser scanning microscopy platform Leica TCS SP8.4.4 Spectroscopic studies
The solvents used to record absorption and emission spectra are of spectroscopic grade. The emission spectra were recorded 10 nm after the excitation wavelength for the standard recording of fluorescence spectra. Quantum yields were determined by comparison with rhodamine B in ethanol (ϕ = 0.33) as a reference using the following equation:where QY is the quantum yield, I is the integrated fluorescence intensity, n is the refractive index, OD is the optical density of that particular excitation wavelength, and R is used for reference.
4.5 Cell culture
Mesenchymal triple-negative breast cancer cell lines (SUM159A) and embryonic mouse fibroblast (MEFs) cells were obtained as a gift from Prof. Ludger Johannes, Institut Curie, Paris, France. SUM159A and MEF cells were cultured in HAM’s F12 and DMEM supplemented with 10% FBS and 1% penicillin–streptomycin. For all the studies, PBS of 1× strength with pH 7.4 was used.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well (100 μL) and incubated for 24 h in a CO2 incubator, maintained at 5% CO2 and 95% humidity, for cell adherence. After that, the cells were washed with PBS and treated with 100 μL of 4 different concentrations of CNPs viz., 10, 20, 30, 50, 100, 200, & 500 μg mL−1, in triplicate and were incubated for 24 h. After the completion of incubation time, a colorimetric 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was performed to quantify the effect of CNPs on cell viability. At the end of the treatment, cells were washed twice with PBS; then 10 μL of MTT solution (5 mg mL−1) was added to each well, and the solutions were further incubated for 3 h at 37 °C. After incubation, the formazan crystals formed were dissolved in 100 μL DMSO and incubated in the dark for 15 min. The intensity of the colour was then measured from the absorbance at 570 nm from the formazan crystals and was representative of the number of viable cells per well. The values thus obtained for the untreated control samples were equated to 100%, and the relative percentage values for CNPs were calculated accordingly. All experiments were performed in triplicate, and the cell viability (%) was calculated using eqn (1).
000 cells per well (100 μL) and incubated for 24 h in a CO2 incubator, maintained at 5% CO2 and 95% humidity, for cell adherence. After that, the cells were washed with PBS and treated with 100 μL of 4 different concentrations of CNPs viz., 10, 20, 30, 50, 100, 200, & 500 μg mL−1, in triplicate and were incubated for 24 h. After the completion of incubation time, a colorimetric 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was performed to quantify the effect of CNPs on cell viability. At the end of the treatment, cells were washed twice with PBS; then 10 μL of MTT solution (5 mg mL−1) was added to each well, and the solutions were further incubated for 3 h at 37 °C. After incubation, the formazan crystals formed were dissolved in 100 μL DMSO and incubated in the dark for 15 min. The intensity of the colour was then measured from the absorbance at 570 nm from the formazan crystals and was representative of the number of viable cells per well. The values thus obtained for the untreated control samples were equated to 100%, and the relative percentage values for CNPs were calculated accordingly. All experiments were performed in triplicate, and the cell viability (%) was calculated using eqn (1).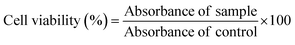 | (1) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well on coverslips in 24 well plate cell culture plates and incubated for 24 h at 37 °C and 5% CO2. After incubation, the cells were washed twice with PBS and treated with 10, 25, 50, and 100 μg mL−1 of CNPs, and all the samples were further incubated for different time points of 5, 15, 30, and 60 min, respectively. After desired incubation time, the cells were washed twice with PBS and then fixed using 4% PFA for 20 min. Afterwards, the cells were washed with PBS thrice, stained with DAPI, and mounted using a mounting medium (moviol). Finally, cellular internalization of CNPs was examined through Leica TCS SP5 confocal microscope with a 63× oil immersion objective. A 405 nm laser was used as the excitation source for DAPI, while a 488 nm argon laser was used as an excitation source for CNPs. The emission bandwidth for DAPI and CNPs was 420–450 nm and 590–700 nm, respectively.
