A continuous flow microfluidic-MS system for efficient OBOC screening†
Weizhi Wanga,
Zewen Weia,
Zihua Wanga,
Huailei Maa,
Xiangli Bua and
Zhiyuan Hu*ab
aCAS Key Laboratory for Biomedical Effects of Nanomaterials & Nanosafety, National Center for Nanoscience and Technology of China, Beijing 100190, China. E-mail: huzy@nanoctr.cn; Fax: +86-10-82545643; Tel: +86-10-82545643
bBeijing Proteome Research Center, Beijing Institute of Radiation Medicine, Beijing 102206, China
First published on 5th November 2014
Abstract
A microfluidic chip based method utilized for effective screening of high-throughput peptide libraries was achieved. Continuous flow bead trapping, sorting, arraying and in situ sequencing was integrated. Peptide library screening with 105 beads was processed within 4 hours and peptide ligands toward the target protein AHA and APN were successfully discovered.
Active peptides can be screened from a large population of randomly generated sequences, which is the essence of lead compound selection in drug discovery.1–4 Combinatorial peptide chemistry has been widely used for affinity ligand identification towards target receptors.5,6 Among them, a one-bead-one-compound (OBOC) library approach has been utilized to discover novel ligands.7,8 However, these methods remain too finicky and inefficient to ensure success with high-throughput on-bead peptide screening. In modern combinational chemistry, several methods have been developed to accelerate peptide screening processes.9–13 With specific conjugation assay, OBOC peptide beads could be trapped by a magnetic field instead of being picked out manually, which have improved the screening efficiency.14,15 High-throughput peptides could also be coded by quantum dots or colloid.11,16,17 Continuous flow beads sorting method is developed to accelerate the screening process.18–20 Also, nucleic acid coding have been emerged.21 However, exist OBOC screening processes still require two tedious steps: one is the manual isolation of individual positive hits from the library. The other is the cleavage and sequencing of the large number of positive peptides. Nowadays, microfluidic and microarray technique provide an integrated strategy for high-throughput sample process.22–25 Based on the previous work,26–29 herein an integrated and automated OBOC screening strategy based on a microfluidic system which embraces the whole peptide ligands screening process was put forward. Using a microfluidic chip with magnetic trapping and sheath flow sorting functions, a peptide library with a large number of beads can be sorted continuously (Fig. 1 and Scheme 1). The sorted positive beads were trapped in the “one-bead-one-well” microarray follows downstream with a high density of 10
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 peptides per cm2 and compatible with in situ matrix assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) sequencing. This system has been applied to the discovery of affinity peptide ligands from OBOC libraries, and also affinity peptide ligands could be identified effectively from the 105 OBOC candidates within 4 hours.
000 peptides per cm2 and compatible with in situ matrix assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) sequencing. This system has been applied to the discovery of affinity peptide ligands from OBOC libraries, and also affinity peptide ligands could be identified effectively from the 105 OBOC candidates within 4 hours.
The integrated microfluidic chip was made of silicon and fabricated by conventional soft lithography techniques (Fig. 1a and Scheme S1 in ESI†).30 It consist of peptide beads sorting channels and the single bead trapping array. To evaluate the effectiveness of this system, proof-of principle experiments were performed. An OBOC library was constructed on the Tentagel beads12 (30 μm) using Fmoc peptide synthesis strategy31 with the known ligand HA towards the protein AHA32 (Scheme S2†). The sequence is YPXXXXXXX with X standing for Y, P, D, V and A randomly so that the capacity of the peptide library was 7 × 104 (57). The redundancy of library was six and there are 105 candidate peptide beads. The target protein AHA was labeled with biotin and the positive beads were given magnetic signal through the bridge of peptide–protein–biotin–streptavidin-magnetic beads.28
In the peptide beads sorting process, the mixture of the peptides beads and magnetic beads was introduced from the inlet, the magnet was set along the channel in the upstream. A sheath flow configuration was used downstream. When the two fluids in the two sheath flow channels differ enough, a “wall” of fluid emerges so that the beads can be directed in different directions. Several flow rates were tested and 60 μL min−1 was found to be the optimized flow rate (Fig. S1a and b†), which is accordance with our simulations (Fig. S1c†). When the positive beads were trapped (Fig. 1b), the sheath flow rate A was set as 10 μL min−1 and the sheath flow rate B was set as 60 μL min−1 so that the negative beads exit through negative outlet. When the two flow rates were switched, the positive beads would flow downstream and get trapped in the microwells.