000 cells per well on coverslips in 24 well plate cell culture plates and incubated for 24 h at 37 °C and 5% CO2. After incubation, the cells were washed twice with PBS and treated with 10, 25, 50, and 100 μg mL−1 of CNPs, and all the samples were further incubated for different time points of 5, 15, 30, and 60 min, respectively. After desired incubation time, the cells were washed twice with PBS and then fixed using 4% PFA for 20 min. Afterwards, the cells were washed with PBS thrice, stained with DAPI, and mounted using a mounting medium (moviol). Finally, cellular internalization of CNPs was examined through Leica TCS SP5 confocal microscope with a 63× oil immersion objective. A 405 nm laser was used as the excitation source for DAPI, while a 488 nm argon laser was used as an excitation source for CNPs. The emission bandwidth for DAPI and CNPs was 420–450 nm and 590–700 nm, respectively.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well. The cells were treated with dynasore (80 μM) and lactose (150 mM) in HAM’s F-12 (serum-free media) in desired wells and incubated for a specific time interval at 37 °C. Dynasore and lactose were used to inhibit clathrin-mediated endocytosis (CME) and clathrin-independent endocytosis (CIE) pathways, respectively. The untreated cells were used as a control. Afterwards, the cells were washed gently and incubated with fluorescently labelled markers, which follow CME (Tf (5 μg mL−1)) and CIE (Gal3 (5 μg mL−1)) pathways and CNPs (25 μg mL−1) for a specific time interval at 37 °C. The cells were washed thrice with PBS and fixed using 4% PFA for 15 min at 37 °C. The coverslips were mounted on Moviol containing Hoechst for imaging. Microscopy imaging of the fixed cells was performed using a confocal laser scanning microscope under 63× resolution. Quantification of data was done using Fiji ImageJ software.
000 cells per well. The cells were treated with dynasore (80 μM) and lactose (150 mM) in HAM’s F-12 (serum-free media) in desired wells and incubated for a specific time interval at 37 °C. Dynasore and lactose were used to inhibit clathrin-mediated endocytosis (CME) and clathrin-independent endocytosis (CIE) pathways, respectively. The untreated cells were used as a control. Afterwards, the cells were washed gently and incubated with fluorescently labelled markers, which follow CME (Tf (5 μg mL−1)) and CIE (Gal3 (5 μg mL−1)) pathways and CNPs (25 μg mL−1) for a specific time interval at 37 °C. The cells were washed thrice with PBS and fixed using 4% PFA for 15 min at 37 °C. The coverslips were mounted on Moviol containing Hoechst for imaging. Microscopy imaging of the fixed cells was performed using a confocal laser scanning microscope under 63× resolution. Quantification of data was done using Fiji ImageJ software.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, respectively. After the transfer to the matrix, the spheroids were subjected to the respective treatments with CNPs. After 24 h, the spheroids were fixed using 4% PFA and actin and nuclei were marked by using the phalloidin–alexa fluor 488 stain and Hoechst. The spheroids were mounted onto slides and further imaged using a Leica confocal microscope.
1 ratio, respectively. After the transfer to the matrix, the spheroids were subjected to the respective treatments with CNPs. After 24 h, the spheroids were fixed using 4% PFA and actin and nuclei were marked by using the phalloidin–alexa fluor 488 stain and Hoechst. The spheroids were mounted onto slides and further imaged using a Leica confocal microscope.
Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
We sincerely thank all the members of the DB group for critically reading the manuscript and their valuable feedback. US and AT thank IITGN-MHRD, GoI for PhD, and SW thanks IITGN-MHRD for the postdoctoral fellowship. PV acknowledges a PhD fellowship from UGC-CSIR, India. We thank Prof. Abhay Raj Goutam for his help with TEM imaging. DB thanks SERB, GoI for the Ramanujan Fellowship, IITGN, for the startup grant, and DBT-EMR, Gujcost-DST, GSBTM, HEFA and BRNS-BARC for research grants. Imaging facilities of CIF at IIT Gandhinagar are acknowledged.References
- N. Malik, T. Arfin and A. U. Khan, Chapter 13 – Graphene Nanomaterials: Chemistry and Pharmaceutical Perspectives, in Nanomaterials for Drug Delivery and Therapy, ed. A. M. Grumezescu, William Andrew Publishing, 2019, pp. 373–402, DOI:10.1016/B978-0-12-816505-8.00002-3.
- K.-K. Liu, S.-Y. Song, L.-Z. Sui, S.-X. Wu, P.-T. Jing, R.-Q. Wang, Q.-Y. Li, G.-R. Wu, Z.-Z. Zhang, K.-J. Yuan and C.-X. Shan, Efficient Red/Near-Infrared-Emissive Carbon Nanodots with Multiphoton Excited Upconversion Fluorescence, Adv. Sci., 2019, 6(17), 1900766, DOI:10.1002/advs.201900766.
- Z. Yu, W. K. Chan and T. T. Y. Tan, Neodymium-Sensitized Nanoconstructs for Near-Infrared Enabled Photomedicine, Small, 2020, 16(1), 1905265, DOI:10.1002/smLl.201905265.
- H. Lu, G. M. Carroll, N. R. Neale and M. C. Beard, Infrared Quantum Dots: Progress, Challenges, and Opportunities, ACS Nano, 2019, 13(2), 939–953, DOI:10.1021/acsnano.8b09815.
- Z. Lv, Y. Wang, J. Chen, J. Wang, Y. Zhou and S.-T. Han, Semiconductor Quantum Dots for Memories and Neuromorphic Computing Systems, Chem. Rev., 2020, 120(9), 3941–4006 CrossRef CAS PubMed.
- M. J. Schnermann, Chemical Biology: Organic Dyes for Deep Bioimaging, Nature, 2017, 551(7679), 176–177, DOI:10.1038/nature24755.
- P.-J. Wu, S.-Y. Kuo, Y.-C. Huang, C.-P. Chen and Y.-H. Chan, Polydiacetylene-Enclosed near-Infrared Fluorescent Semiconducting Polymer Dots for Bioimaging and Sensing, Anal. Chem., 2014, 86(10), 4831–4839, DOI:10.1021/ac404237q.
- S. Y. Lim, W. Shen and Z. Gao, Carbon Quantum Dots and Their Applications, Chem. Soc. Rev., 2014, 44(1), 362–381, 10.1039/C4CS00269E.
- Z.-F. Pu, Q.-L. Wen, Y.-J. Yang, X.-M. Cui, J. Ling, P. Liu and Q.-E. Cao, Fluorescent Carbon Quantum Dots Synthesized Using Phenylalanine and Citric Acid for Selective Detection of Fe3+ Ions, Spectrochim. Acta, Part A, 2020, 229, 117944, DOI:10.1016/j.saa.2019.117944.
- D. Qu and Z. Sun, The Formation Mechanism and Fluorophores of Carbon Dots Synthesized via a Bottom-up Route, Mater. Chem. Front., 2020, 4(2), 400–420, 10.1039/C9QM00552H.
- W. Liu, C. Li, Y. Ren, X. Sun, W. Pan, Y. Li, J. Wang and W. Wang, Carbon Dots: Surface Engineering and Applications, J. Mater. Chem. B, 2016, 4(35), 5772–5788, 10.1039/C6TB00976J.
- M. Zheng, S. Ruan, S. Liu, T. Sun, D. Qu, H. Zhao, Z. Xie, H. Gao, X. Jing and Z. Sun, Self-Targeting Fluorescent Carbon Dots for Diagnosis of Brain Cancer Cells, ACS Nano, 2015, 9(11), 11455–11461, DOI:10.1021/acsnano.5b05575.
- H. Ding, J.-S. Wei, N. Zhong, Q.-Y. Gao and H.-M. Xiong, Highly Efficient Red-Emitting Carbon Dots with Gram-Scale Yield for Bioimaging, Langmuir, 2017, 33(44), 12635–12642, DOI:10.1021/acs.langmuir.7b02385.
- H. Ding, S.-B. Yu, J.-S. Wei and H.-M. Xiong, Full-Color Light-Emitting Carbon Dots with a Surface-State-Controlled Luminescence Mechanism, ACS Nano, 2016, 10(1), 484–491, DOI:10.1021/acsnano.5b05406.
- X.-W. Hua, Y.-W. Bao, J. Zeng and F.-G. Wu, Ultrasmall All-in-One Nanodots Formed via Carbon Dot-Mediated and Albumin-Based Synthesis: Multimodal Imaging-Guided and Mild Laser-Enhanced Cancer Therapy, ACS Appl. Mater. Interfaces, 2018, 10(49), 42077–42087 CrossRef CAS PubMed.
- X.-W. Hua, Y.-W. Bao and F.-G. Wu, Fluorescent Carbon Quantum Dots with Intrinsic Nucleolus-Targeting Capability for Nucleolus Imaging and Enhanced Cytosolic and Nuclear Drug Delivery, ACS Appl. Mater. Interfaces, 2018, 10(13), 10664–10677, DOI:10.1021/acsami.7b19549.
- D. Li, J. Huang, R. Li, P. Chen, D. Chen, M. Cai, H. Liu, Y. Feng, W. Lv and G. Liu, Synthesis of a Carbon Dots Modified G-C3N4/SnO2 Z-Scheme Photocatalyst with Superior Photocatalytic Activity for PPCPs Degradation under Visible Light Irradiation, J. Hazard. Mater., 2021, 401, 123257, DOI:10.1016/j.jhazmat.2020.123257.
- S. Ye, H. Huang, C. Yuan, F. Liu, M. Zhai, X. Shi, C. Qi and G. Wang, Thickness-Dependent Strain Effect on the Deformation of the Graphene-Encapsulated Au Nanoparticles, J. Nanomater., 2014, 2014, e989672, DOI:10.1155/2014/989672.