The positive beads were trapped in the microwell array in the downstream. Each microwell was a cube shape of 40 μm (L) × 40 μm (W) × 45 μm (D), which is a suitable size to trap individual peptide beads in a one-well-one-bead manner (Fig. 1c). To prevent the loss of positive beads, microwell dimensions had to be optimized (Fig. S2†). The microwell array was utilized for in situ MALDI-TOF-MS detection (Fig. 1c–e). A photo cleavable linker, ANP (3-amino-3-(2-nitrophenyl) propionic acid), was designed for in situ cleavage of the peptides from the beads by the laser equipped in MS instrument. The microfluidic chip was inserted into a modified MALDI target for in situ single bead analysis. To provide the necessary conductivity for ionization, a gold layer of 300 μm thickness was sputtered on the surface (Fig. 1f). External calibration wells with the same 3D dimension were designed and fabricated in the adjacent positions. In our procedure, the trapping rate of the mircowell is up to 70% and the in situ MS sequencing was achieved.
We obtained more than 400 sequences and 10 of them have hit at the highest frequency which are aligned as shown in Fig. 2a. Using the software ClustalX2,33 conserved sequences were determined which is in accordance with the wild type ligand HA with the sequence of YPYDVPDYA (Fig. 2b). It suggested the motif with the highest affinity (Fig. 2c, KD = 7.12 × 10−9 calculated by Surface Plasmon Resonance image (SPRi) detection) towards target protein AHA could be screened out efficiently. It is also notable for this high-throughput screening system that promising sequences could be identified quickly by sequence alignment.
Speed, throughput, and efficiency is important in OBOC peptide screening. With the automated and integrated microfluidic chip system, a peptide library screened with 105 beads could be processed within 4 hours up to 4 × 104 high density microwell array for positive beads displaying and in situ MALDI-TOF-MS sequencing was achieved.
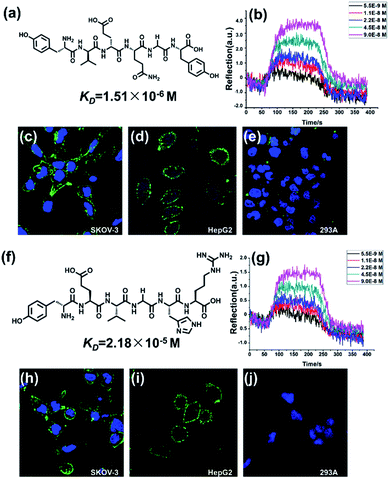
We next applied this system to screen cancer target peptide ligands. APN (aminopeptidase N), a membrane protein that plays a key role in tumor angiogenesis was chosen as the target protein.34 Peptide library was constructed with a library capacity of 8000. The peptide library was constructed with the sequence of X1X2X3X4X5X6 in which X1 represents either F, Y, A or L residues. X2 and X3 represent V, E, I or K residues and X4–6 represent N, R, G, H or Y residues. Using the microfluidic system, APN affinity peptide ligand was screened and conserved sequences were determined. More than 60 sequences were obtained and it suggested that the motif Tyr-Val(Glu)-Glu(Val)-hit at the highest probability. The conserved sequence that emerged is YVENGY (Y-1, Fig. 3a) and YEVGHR (Y-2, Fig. 3f). We next tested the candidate ligand peptides as affinity probes for the recognition of cancer cells. The APN-expressing human ovarian carcinoma cell line SKOV-3 and hepatoma cell line HepG2 were chosen as positive cells in vitro. Human embryonic kidney (HEK) 293A cells, which express little APN were chosen as negative cells. All the cells were cultured in the medium with approximately 1 × 105 mL−1 cells per dishes. Fluorescein isothiocyanate (FITC) labelled Y-1 and Y-2 (1 mg mL−1 in PBS, 200 μL) and Hoechst 33342 (nuclear localization reagent) were used to image all the cells. Binding was achieved by incubating the cells in the solution for 40 min at 4 °C. Confocal fluorescence imaging was performed on an Olympus FV1000-IX81 confocal-laser scanning microscope.
As shown in Fig. 3, incubation of SKOV-3 cells with both the peptides resulted in strong binding (Fig. 3c and h). Also, incubation of HepG2 cells also shows strong binding (Fig. 3d and i). For the affinity probes, the fluorescence distributed evenly across the cell membrane while no positive signal was detected in nucleus or cytoplasm. This suggested that APN might exist mainly on extracellular domain which is consistent with previous reports.35 Meanwhile, neither of peptide shows binding affinity towards t 293A cells (Fig. 3e and j). It is indicated that the conserve sequence Tyr in the N terminal has made a hydrophobic structure which can interact with hydrophobic elements in the APN, which leads to the specific recognition of peptides towards the extracellular domains of APN. These data indicated that cells expressing APN specifically bind to the peptide in a ligand and receptor dependent manner. The dissociation constants (KD) of the Y-1 and Y-2 were calculated to be 1.51 μmol L−1 and 21.8 μmol L−1, respectively by SPRi detection (Fig. 3b and g). These SPRi results agree nicely with the fluorescence results. Though specific optimization method, the two peptide ligand may developed into cancer target probes prospectively.
Conclusions
A rapid and efficient microfluidic system for screening peptide ligands was provided. Affinity peptides could be screened out in a high-throughput manner of which the screening process is simplified and accelerated. Our system has been applied to the identification of the wild type sequence towards protein AHA and was also successfully utilized in discovery of the peptide probe for tumor marker APN. Our work provides a new insight into the establishment of effective and universal strategy for screening peptide probes for different biological system.Acknowledgements
We acknowledge funding from the National Natural Science Foundation of China (21305023), Beijing Municipal Natural Science Foundation (2144058), National Natural Science Foundation of China (31270875, 31470049) and Project of Chinese Academy of Science (YZ201217). We express our gratitude to Dr Yu Gao of Scripps Research Institute, Dr Zongxiu Nie and Dr Rui Zhao from Institute of Chmistry, Chinese Academy of Science and Dr Zhihong Li from Peking University for their valuable help.References
- P. Picotti, O. Rinner, R. Stallmach, F. Dautel, T. Farrah, B. Domon, H. Wenschuh and R. Aebersold, Nat. Methods, 2010, 7, 43–U45 CrossRef CAS PubMed.
- B. P. Gray and K. C. Brown, Chem. Rev., 2014, 114, 1020–1081 CrossRef CAS PubMed.
- Y. Choi, J. Lee, K. Kim, H. Kim, P. Sommer and R. Song, Chem. Commun., 2010, 46, 9146–9148 RSC.
- H. Wu, J. Ge, M. Uttamchandani and S. Q. Yao, Chem. Commun., 2011, 47, 5664–5670 CAS.
- O. H. Aina, R. Liu, J. L. Sutcliffe, J. Marik, C.-X. Pan and K. S. Lam, Mol. Pharm., 2007, 4, 631–651 CrossRef CAS PubMed.
- J. Kofoed and J.-L. Reymond, Chem. Commun., 2007, 4453–4455 CAS.
- A. R. Kunys, W. Lian and D. Pei, Current protocols in chemical biology, 2012, vol. 4, pp. 331–355 Search PubMed.
- T. M. Doran, Y. Gao, K. Mendes, S. Dean, S. Simanski and T. Kodadek, ACS Comb. Sci., 2014, 16, 259–270 CrossRef CAS PubMed.
- J. M. Astle, L. S. Simpson, Y. Huang, M. M. Reddy, R. Wilson, S. Connell, J. Wilson and T. Kodadek, Chem. Biol., 2010, 17, 38–45 CrossRef CAS PubMed.
- C.-F. Cho, G. A. Amadei, D. Breadner, L. G. Luyt and J. D. Lewis, Nano Lett., 2012, 12, 5957–5965 CrossRef CAS PubMed.
- J.-H. Kim, H. Kang, S. Kim, B.-H. Jun, T. Kang, J. Chae, S. Jeong, J. Kim, D. H. Jeong and Y.-S. Lee, Chem. Commun., 2011, 47, 2306–2308 RSC.
- R. W. Liu, J. Mark and K. S. Lam, J. Am. Chem. Soc., 2002, 124, 7678–7680 CrossRef CAS PubMed.
- M. M. Reddy, R. Wilson, J. Wilson, S. Connell, A. Gocke, L. Hynan, D. German and T. Kodadek, Cell, 2011, 144, 132–142 CrossRef CAS PubMed.
- X. Qi, J. Astle and T. Kodadek, Mol. BioSyst., 2010, 6, 102–107 RSC.
- D. Pei, Chem. Biol., 2010, 17, 3–4 CrossRef CAS PubMed.
- B. J. Battersby, D. Bryant, W. Meutermans, D. Matthews, M. L. Smythe and M. Trau, J. Am. Chem. Soc., 2000, 122, 2138–2139 CrossRef CAS.
- C. Heinis, T. Rutherford, S. Freund and G. Winter, Nat. Chem. Biol., 2009, 5, 502–507 CrossRef CAS PubMed.
- M. Hintersteiner, T. Kimmerlin, F. Kalthoff, M. Stoeckli, G. Garavel, J.-M. Seifert, N.-C. Meisner, V. Uhl, C. Buehler, T. Weidemann and M. Auer, Chem. Biol., 2009, 16, 724–735 CrossRef CAS PubMed.
- A. R. Burns, T. C. Y. Kwok, A. Howard, E. Houston, K. Johanson, A. Chan, S. R. Cutler, P. McCourt and P. J. Roy, Nat. Protoc., 2006, 1, 1906–1914 CrossRef CAS PubMed.
- A. K. Price, A. B. MacConnell and B. M. Paegel, Anal. Chem., 2014, 86, 5039–5044 CrossRef CAS PubMed.
- W. R. J. D. Galloway and D. R. Spring, Chem. Biol., 2011, 18, 1209–1210 CrossRef CAS PubMed.
- M. Xiong, N. Hao, T. Yu, J. J. Xu and H. Y. Chen, Chem. Commun., 2014, 50, 10303–10306 RSC.
- S. Xu, Y. Zhang, L. Jia, K. E. Mathewson, K.-I. Jang, J. Kim, H. Fu, X. Huang, P. Chava, R. Wang, S. Bhole, L. Wang, Y. J. Na, Y. Guan, M. Flavin, Z. Han, Y. Huang and J. A. Rogers, Science, 2014, 344, 70–74 CrossRef CAS PubMed.
- P. N. Nge, C. I. Rogers and A. T. Woolley, Chem. Rev., 2013, 113, 2550–2583 CrossRef CAS PubMed.
- A. K. Yetisen, M. S. Akram and C. R. Lowe, Lab Chip, 2013, 13, 2210–2251 RSC.
- W. Wang, Y. Huang, Y. Jin, G. Liu, Y. Chen, H. Ma and R. Zhao, Analyst, 2013, 138, 2890–2896 RSC.
- W. Wang, Y. Huang, J. Liu, Y. Xie, R. Zhao, S. Xiong, G. Liu, Y. Chen and H. Ma, Lab Chip, 2011, 11, 929–935 RSC.
- W. Wang, M. Li, Z. Wei, Z. Wang, X. Bu, W. Lai, S. Yang, H. Gong, H. Zheng, Y. Wang, Y. Liu, Q. Li, Q. Fang and Z. Hu, Anal. Chem., 2014, 86, 3703–3707 CrossRef CAS PubMed.
- H. Zheng, W. Wang, X. Li, Z. Wang, L. Hood, C. Lausted and Z. Hu, Lab Chip, 2013, 13, 3347–3350 RSC.
- C. L. Cheung, R. J. Nikolic, C. E. Reinhardt and T. F. Wang, Nanotechnology, 2006, 17, 1339–1343 CrossRef CAS.
- G. B. Fields and R. L. Noble, Int. J. Pept. Protein Res., 1990, 35, 161–214 CrossRef CAS PubMed.
- K.-F. Lechtreck, S. Luro, J. Awata and G. B. Witman, Cell Motil. Cytoskeleton, 2009, 66, 469–482 CrossRef CAS PubMed.
- R. Chenna, H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins and J. D. Thompson, Nucleic Acids Res., 2003, 31, 3497–3500 CrossRef CAS PubMed.
- A. H. M. Wong, D. Zhou and J. M. Rini, J. Biol. Chem., 2012, 287, 36804–36813 CrossRef CAS PubMed.
- S. V. Bhagwat, J. Lahdenranta, R. Giordano, W. Arap, R. Pasqualini and L. H. Shapiro, Blood, 2001, 97, 652–659 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: The fabrication and optimization details of the microfluidic system. The experimental details of SPRi detection. See DOI: 10.1039/c4ra12911c |
| This journal is © The Royal Society of Chemistry 2014 |