- S. Walia, A. K. Shukla, C. Sharma and A. Acharya, Engineered Bright Blue- and Red-Emitting Carbon Dots Facilitate Synchronous Imaging and Inhibition of Bacterial and Cancer Cell Progression via 1O2-Mediated DNA Damage under Photoirradiation, ACS Biomater. Sci. Eng., 2019, 5(4), 1987–2000, DOI:10.1021/acsbiomaterials.9b00149.
- R. Sato, Y. Iso and T. Isobe, Fluorescence Solvatochromism of Carbon Dot Dispersions Prepared from Phenylenediamine and Optimization of Red Emission, Langmuir, 2019, 35(47), 15257–15266, DOI:10.1021/acs.langmuir.9b02739.
- X. Ma, S. Li, V. Hessel, L. Lin, S. Meskers and F. Gallucci, Synthesis of N-Doped Carbon Dots via a Microplasma Process, Chem. Eng. Sci., 2020, 220, 115648, DOI:10.1016/j.ces.2020.115648.
- Carbon Quantum Dots – Synthesis, Properties and Applications|Raz Jelinek|, Springer, https://www.springer.com/gp/book/9783319439099, accessed 2021/07/08 Search PubMed.
- C. Liu, F. Zhang, J. Hu, W. Gao and M. Zhang, A Mini Review on PH-Sensitive Photoluminescence in Carbon Nanodots, Front. Chem., 2021, 8 DOI:10.3389/fchem.2020.605028.
- W. U. Khan, L. Qin, A. Alam, P. Zhou, Y. Peng and Y. Wang, Water-Soluble Green-Emitting Carbon Nanodots with Enhanced Thermal Stability for Biological Applications, Nanoscale, 2021, 13(7), 4301–4307, 10.1039/D0NR09131F.
- M. Sousa de Almeida, E. Susnik, B. Drasler, P. Taladriz-Blanco, A. Petri-Fink and B. Rothen-Rutishauser, Understanding Nanoparticle Endocytosis to Improve Targeting Strategies in Nanomedicine, Chem. Soc. Rev., 2021, 50(9), 5397–5434, 10.1039/d0cs01127d.
- P. Hivare, A. Rajwar, S. Gupta and D. Bhatia, Spatiotemporal Dynamics of Endocytic Pathways Adapted by Small DNA Nanocages in Model Neuroblastoma Cell-Derived Differentiated Neurons, ACS Appl. Bio Mater., 2021, 4(4), 3350–3359, DOI:10.1021/acsabm.0c01668.
- I. Sathisaran, D. D. Bhatia and S. V. Dalvi, New Curcumin-Trimesic Acid Cocrystal and Anti-Invasion Activity of Curcumin Multicomponent Solids against 3D Tumor Models, Int. J. Pharm., 2020, 587, 119667 CrossRef CAS PubMed.
- H. Dong, M. Roming and C. Feldmann, Unexpected Fluorescence of Polyols and PEGylated Nanoparticles Derived from Carbon Dot Formation, Part. Part. Syst. Charact., 2015, 32(4), 467–475 CrossRef CAS.
- H. Ding, Y. Ji, J.-S. Wei, Q.-Y. Gao, Z.-Y. Zhou and H.-M. Xiong, Facile Synthesis of Red-Emitting Carbon Dots from Pulp-Free Lemon Juice for Bioimaging, J. Mater. Chem. B, 2017, 5(26), 5272–5277 RSC.
- K. Hola, M. Sudolska, S. Kalytchuk, D. Nachtigallova, A. L. Rogach, M. Otyepka and R. Zboril, Graphitic Nitrogen Triggers Red Fluorescence in Carbon Dots, ACS Nano, 2017, 11(12), 12402–12410 CrossRef CAS PubMed.
- L. Yue, H. Li, Q. Sun, J. Zhang, X. Luo, F. Wu and X. Zhu, Red-Emissive Ruthenium-Containing Carbon Dots for Bioimaging and Photodynamic Cancer Therapy, ACS Appl. Nano Mater., 2020, 3(1), 869–876 CrossRef CAS.
- Y. Hu, Z. Yang, X. Lu, J. Guo, R. Cheng, L. Zhu, C.-F. Wang and S. Chen, Facile Synthesis of Red Dual-Emissive Carbon Dots for Ratiometric Fluorescence Sensing and Cellular Imaging, Nanoscale, 2020, 12(9), 5494–5500 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1na00813g |
| ‡ US, AT, and SW contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |