Emerging nanobiotechnology-encoded relaxation tuning establishes new MRI modes to localize, monitor and predict diseases
Taixia
Wang
ab,
Xueni
Zhang
a,
Yuan
Xu
 a,
Yingchun
Xu
a,
Yifeng
Zhang
*ab and
Kun
Zhang
a,
Yingchun
Xu
a,
Yifeng
Zhang
*ab and
Kun
Zhang
 *ab
*ab
aCentral Laboratory and Ultrasound Research and Education Institute, Shanghai Tenth People's Hospital, Tongji University School of Medicine, No. 301 Yan-chang-zhong Road, Shanghai 200072, China. E-mail: zhangyifeng@tongji.edu.cn; zhang1986kun@126.com
bShanghai Engineering Research Center of Ultrasound Diagnosis and Treatment, National Clinical Research Center for Interventional Medicine, Tongji University School of Medicine, No. 301 Yan-chang-zhong Road, Shanghai 200072, China
First published on 17th June 2022
Abstract
Magnetic resonance imaging (MRI) is one of the most important techniques in the diagnosis of many diseases including cancers, where contrast agents (CAs) are usually necessary to improve its precision and sensitivity. Previous MRI CAs are confined to the signal-to-noise ratio (SNR) elevation of lesions for precisely localizing lesions. As nanobiotechnology advances, some new MRI CAs or nanobiotechnology-enabled MRI modes have been established to vary the longitudinal or transverse relaxation of CAs, which are harnessed to detect lesion targets, monitor disease evolution, predict or evaluate curative effect, etc. These distinct cases provide unexpected insights into the correlation of the design principles of these nanobiotechnologies and corresponding MRI CAs with their potential applications. In this review, first, we briefly present the principles, classifications and applications of conventional MRI CAs, and then elucidate the recent advances in relaxation tuning via the development of various nanobiotechnologies with emphasis on the design strategies of nanobiotechnology and the corresponding MRI CAs to target the tumor microenvironment (TME) and biological targets or activities in tumors or other diseases. In addition, we exemplified the advantages of these strategies in disease theranostics and explored their potential application fields. Finally, we analyzed the present limitations, potential solutions and future development direction of MRI after its combination with nanobiotechnology.
 Xueni Zhang | Xueni Zhang is currently an undergraduate at Tongji University. Her research interests include the applications of nanomaterials in medicine and molecular medicine. |
 Yuan Xu | Yuan Xu is currently an undergraduate student and about to attain his Bachelor's Degree from Tongji University in 2023. His research interest is focused on nano-biomedical materials. |
1. Introduction
Magnetic resonance imaging (MRI), due to its noninvasiveness and nonionizing radiation, is one of the most commonly utilized imaging techniques in both clinical and fundamental research.1,2 In addition to the excellent penetration depth of MRI for tomography, it exhibits high spatial resolution in soft tissues, which has achieved significant progress in the diagnosis, treatment monitoring and prognosis evaluation of many diseases, such as cancers.3 It has been documented that many atom nuclei can be used in MRI, such as 1H, 2H, 11B, 13C, 19F, 23Na, 31P, and 129Xe,4 among which 1H-based MRI is the most prevalent imaging technique in clinical practice and biomedical research. The high sensitivity and abundance of 1H lead to strong signals in 1H MRI, thus offering precise anatomical and pathological information. However, 1H also causes strong background signals, making 1H MRI difficult to discern the nuance differences between the scarce critical cells and surrounding irrelevant cells.In 1H MRI, the relaxation process of hydrogen protons follows two distinct, independent and simultaneous pathways, i.e., T1 and T2 relaxation.5 To differentiate this nuance difference, contrast agents (CAs) are usually used to improve the precision and sensitivity of 1H MRI, which shorten either the longitudinal (T1) or transverse (T2) relaxation times of water protons.6T1-weighted MRI CAs result in a positive/brighter contrast enhancement, while T2-weighted MRI CAs result in a negative/dark contrast enhancement.7 The basic imaging principle of heteroatom MRI is approximately identical to that of 1H MRI.8 Among the nuclei, 19F, which has relatively high sensitivity and extremely low biological signal background, is accepted to be an exceptional complement to 1H in MRI;9,10 however, its low sensitivity is still the major obstacle in the widespread application of 19F MRI.
Empowered by the inspiring advancement of nanotechnology, recently some new nanobiotechnology-enabled MRI modes with specific applications have been established to vary the longitudinal or transverse relaxations of MRI CAs. Driven by the development of nanobiotechnology, novel concepts and advanced tools have been applied to overcome the shortcomings of conventional MRI CAs, and simultaneously establish new MRI modes to widen the application scope of MRI.11 Besides exhibiting unique properties, these emerging nanobiotechnology-enabled MRI modes also exhibit unexpected capabilities including lesion target detection, lesion imaging localization, curative effect and prognosis evaluations and even imaging-guided treatment, which render the corresponding MRI CAs multi-functional nanoplatforms.12 In the few past decades, a series of new design and synthesis strategies for MRI CAs based on MRI principles have been reported with in-depth and distinctive insights.13 Nanoparticles for fabricating novel MRI CAs are burgeoning into an advanced field, and many reviews have been published. Among them, some focused on special nanomaterials such as superparamagnetic iron oxide nanoparticles (SPIONs)14,15 and SPION-embedded polymeric nanoparticles16 and discussed their biological applications including MRI CAs. Some reviews summarized the crucial preparation parameters and indispensable pre-treatment conditions of magnetic nanoparticles needed in MRI.17 However, to the best of our knowledge, few reviews have focused on the advances in the design strategies of nanobiotechnology in smart relaxation tuning in response to the lesion microenvironment or specific targets, while their applications in the detection of biological targets or activity or monitoring in other diseases besides tumors have not been comprehensively discussed to date. Hence, in this review, we systematically summarize the design principles and potential applications of these new nanotechnology-originated MRI modes and the corresponding MRI CAs (Scheme 1), hopefully offering some guidance and distinctive insights into this field. Initially, the basic principles and classifications of current MRI and MRI CAs are outlined to give an initial impression, followed by the systematic elucidation of the cutting-edge progress in the design strategies of nanobiotechnologies and corresponding MRI CAs based on relaxation tuning. Subsequently, the advantages of these strategies over traditional MRI modes in biomedical and other potential applications will be exemplified with convincing and typical cases, among which their design principles are highlighted. Finally, the current challenges, potential solutions and future development direction of nanobiotechnology-enabled MRI modes and corresponding MRI CAs will be analyzed and deeply discussed.
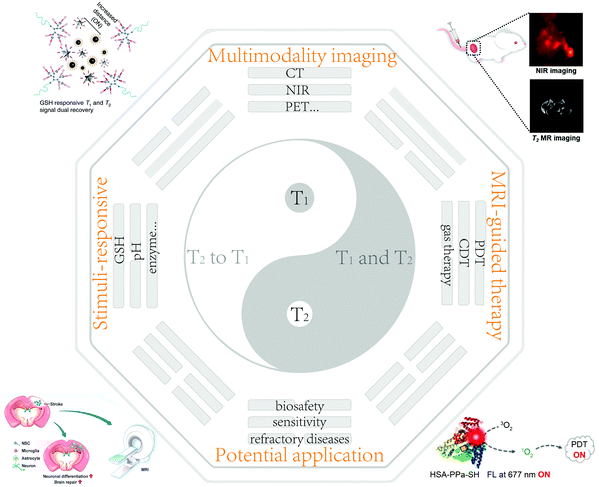 | ||
| Scheme 1 Schematic of the design principles of nanobiotechnology and corresponding MRI CAs with their potential applications. Reproduced with permission from ref. 51. Copyright 2021, the American Chemical Society. Reproduced with permission from ref. 59. Copyright 2020, Wiley-VCH. Reproduced with permission from ref. 107. Copyright 2020, Springer Nature. Reproduced with permission from ref. 128. Copyright 2021, the American Chemical Society. | ||
2. Basic principles of MRI and common MRI CAs
Here, to understand MRI, its basic principles and common MRI CAs are introduced. MRI CAs were developed based on MRI, which can be used to enhance the contrast difference between the target and adjacent tissues.2.1 Basic principles of MRI
The basic principles of MRI are shown in Fig. 1. In detail, as an external magnetic field (B0) is applied, the magnetic spin moments of water protons tend to align with B0, resulting in a net magnetization vector (M0). When an orthogonal radiofrequency pulse is applied, the system can be energized and tends to “flip” the magnetic moments of proton nuclei towards the transversal plane. When the radiofrequency pulse is turned off, the magnetization returns to equilibrium through two distinct processes, i.e., T1 and T2 relaxation.5,18,19T1 measures the recovery time of the initial longitudinal magnetic moments (M0), while T2 measures the loss level of transverse magnetic moments due to dephasing.20,21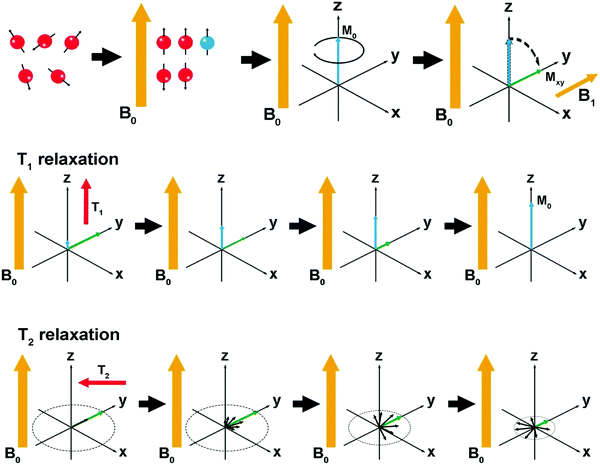 | ||
| Fig. 1 Schematic representation of the basic principles of MRI. Reproduced with permission from ref. 19. Copyright 2019, Springer Nature. | ||
2.2 Conventional MRI CAs
Based on the principles of MRI, various MRI CAs have been developed and their electronic configuration has unpaired electrons, which means that they can change the local relaxation time of H nuclei. T1 MRI CAs can shorten the T1 relaxation time, finally leading to brighter images, which contain paramagnetic complexes or nanoparticles (NPs), e.g., lanthanide ion gadolinium (Gd3+) complexes and transition metal ion manganese (Mn2+)-based NPs.22–25T2 MRI CAs possess stronger ability to shorten the T2 relaxation, leading to a negatively enhanced MR signal and darker images. T2 MRI CAs specially refer to superparamagnetic iron oxide nanoparticles (SPIONs). MRI CAs are applied to shorten the relaxation rate (r1,2 = 1/T1/2) of protons in specific organs, tissues, and disease progression, thus positively or negatively enhancing their contrast and showing enhanced tumbling rates and increased relaxivity.26,27Actually, Fe3+ ions have remarkable T1-weighted contrast enhancement effects. The unpaired spins of Fe3+ ions originate from their outer unpaired electrons. Based on this principle, Zhang and coworkers prepared a tumor microenvironment (TME)-activated nanoprobe for boosting the Fe-based T1 MRI by rearranging the d-electron spin direction of Fe3+ ions once they were released in the TME.36 Also, surface modification could bind more water molecules adjacent to extremely small SPIONs via hydrogen bonding with coordinating phenoxide oxygens after binding with human serum albumin, which also resulted in an outstanding relaxation enhancement.37
Inspired by this, as the size of IONPs decreases, their magnetic moments will be reduced rapidly given that their surface area is enhanced and their volume magnetic anisotropy is reduced, which cooperatively suppress the T2 effect of IONPs, and simultaneously elevate their r1 value,38 eventually heightening the r1/r2 value, which is a key parameter in distinguishing T1 and T2 MRI CAs.25 In this case, these IONPs, which are called extremely ultrasmall superparamagnetic iron oxide nanoparticles (USPIO NPs), have gained increasing interest.39,40 Especially after appropriate modification (e.g., Pepstatin A), USPIO NPs showed incomparable T1 MRI ability with r1 = 4.16, which is close to that of the clinical Gd-DTPA (4.43),41 whose r2/r1 decreased to 5.35, indicating the high potential of T1-weighted MRI. Intriguingly, the stabilization of LDH could increase the space between ultrasmall Fe3O4 nanoparticles and reduce their magnetic coupling, which allowed the LDH-stabilized ultrasmall Fe3O4 to performance much better than free Fe3O4 in enhancing the T1 MRI effect.42
Biosafety is an important concern considering clinical translation, which determines that designing USPIO NPs with excellent biosafety is another direction or hotspot. Actually, due to their extremely small size (below 5 nm), USPIO NPs can cross the kidney filtration system, which allows their rapid excretion in the urine, guaranteeing their easy clearance and favorable biocompatibility in biological applications.43 Xue et al.44 developed nano-enabled, iron-based T1 MRI CAs without nephrotoxicity, which produced a significantly high signal-to-noise ratio (SNR) and rapid urine excretion, making them desirable for precisely localizing tumors in vivo. Thus, these T1-weighted MRI CAs are anticipated to be alternative clinical Gd-based CAs for patients with kidney dysfunction.
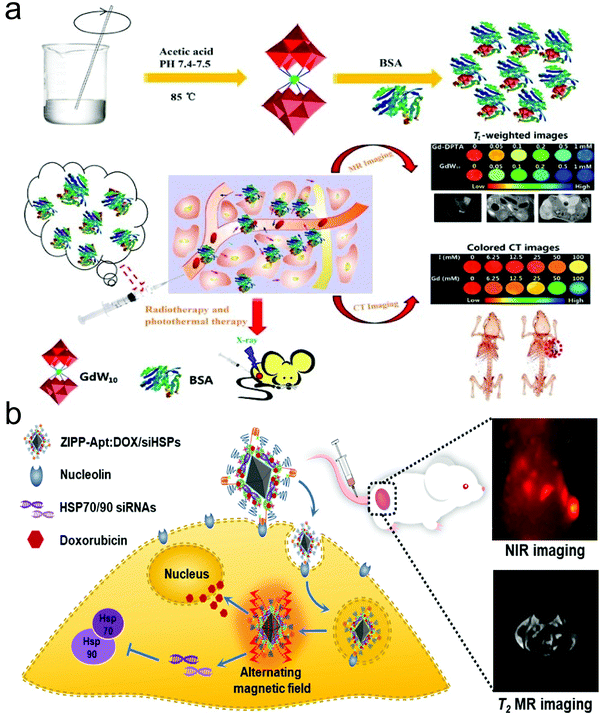 | ||
| Fig. 2 (a) Schematic illustration of GdW10@BSA NCs for dual-modal MR/CT imaging-guided photothermal therapy (PTT)/radiotherapy against cancer. Reproduced with permission from ref. 50. Copyright 2016, Springer Nature. (b) Schematic illustration of sensitized magneto-chemo theranostic agents. Reproduced with permission from ref. 51. Copyright 2021, the American Chemical Society. | ||
Under the assistance of these molecular probes, imaging-guided therapy has been established to improve the treatment efficacy, precision and biosafety.54 Typically, Zou et al. fabricated a dual-modal imaging theranostic system using hollow mesoporous Mn-doped silica shells modified with NIR-FLI molecules as carriers to load anti-tumor drugs.55 The biodegradation of the Mn-doped shell in the acidic TME enabled TME-responsive dual-modal MR/NIR-persistent luminescence (PL) imaging, and concurrently efficiently promoted the controlled release of the loaded anti-tumor drugs for killing tumor cells without side effects.
3. MRI-guided cancer treatment
Imaging-guided treatment is also prevalent and attractive, which can elevate the biosafety and precision of disease treatment especially for interventional ablation and surgery, and meanwhile can effectively alleviate side effects and complications and repress tumor metastasis or reucrrence.56 In the laboratory, the development of various MRI CAs has flourished in this field, laying a solid foundation for clinical translation.3.1 MRI-guided photodynamic therapy
Photodynamic therapy (PDT) is a noninvasive therapeutic method, which employs photosensitizers (PSs) to convert oxygen into highly cytotoxic singlet oxygen (1O2) under light irradiation.57,58 Although most photosensitizers can spontaneously emit fluorescence for FLI-guided PDT, these stimuli-responsive smart photosensitizers in imaging-guided PDT are also impeded by low-sensitivity and low spatial-resolution signals. Thus, to address this issue, An et al.59 developed magnetic and fluorogenic nano-assemblies (NP-RGD) encapsulating PSs via the assembly of cRGD- and disulfide-containing paramagnetic small molecules (1RGD). As illustrated in Fig. 3a, NP-RGD exhibited a high MRI signal with quenched fluorescence and inhibited the PDT effect. Upon entering tumor cells via αvβ3 integrin-mediated targeting delivery, the NP-RGD NPs were reduced by glutathione (GSH) and disassembled into small-molecule probes (2RGD) and organic PSs (PPa-SH). PPa-SH could bind with intracellular albumin and trigger the cascade activation of fluorescence and PDT activity. NIR-FLI and MRI exhibited a synergistic effect to provide accurate bi-modal imaging information with the strongest FLI/MRI signal, highest SNR and considerably prolonged retention time to guide PDT (Fig. 3b and c), consequently resulting in the largest inhibitory rate (Fig. 3d). Tang's group reported the use of upconversion nanoparticles (UCNPs) and aggregation-induced emission (AIE)-active photosensitizers to fabricate a triple-jump photodynamic theranostic nanoplatform, exhibiting an unprecedented performance in FLI-MRI-guided PDT with high efficacy.60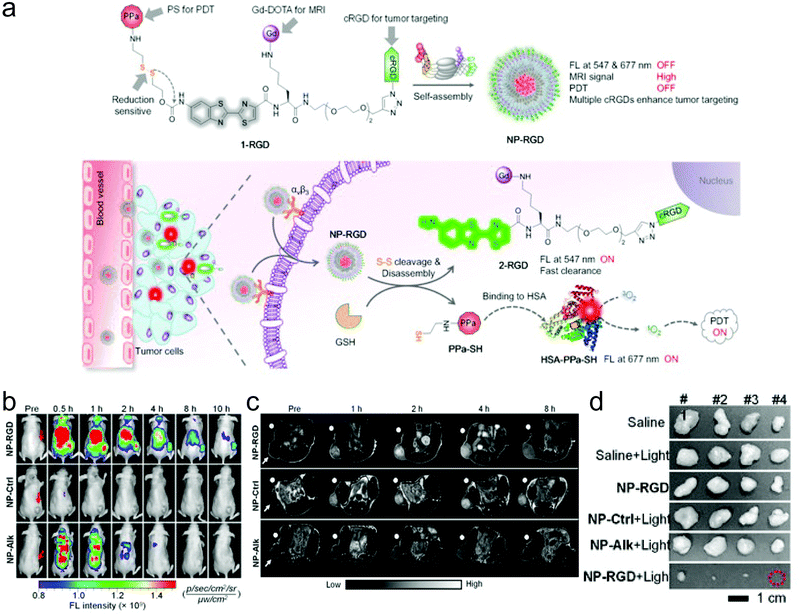 | ||
| Fig. 3 (a) Schematic on how NP-RGD realized on-demand tumor PDT via cascade activation by intracellular GSH and albumin. (b) Fluorescence (FL) images and (c) FL intensities of U87MG tumor in mice receiving i.v. injection of NP-RGD, NP-Ctrl, or NP-Alk (200 μM, 200 μL) at 0 (Pre), 0.5, 1, 2, 4, 8, and 10 h. (d) Photographs of harvested U87MG tumors on day 16 after indicated treatments. Reproduced with permission from ref. 59. Copyright 2020 Wiley-VCH. | ||
The PDT efficacy is still hampered by the hypoxic tumor microenvironment and antioxidant-like GSH, which is another intractable concern.61 Thus, great efforts have been devoted to improving the production of reactive oxygen species (ROS) under the guidance of MRI for precisely and highly efficiently improved PDT-based anti-tumor effects, e.g., uniting chemotherapy and Fenton-like reactions enabled by Fe-based NPs to increase ROS production and alleviate hypoxia.62 Also, catalase (CAT) and MnO2 could be used to produce Mn2+ and deplete GSH to break the ROS and GSH-sustained redox balance, which are available for elevating the net content of PDT-provoked ROS and improving T1-weighted MRI.63
3.2 MRI-guided chemodynamic therapy
Chemodynamic therapy (CDT) is an emerging field, which has been systematically explored in cancer treatment. Different from PDT or sonodynamic therapy (SDT), which needs an external laser or ultrasound source to trigger ROS, the principle of CDT is to harness injected Fenton or Fenton-like agents to react with intratumoral H2O2 for the production of ROS and executing anti-tumor actions. Therefore, imaging guidance is more important for localizing, tracing and evaluating the retention of CDT agents in tumors, wherein MRI alone and MRI-containing multi-modal imaging have been used.64 Typically, Wang et al. reported a novel in situ activatable CDT agent capable of inducing Fe2+ release to catalyze the oxidation of ascorbate monoanion (AscH−) and produce H2O2 with improved efficacy, hence resulting in enhanced Fe-mediated T1-weighted MRI.653.3 MRI-guided hyperthermia therapy
Magnetic hyperthermia therapy (MHT) relies on the heat dissipation by magnetic nanoparticles upon exposure to an alternating magnetic field (AMF).66 The contactless and localized heat production endow MHT with extraordinary advantages in the treatment of deep tumors.67 MRI-guided MHT is easily accessible given that the existing magnetic IONPs, which are usually used as T2-weighted MRI CAs, can be harnessed to enhance MHT.68–70 Shen et al.71 functionalized MnFe2O4 NPs with Ir(III) complexes to construct Ir@MnFe2O4 nanoparticles to elevate the local temperature and induce mitochondrial damage and cell death upon exposure to AMF, during which Fe(III) was reduced to Fe(II) by the overexpressed GSH in the mitochondria of tumor cells, hence offering precise and effective treatment against cancer. Significantly, to address the failure of magnetic nanoparticles in providing real-time thermal feedback in MHT, Erving Ximendes and coworkers72 integrated magnetic nanoparticles with infrared luminescent nanothermometers (LNTh; Ag2S nanoparticles) in optomagnetic nanocapsules. In this strategy, the novel nanocapsules acted as multimodal CAs, providing accurate and real-time thermal feedback during in vivo controllable MHT and avoiding the collateral damage caused by overheating. Besides MHT, MRI-guided photothermal therapy (PTT) was also explored, e.g., polydopamine shell-coated Fe3O4 (i.e., Fe3O4@PDA) could lead to T2-weighted MRI-guided PTT against tumors.73Our group used porous Fe3O4 as carriers to load DL-menthol and constructed a radiofrequency ablation (RFA)-enhancement agent (DLM@Fe3O4).74 DL-Menthol (DLM)-induced inertial cavitation cooperated with Fe3O4-mediatd magnetothermal transformation to significantly enhance the RFA outcome against tumors given that cavitation has been widely accepted to elevate the transformation energy.75–77 However, cavitation is uncontrollable, which probably results in injury to adjacent normal tissues. Fortunately, the loaded DLM made the surface of the Fe3O4 carriers hydrophobic and prevented H2O from approaching to the paramagnetic Fe centers, receiving the quenched MRI. As DLM cavitation proceeded, the gradually decreased DLM in DLM@Fe3O4 increased the contact probability of Fe3O4 with H2O and caused the gradual recovery of MRI. Therefore, the MRI signal variation could be used to monitor DLM cavitation and controllably and precisely ablate tumor without injures to adjacent normal tissues.
4. Nanobiotechnology-encoded relaxation tuning for localizing, monitoring and predicting diseases
Although the above-mentioned MRI CAs are empowered with robust T1 or T2 MRI with large r1 or r2 values, these conventional MRI CAs are local-concentration dependent and suffer from inherently low sensitivity given that an increase in dose cannot elevate the SNR, while facilely posing a high biosafety risk.5 Inspiringly, great advances in constructing CAs to achieve higher relaxivity and/or enhanced accumulation in tissues of interest have been made to overcome these concerns. Especially, the stimulus-activated MRI CAs in response to various lesion microenvironments are favorable to significantly vary the relaxation time and establish new and distinctive MRI modes for directly improving the SNR and magnifying and localizing lesions.78 Conversely, they can reflect and map the stimulated or activated microenvironment such as pH, hypoxia, specific species and targets (e.g., lactate acid and enzymes). These biological targets or events (e.g., pH, GSH, protease activity) have been quantitatively detected and even traced by FLI and PAI after designing rational molecular probes or nanoparticles,79–81 which is a growing field deserving deep exploration. Here, we summarize the nanobiotechnology or MRI CAs-encoded relaxation tuning strategies after rational designs with emphasis on the created novel MRI modes and discuss their merits and properties in reflecting specific lesion microenvironments and localizing, monitoring, predicting and evaluating diseases.4.1 Acidic microenvironment-induced generation of Mn2+ for tuning longitudinal relaxation
Mn-based NPs usually serve as T1 MRI CAs, but they are confronted with poor T1-weighted MRI ability due to their low r1 value (<1). Instead, Mn2+ has a high magnetic moment and long electron spin relaxation time, which make Mn2+ ions ideal T1 MRI CAs with a much larger r1 value (>5).82 Especially after chelation and modification with ligands, the adsorbed water molecules on the outer sphere of Mn2+ complexes allowed molecule rotation to decrease and reinforce the T1 relaxivity of Mn2+ with a considerable increase in r1 value.83In particular, Mn-based NPs can respond to H+ and release Mn2+ after the decomposition of NPs,84,85 enabling intratumoral signal magnification with a high SNR, precision tumor localization, intratumoral pH detection to predict tumor progression and realizing prognosis evaluation.84,86 It has been reported that Mn2+ can amplify the MR signals by interacting with proteins.87,88 As a paradigm, Li and coworkers conjugated ultrasmall manganese ferrite nanoparticles (UMFNPs) with a class of pentapeptide Cys-Arg-Glu-Lys-Ala (CREKA) to synthesize an ultrasensitive T1-weighted MRI probe (UMFNP-CREKA) (Fig. 4a).89 In response to the unique pathological environments of metastases (low pH = 6.5 and rich H2O2), Mn2+ ions were released from the nanoprobe, successfully contributing to the increase in r1 value, especially after binding with 10% fetal bovine serum (FBS) (Fig. 4b). This acidic TME-activated Mn2+ release successfully elevated the SNR of T1-weighted MRI images, as shown in 3D T1 MRI (Fig. 4c), wherein the whole tumor was illuminated after administrating UMFNP-CREKA, whereas the surrounding normal tissues had no significant contrast enhancement.
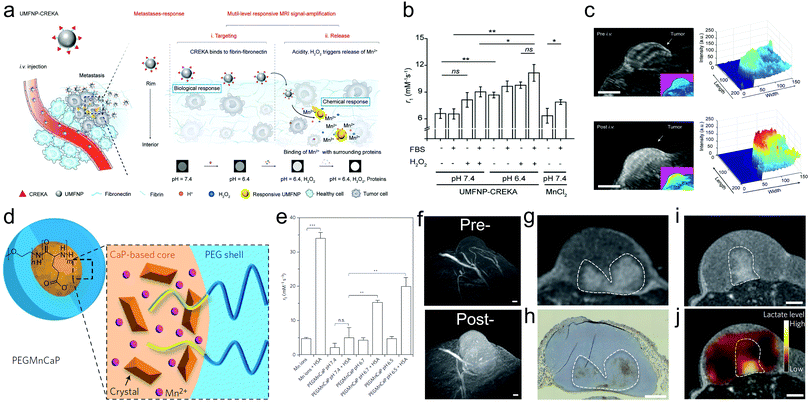 | ||
| Fig. 4 (a) Schematic illustration of the bioinspired UMFNP-CREKA nanoprobe with multilevel responsive T1-weighted MR signal-amplification capabilities for illuminating ultrasmall metastases. (b) r1 values from T1-weighted MR images of UMFNP-CREKA incubated in the indicated conditions for 12 h. (c) 3D MR images of the subcutaneous breast tumor before and at 50 min after intravenous injection of UMFNP-CREKA. Reproduced with permission from ref. 89. Copyright 2019, Wiley-VCH. (d) Structural depiction of the PEG-modified and Mn-doped CaP NPs (PEGMnCa). (e) r1 value of PEG-MnCaP NPs in human serum albumin (HSA)-free or HSA-contained dispersion with varying pH values. (f) 3D MRI images of C26 tumors at pre- and 1 h later post-injection of PEG-modified MnCaP NPs under the 7 T magnetic field, scale bar: 50 μm. MRI image (g) and optical hypoxia-stained image (h) of C26 tumor 4 h after 4 h post-injection of PEG-modified MnCaP NPs under a 1 T magnetic field, scale bar: 1 mm. MRI image (i) and chemical-shift image (j) of C26 tumors after injecting PEG-modified MnCaP and (g) under 7 T magnetic field, scale bar: 1 mm. Reproduced with permission from ref. 90. Copyright 2016, Springer Nature. | ||
Also, Mi and coworkers designed Mn2+-doped calcium phosphate (CaP) nanoparticles,90 followed by PEG modification (Fig. 4d). These MnCaP-PEG nanoparticles were accessible to subject them to H+ erosion for releasing Mn2+, which could be used to tune the longitudinal relaxation and achieve a larger r2 value after chelating with proteins (Fig. 4e). Based on this, these types of nanoparticles can be designed to respond to the acidic TME and specifically magnify the T1-weighted MRI signal of tumors for localizing undetectable metastatic tumors and indirectly reflecting the pH distribution after intravenous injection (Fig. 4f). Considering the fact that the tumor acidity positively correlates with the lactate acid level, which is manipulated by anaerobic metabolism (hypoxia), these nanoparticles can also indirectly map the distributions of hypoxia (Fig. 4g and h) and lactate acid (Fig. 4i and j).
4.2 MRI CA aggregation or disintegration-enabled relaxation tuning for brightening tumor and detecting targets
MRI CAs have aroused considerable interest given that they can serve as magnetic signal transducers for magnetic sensing.91,92 However, due to the dependence on the state change of magnetic nanoparticles, traditional T2-based magnetic immunosensors exert signals that are not sensitive enough to detect the trace amounts of targets, and meanwhile could be interfered by nonspecific aggregation easily.93 It has been accepted that the MRI property is dependent on the structure and surface state of MRI CAs, based on which the aggregation or disintegration of MRI CAs is envisioned to switch ‘on’ or ‘off’ the T1 or T2 MRI signals or vary their longitudinal and transverse relaxations (r1/r2) in response to biological targets or events including acidity, GSH, hypoxia, specific species and targets (e.g., lactate acid and protease) in the disease microenvironment.94 The alteration in on/off or off/on switching in T1 or/and T2 MRI signals can reflect or even track these biological targets or events. Long and coworkers synthesized two sets of IONPs,95 which allowed the transformation of monodispersed sate to aggregate to enhance the T2 signal upon sensing matrix metalloproteinase (MMP) enzymes (Fig. 5a). Once they entered cells that expressed MMP2/9 and CXCR4, the nanoengineered novel IONPs would be activated into a self-assembled superparamagnetic nanocluster network. Consequently, a T2 signal enhancement with around 160% enhancement from the ‘off’ state to ‘on’ state was acquired, providing an opportunity to detect MMP activity at the cellular level (Fig. 5b). Notably, the disaggregation transformation into self-aggregation for altering the relaxation or switching on the MRI signal can be also activated by H+, which indicates the feasibility of H+ detection using this strategy.96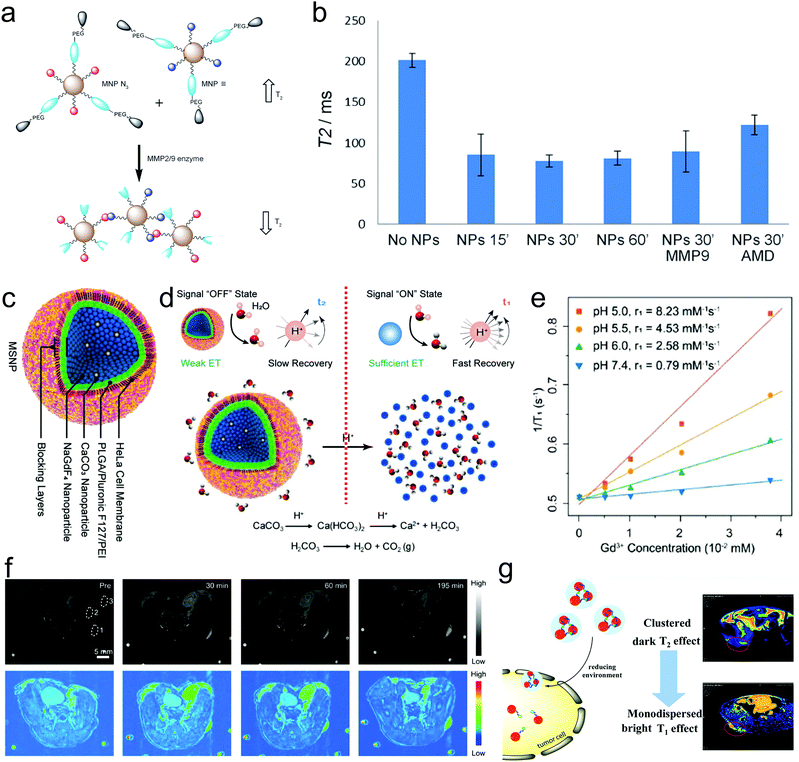 | ||
| Fig. 5 (a) Schematic illustration of MMP-activated particle aggregation. (b) Transverse (T2) relaxation time of U87 cells highly expressing CD4 and CXCR4 and embedded in 1% agarose gel containing Fe3O4 nanoparticles with or without exogenous MMP9 or the CXCR4 blockading agent AMD3100. Reproduced with permission from ref. 95. Copyright 2014, Wiley-VCH. (c) Structural depiction of NaGdF4&CaCO3 co-loaded nanospheres coated with polymeric shells and cell membrane in sequence. (d) Schematic illustration explaining the principles of ‘off’ and ‘on’ states of T1-weighted MRI signals in response to H+. (e) Gd3+ concentration-dependent longitudinal relaxivity profiles at different pH values for calculating the r1 values and (f) T1 MRI images and pseudo-color images of one tumor-implanted mouse before (pre-) and after different time points post-administering of these probes. Reproduced with permission from ref. 97. Copyright 2019 Wiley-VCH. (g) Schematic illustration of the switching of USPIO NP-assembled nanoclusters to monodispersed sate in response to reducing agent for brightening tumor via switching T2 and T1 MRI signals. Reproduced with permission from ref. 98. Copyright 2019, the American Chemical Society. | ||
Although the above-mentioned transformation of the monodispersed state to aggregation can enhance MRI, most cases highlight the aggregation transformation to disintegration activated by some specific tumor targets for realizing the switching of MRI signals from ‘on’ to ‘off’ or from T1 to T2, and vice versa. Typically, Yi constructed NaGdF4&CaCO3 co-loaded nanospheres coated with polymeric shells and cell membrane in sequence (Fig. 5c), wherein the polymeric shells served as blocking layers to prevent H+ erosion and sustain the ‘off’ T1 MRI signal of Gd3+ in NaGdF4 by the confinement-blockaded interaction between H2O and Gd3+ (Fig. 5d).97 As the nanospheres were engulfed by cells, the H+ in the acidic microenvironment could erode and decompose the CaCO3 NPs into CO2 bubbles and destroy the nanospheres, consequently making the NaGdF4 nanoparticles exposed and changing the aggregation state into a disintegrated state (Fig. 5d). This promoted the interaction of Gd3+ in NaGdF4 and H2O and led to T1 MRI signal recovery (‘on’ state) (Fig. 5e). This work not only magnified and localized the tumor with a considerably elevated SNR (Fig. 5f), but also traced or detected the acidity or pH value in the tumor.
Intriguingly, the T1/T2 signal exchange by varying their aggregation or disaggregation state to simultaneously tune their transverse and longitudinal relaxations is also available for detecting specific tumor targets. As a paradigm, Ma et al. reported the preparation of USPIO NP-assembled nanoclusters, and cystamine dihydrochloride (Cys) was used to connect these USPIO NPs, endowing the nanoclusters with robust T2-weighted MRI property (Fig. 5g).98 Under the reducing agent (e.g., GSH) in tumor cells, the disulfide bonds in Cys could be broken, and the USPIO NP-assembled nanoclusters disintegrated and became monodispersed. At this time, the T2-weighted MRI was impaired and the T1-weighted MRI recovered, resulting in ‘off’ T2 and ‘on’ T1 MRI (Fig. 5g), which was available for detecting the GSH level.
4.3 Aggregated MRI CA disintegration for monitoring drug release
Actually, aggregation transformation into the monodispersed sate also favors drug release monitoring, which is beneficial for evaluating and predicting treatment outcomes. Enlightened by the IONP aggregate disintegration-enhanced MRI, anti-tumor drugs can be co-loaded with particle aggregates in pH-sensitive carriers. Upon entry in the acidic TME, the carriers are degraded to release IONPs and anti-tumor drugs, and the switch of the IONPs state from aggregation to disintegration can enhance the MRI signal for reflecting anti-tumor drug release.99 Typically, Zhao et al. loaded manganese arsenite complexes that served as the prodrug of arsenic trioxide (ATO) in hollow silica vehicles, wherein the loaded manganese arsenite complexes could respond to the acidic microenvironment and generate Mn2+ after their decomposition and release ATO. Mn2+ can enhance T1-weighted MRI, enabling the real-time monitoring of the ATO release.100 Besides shaping into pro-drugs via chelation with anti-tumor drugs, Mn-based NPs could be directly utilized with anti-tumor drugs on MSN, and Mn2+ release in response to acid, oxidative stress and redox elevated T1 MRI visualization to monitor anti-tumor drug release.1014.4 Distance-dependent relaxation tuning for detecting targets
Traditional MRI CAs are usually effective in either the T1 or T2 imaging mode. However, the drawbacks such as the short circulation time of T1 MRI CAs and the susceptibility artifacts of T2 MRI CAs are always inevitable.102,103 The challenges in combining the strong T1 and T2 contrast effects in a single CA are mostly ascribed to the strong magnetic coupling between the CAs of T1 and T2.104 Thus strategies to overcome these setbacks have been put proposed. Herein, a distance-dependent magnetic resonance tuning (MRET) strategy was established and the corresponding MRI CAs comprising a paramagnetic T1 MRI ‘enhancer’ (e.g., Gd-DOTA) and a superparamagnetic T2 MRI ‘quencher’ (e.g., Zn0.4Fe2.6O4) were developed.105 The MRET principle was based on the fact that the Gd-DOTA-mediated T1 MRI signal was ‘off’ as the separation distance between Zn0.4Fe2.6O4 and Gd-DOTA was close due to the slow water proton spin relaxation and the slow electron spin fluctuation in Zn0.4Fe2.6O4. In contrast, as they were separated from each other, the water proton spin relaxation and electron spin fluctuation in Zn0.4Fe2.6O4 were concurrently accelerated, leading to ‘on’ T1-weighted MRI (Fig. 6a and b). According to the cleavage or binding with different molecules on the enhancer and quenchers, respectively, this MRET strategy could serve as an MRI-based sensor for detecting both molecular interactions (e.g., binding, cleavage, and reversible protein folding/unfolding) and biological targets (e.g., H2O2, MMP2, Cu+, DNA, H+, Hg+ and ethylene diamine tetraacetic acid (EDTA)) (Fig. 6c). Besides biological activities and disease targets, MRET could be used to detect biological electricity. Typically, Wang et al. reported the preparation of injectable USPIO NP-aggregated clusters (enhancer) entrapped in paramagnetic polymers (quencher), wherein the T1 MRI signal of the clusters was quenched by polymers.106 Once they entered the epilepsy zone, the abnormal electric discharging during epilepsy seizure could widen the distance between the polymer and clusters, restoring the T1 MRI signal and realizing epilepsy visualization with high sensitivity and SNR (Fig. 6d).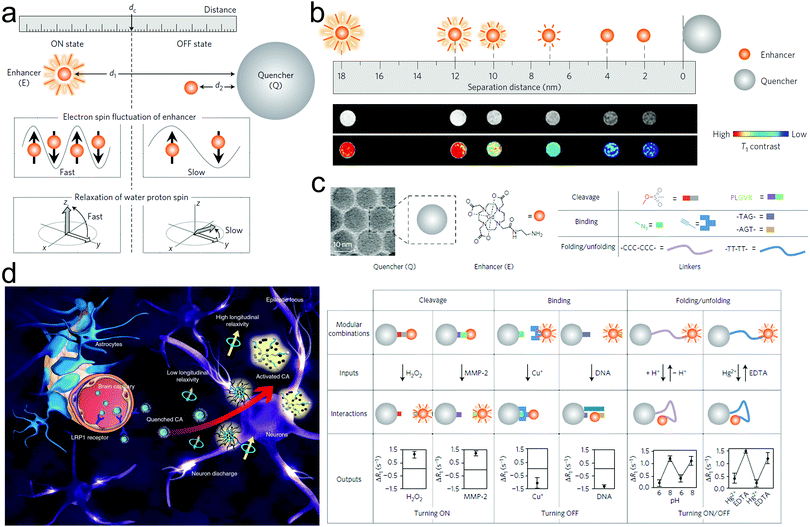 | ||
| Fig. 6 (a) Schematic of MRET principle for illustrating how the distance variation between the enhancer and quencher influences the electron spin and proton spin relaxation. (b) Distance expanding schematic (upper) and corresponding MRI images (bottom) of these enhancer–quencher conjugates in response to their distance variation. (c) Types of quenchers, enhancers, inputs, interactions and outputs in distance-dependent MRET-based probes for distinguishing biological interactions including cleavage, binding and folding/unfolding and detecting various targets. Reproduced with permission from ref. 105. Copyright 2019, Springer Nature. (d) Schematic illustration of electrical field-activated MRET for monitoring abnormal electricity in epilepsy seizure and illuminating the epilepsy zone using T1-MRI. Reproduced with permission from ref. 106. Copyright 2021, Springer Nature. | ||
Actually, based on the magnetism-based nanoscale MRET, dynamical “T2 and T1” dually-switchable MRI CAs could be developed to simultaneously activate T1 and T2 MRI signals for detection.43 Wang et al. introduced new two-way MERT (TMRET) nanotechnology with dually-activatable T1 and T2 signals.107 TMRET nanotechnology was constructed via the co-encapsulation of paramagnetic Mn2+ (P–Mn) chelates and SPIONs in a disulfide-crosslinked micelle (Fig. 7a). Both the T1 and T2 MRI signals were turned ‘off’ when the TMRET pair was ‘locked’ closely in the micelle core. After interacting with biological stimuli (e.g., GSH) to modulate the integrity of the micelles (Fig. 7b), both the T1 and T2 MRI signals were turned ‘on’ as the distance between Mn2+ and SPION increased, enabling the detection of GSH in vitro and in vivo. Pan and coworkers proposed an in situ stimuli-triggered T1–T2 switchable MRI nanoprobe, with IONPs modified with HA,108 and the underlining mechanism is illustrated in Fig. 7c. Before modification with hyaluronic acid (HA), the monodispersed IONPs chelated with polyacrylic acid showed a strong T1 contrast enhancement effect with r1 = 2.227 mM−1 s−1 and r2/r1 ratio = 3.211 (Fig. 7d). After stimulation in an acid environment, the HA-modified IONP-polyacrylic acid nanoparticles (IONP-HP) underwent self-assembly to form nanoclusters, leading to a T2 MRI enhancement (Fig. 7e).
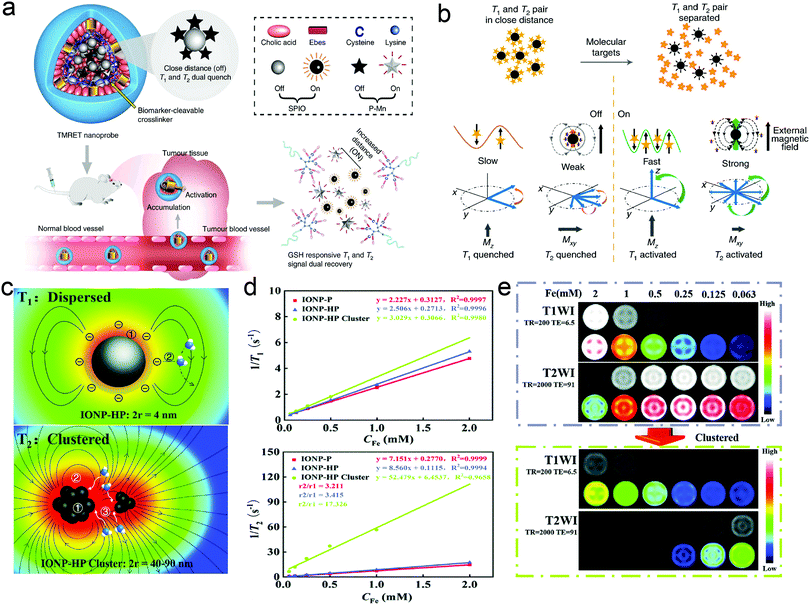 | ||
| Fig. 7 (a) Schematic illustration of TMRET nanotechnology. (b) Illustration of the mechanism of T1 and T2 quenching and recovery in the TMRET nanoprobe. Reproduced with permission from ref. 107. Copyright 2020, Springer Nature. (c) Schematic illustration of the correlation between T1/T2 relaxation and the associated parameters. (d) T1 and T2 relaxation rates of IONP-P, IONP-HP, and IONP-HP clusters at a magnetic field of 1.5 T. (e) T1- and T2-weighted images of IONP-HP before and after clustering at different Fe concentrations on a 1.5 T MRI scanner. Reproduced with permission from ref. 108. Copyright 2021, The Royal Society of Chemistry. | ||
4.5 TME-activatable relaxation tuning for monitoring targets and biological events
Although the MRET and its derived TMRET strategies mentioned above offer unique approaches to improve the sensitivity in tumor imaging,105,107 the special structure consisting of an enhancer and quencher whose selection and connection need precise and rational design to desired targets or biological process, limits its application domain. Herein, various microenvironment-activatable magnetic resonance relaxation tuning strategies have been developed with the facile synthesis and design of the corresponding MRI CAs, showing widespread application potential.Firstly, a new MRI technique, i.e., chemical exchange saturation transfer (CEST), was established to indirectly detect non-labelled and native molecules, wherein the water proton signal through selective saturation of exchangeable protons could be manipulated.109 Based on a furin-mediated 2-cyanobenzothiazole (CBT)-Cys click reaction, Yuan et al. constructed an intracellular conjugate molecule consisting of the anticancer agent Olsalazine (Olsa) and cell-penetrating peptide RVRR (Olsa-PVRR).110 As Olsa-PVRR molecules entered tumor cells, they further self-assembled into Olsa-PVRR NPs under the activation of tumor-associated enzyme furin, consequently varying the CEST MRI signals for assessing tumor aggressiveness and predicting therapeutic outcomes (Fig. 8a). Interestingly, this modality could be reinforced and enriched after incorporating imaging/diagnosis units (e.g., Gd3+ doping).111
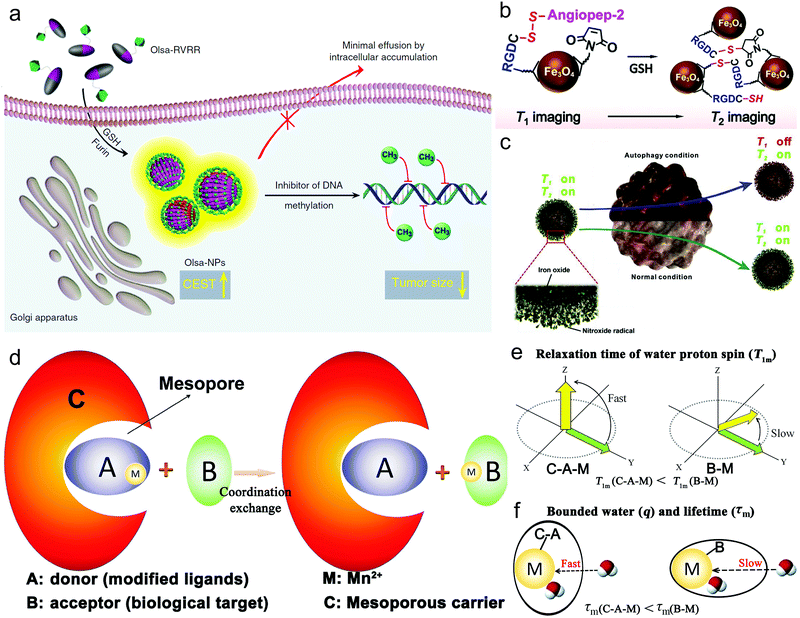 | ||
| Fig. 8 (a) Schematic depicting the GSH and furin-responsive Olsa-RVRR cleavage and self-assembly for enhancing CEST and shrinking tumor in HCT116 cells that highly expressed furin. Reproduced with permission from ref. 110. Copyright 2019, Springer Nature. (b) Schematic depictions on the principle of GSH-activated probe aggregation for enhancing T2-MRI. Reproduced with permission from ref. 113. Copyright 2021, Wiley-VCH. (c) Schematic showing how Fe3O4–NO˙ probe monitors ROS and reflects autophagy. Reproduced with permission from ref. 123. Copyright 2018, Elsevier Ltd. (d) Schematic to shed light on the CSRT strategy. Schematic on the relaxation time of water proton spin (e) and the bonded number of water and the exchange lifetime (f) in CRST strategy. Reproduced with permission from ref. 124. Copyright 2018, Wiley-VCH. | ||
A redox-activatable relaxation tuning strategy for the detection of GSH was also established and various imaging designs and corresponding CAs were designed. Typically, Mn-based nanoparticles with a high valence (e.g., +4) can be reduced to Mn2+ by intratumoral GSH, which can also specifically brighten tumors and reversibly reflect the GSH level.112 Zhang et al. also reported an Fe3O4 nanoparticle-based GSH-responsive MRI probe with simple structure and small dimensions,113 which exhibited interlocked responses of T1 and T2 signals upon reaction with GSH. Moreover, according to their theoretical analysis, they established a mathematic model and realized quantitative mapping of the GSH distribution in brain gliomas through MRI (Fig. 8b). In another study, it was reported that the marriage of concurrent widefield optical imaging and functional MRI could simultaneously realize the fluorescent measurement of the whole brain blood oxygen level and whole-cortex calcium ions for monitoring brain activity, which is beneficial for a comprehensive understanding of brain function.114
Reactive oxygen species or reactive nitrogen species that sustain the redox balance with reducing species have been demonstrated to be correlated with many biological processes, e.g., vascular homeostasis, cancer evolution, stem cell differentiation, bacterial infection, and inflammation.115–118 Their levels decide the different application categories, e.g., tissue repairment or regeneration, tumor treatment, and inflammation alleviation. Considering this, elevating, reducing or sustaining the levels of ROS or reactive nitrogen species (RNS) has been harnessed using different methods (e.g., PDT, CDT, SDT, GSH synthesis inhibition, GSH depletion and nano-enzyme-mediated ROS scavenging) to target these different applications,119,120 which inevitably demand the precise monitoring of ROS or RNS. MRI also provides a candidate for tracing the reactive species. Autophagy plays an extraordinary role in tumor progression by selectively removing damaged organelles under cellular stress, indicating that the in vivo read-out of autophagy is of great significance to illustrate the therapeutic and fundamental effect.121,122 Intriguingly, tracking cellular stress can monitor the autophagy flux. Therefore, Zhang et al. synthesized NO˙ radical-conjugated Fe3O4 nanoparticles (Fe3O4–NO˙ NPs), acting as a dual-modal CA capable of analyzing autophagy dynamics under MR guidance.123 In their investigation, the onset of autophagy causing the mass production of ROS would quench the NO˙-induced T1-weighted signal, whereas the T2-weighted signal was condition-insensitive and remained constant due to the stability of the Fe3O4 nanoparticles upon autophagy, as shown in Fig. 8c. Therefore, the T1-signal variation could reflect the ROS level in a quantitative manner by benchmarking the T2-weighted signal in the autophagy dynamics.
Our group established a coordination-substitution-based relaxation tuning (CSRT) strategy for monitoring disease targets,124 and its principle was shown in Fig. 8d. In detail, as Mn2+-containing MRI CAs (C–A–M) touched biological targets (B), Mn2+ (M) ions were captured via coordination exchange by B to generate B–M (Fig. 8d), resulting in alterations in the water proton spin relaxation time and the number of bonded water protons and proton residence lifetime compared to C–A–M (Fig. 8e and f). By utilizing CSRT and designing the corresponding MRI CAs (C–A–M), GSH and hyaluronic acid (HA) were successfully detected and mapped.
Hypoxia is a specific hallmark of tumor progression, emerging in most solid tumors. It also severely resists ROS-based anti-tumor methods, e.g., chemotherapy, SDT, PDT, and CDT, and thus great efforts have been devoted to alleviating hypoxia and improve anti-tumor outcomes.120,125 In this regard, mapping hypoxia and detecting its level are also important for predicting tumor progression and evaluating curative and prognostic effects. Gregorio et al. utilized red blood cells (RBC) as vehicles to simultaneously conjugate with Gd-DOTP and Gd-HPDO3A, wherein Gd-DOTP and Gd-HPDO3A served as a hypoxia-responsive agent and RBC concentration reporter.126 Relying on the hypoxia-activated longitudinal relaxation variation, these biomimetic probes were equipped with the ability to map hypoxia in a xenografted tumor-bearing mouse model.
Neural activity is closely correlated with intracellular calcium ions, and thus monitoring calcium ions is of great importance for assessing Ca2+ abnormality-related nerve diseases, similar to the aforementioned abnormal electric discharging. Alan Jasanoff et al. synthesized ManICS1-acetomethoxyl MRI CAs.127 After entering cells, these MRI CAs underwent esterase cleavage, eventually enabling intracellular calcium levels to be monitored by MRI. As shown in Fig. 9a, the entrance of MRI CAs into cells led to the cleavage of AM esters, further liberating the calcium-free “off” state and eliciting a low MRI signal. Upon binding with calcium ions, the MRI signal could be enhanced again due to the blockade interactions between BAPTA and the paramagnetic platform. Besides monitoring the dynamic level of Ca2+, neural stem cell (NSC) tracking is also favorable for evaluating the repairing progression of structural and functional damage after stroke and predicting the prognosis by co-delivering SPIONs and small interfering RNA/antisense oligonucleotides (siRNA/ASO) against Pnky long noncoding RNA (lncRNA) (Fig. 9b).128 In detail, in vivo histologic analysis revealed that the neuronal differentiation (red) of the green fluorescence protein (GFP)-NSCs (green) indeed occurred at the transplantation site in stroke mice at 2 and 4 weeks after receiving Pnky-SPIO-PALC- or SCR-SPIO-PALC-transfected GFP-NSCs (Fig. 9c). Based on the cerebral infarct volume measured in the T2-weighted images, this multifunctional nanomedicine targeting lncRNA harvested a considerably-elevated treatment outcome in shrinking the cerebral infarct volume over time (Fig. 9d).
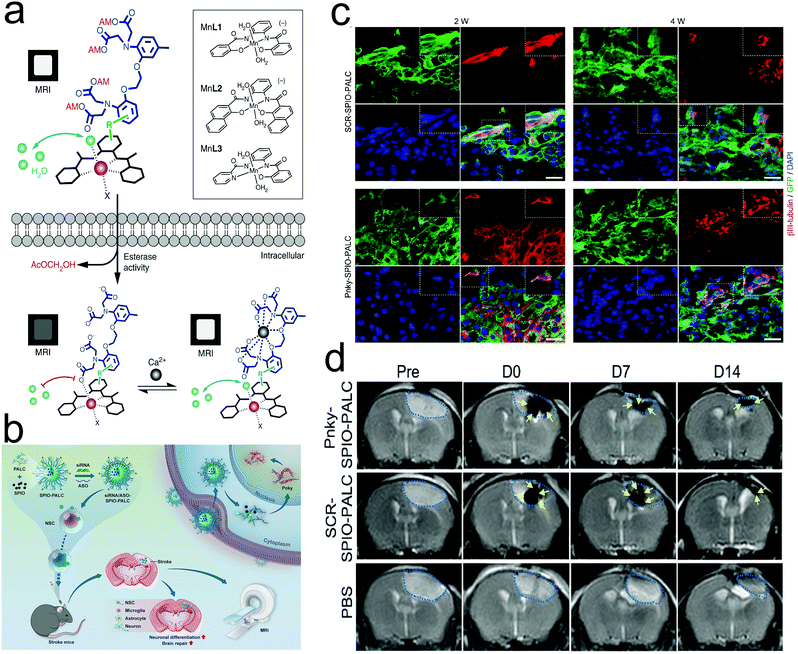 | ||
| Fig. 9 (a) Design of cell-permeable sensors for calcium-dependent molecular fMRI. Reproduced with permission from ref. 127. Copyright 2019, Springer Nature. (b) Schematic depicting the directed neuronal differentiation of transplanted NSCs by silencing Pnky lncRNA and MRI tracking of NSCs in a mouse stroke model. (c) Confocal micrographs showing neuronal differentiation (red) of GFP-NSCs (green) in the transplantation site in stroke mice at 2 and 4 weeks. (d) Coronal T2-weighted MR images of stroke mice transplanted with Pnky-SPIO-PALC- or SCRSPIO-PALC-transfected NSCs and stroke mice. Reproduced with permission from ref. 128. Copyright 2021, the American Chemical Society. | ||
4.6 TME-activatable relaxation tuning for therapeutic gas monitoring
Gas therapy is another cancer therapeutic modality that utilizes gaseous molecules (e.g., carbon monoxide (CO), nitric oxide (NO), and hydrogen sulfide (H2S)) to elicit a therapeutic effect with negligible side effects.129–131 Nevertheless, the precise delivery and controllable release of these therapeutic gases are still challenging, indicating that imaging-guided gas therapy is desirable and urgently demanded. Wu et al.132 proposed a “TME remodeling” strategy, which could achieve the controllable and in situ release of CO. As reported in their research, the decomposition of manganese carbonyl could be caused by acidic TME remodeling, under which the H2O2-enriched TME could also stimulate CO gas release and paramagnetic Mn2+ ion generation for enhancing T1-weighted MRI. Therefore, the treatment process could be real-time monitored by T1-weighted MRI, avoiding the undesirable CO leakage.H2S plays an important role in various physiological and pathological processes, thereby arousing interest in qualitative and quantitative detection techniques for assessing the H2S level in vivo. Based on the fact that Cu2O can in situ respond to endogenous H2S in colon tumors, Yan's group71 constructed a core-shell-structured MRI/PAI dual-modal probe (Fe3O4@Cu2O) for the detection of H2S. In detail, the outer Cu2O shell could adsorb H2S on its surface, thus decreasing the distance between hydrogen protons and Fe, which resulted in a T1-weighted MRI enhancement, consequently visualizing H2S in colorectal cancer. Similarly, a new type of FeS@BSA nanoclusters for MRI-guided synergetic tumor therapy was fabricated.133 In the acidic TME, the FeS@BSA nanoclusters degraded to simultaneously release H2S gas and Fe2+, where the released Fe2+ not only could induce ROS production effectively via Fenton reaction, but also enhance the on-site T2-weighted MRI signals.
4.7 Activatable MRI for tracking cell trajectory and change
Due to the high spatial resolution, inherent soft-tissue contrast and ion-free radiation, MRI is a promising visualization tool that has been utilized to qualitatively track large numbers of cells in vivo,134 among which immune effector cells and immunosuppressive cells are highlighted because their distribution and infiltration directly decide the success or failure of anti-tumor immunotherapy.135–137 Luo et al. developed a bioactivated in vivo assembly method and synthesized a CD206-targeting molecular MRI probe with cathepsin B-sensitive linker. As the probe entered macrophages, the intracellular cathepsin B degraded the linker and induced the probe to assemble into nanofibers with enhanced T1-weighted MRI, which allowed the real-time tracking of macrophage migration (Fig. 10a).138 In another study that focused on dendritic cell (DCs)-activating magnetic nanoparticles,139 researchers revealed that the inclusion of iron oxide could improve DC transfection, and also enable tracking the migration of DCs with MRI. As illustrated in Fig. 10b, researchers successfully fabricated aIO-RNA-NPs by combining IONPs and mRNA encoding tumor antigens in translated lipids. In the presence or absence of a magnetic field, the co-incubation of aIO-RNA-NPs with DCs resulted in profound DC activation and enabled the MRI-based detection of DC migration. In brief, this nanoplatform furnished a powerful tool to activate DCs and predict individual treatment responses to DC vaccines before tumor progression-related signs emerge. Furthermore, myeloid cells including immunosuppressive myeloid-derived suppressor cells (MDSCs) were also successfully tracked using 19F MRI after developing the corresponding CAs.140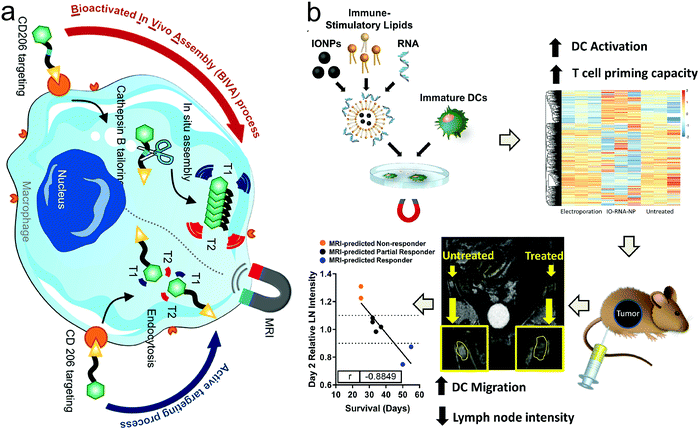 | ||
| Fig. 10 (a) Schematic showing the principle of the bioactivated in vivo assembly method. Reproduced with permission from ref. 138. Copyright 2022, the American Chemical Society. (b) Schematic illustration of aIO-RNA-NPs. Reproduced with permission from ref. 139. Copyright 2019, the American Chemical Society. | ||
Stem cells are prevalent in damage repair, indicating that monitoring the viability, migration, homing and differentiation of stem cells is of importance. The NIR-FLI/MRI dual-modal imaging-guided tracking of labeled mesenchymal stem cells in the brain of a mouse photothrombotic stroke model was reported to achieve noninvasive and real-time tracking for up to 14 days.141 To reveal the in vivo fates of stem cell transplants, which is a promising treatment option for brain injuries, Deng et al. constructed a T2 MRI CA for labeling transplanted stem cells via electroporation.142 As exocytosis or cell death occurred, the released MRI CAs could induce a signal enhancement in the surrounding tissue, and thus the labeled cells could be distinguished from hot tissue, concurrently providing abundant information on the in vivo fates of stem cell transplants.
Specific molecular MRI probes enable the characterization and visualization of molecular and cellular processes in diseases, thereby enhancing the therapeutic effect and monitoring disease progression, e.g., thrombosis.143 In a preclinical study, MRI uncovered the presence of multiple brain metastases in 15 patients bearing melanoma, lung cancer, colon cancer, and breast cancer in the first-in-human phase 1 clinical trial that received a single intravenous administration of Gd-based AGuIX nanoparticles.144 The diagnostic value of Gd-based AGuIX nanoparticles for detecting brain metastases was measured to be equivalent to that of commercial clinically used CAs.
4.8 Activatable 19F-based nanoprobe for MRI and target detection
As stated above, 19F is an exceptional complement to 1H in MRI given that it has relatively high sensitivity, low biological background, and broad chemical shift range compared with other competitors. Modulating the relaxation of 19F nuclei is the major design strategy of 19F MRI nanoprobes. As stated above, the 19F sources that can be utilized in 19F MRI include perfluorocarbons (PFCs), trifluoromethyl compounds, fluorinated ionic liquids, and other fluorine-containing compounds.145 Tang et al.146 synthesized a cascaded multi-responsive 19F-bearing nanoprobe, where the 19F signals could be firstly activated in the reducing TME, and subsequently the photothermal process could amplify the signal (Fig. 11a). The experimental results indicated that the external NIR light irradiation treatment indeed led to higher and sharper 19F nuclear magnetic resonance (NMR) peaks (Fig. 11b) and elevated SNR as the irradiation time or the power density increased (Fig. 11c and d). In vitro and in vivo imaging experiments also demonstrated the feasibility of 19F signal activation/amplification in monitoring the reducing TEM and the photothermal process. Fluorinated ion liquids (ILs) is a new type of fluorine MRI CAs, which could be engineered into activatable 19F MRI nanoplatforms.147 Upon exposure to biological stimuli such as low pH, the 19F signals would be obviously elevated given that the trapped ILs are released to switch on the 19F signal (Fig. 11e). When acidity-activatable 19F MRI was integrated with stable 1H MRI in a MRI nanoconjugate, it provided a ratiometric dual-modal imaging probe for quantitatively detecting pH in tumors (Fig. 11f) using the pH-sensitive variation in the 19F MRI signal as a pH indicator and 1H MRI as the reference tag.148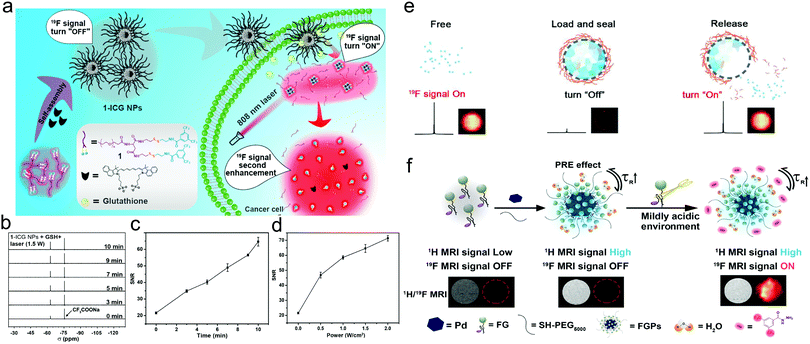 | ||
| Fig. 11 (a) Schematic illustration of self-assembled multi-responsive probes for cascaded 19F signal activation/amplification. (b) 19F NMR spectra of 1-ICG nanoparticles in 500 μL of 10% D2O/H2O (after incubation with GSH for 2 h) upon 808 nm laser irradiation. (c) SNRs of 19F NMR of 1-ICG nanoparticles in 500 μL of 10% D2O/H2O (after incubation with GSH for 2 h) upon 808 nm laser irradiation. (d) SNRs of 19F NMR of 1-ICG nanoparticles (1368 μM; ICG, 21 μg mL−1) with 808 nm laser irradiation at different power intensities for 10 min after incubation with GSH for 2 h. Reproduced with permission from ref. 146. Copyright 2020, the American Chemical Society. (e) Schematic illustration of FILAMP for stimuli-responsive 19F MRI. Reproduced with permission from ref. 147. Copyright 2020, Elsevier. (f) Schematic illustration of fluorinated gadolinium chelate-grafted nanoconjugates for contrast-enhanced T1-weighted 1H and pH-activatable 19F dual-modal MRI. Reproduced with permission from ref. 148. Copyright 2020, the American Chemical Society. | ||
Intriguingly, 19F MRI could be also engineered into a theranostic nanoplatform capable of monitoring protease activity. As a paradigm, Ding and coworkers reported a protease-controlled strategy, which combined simultaneous T2 1H MRI enhancement and 19F MRI “turn-on” effect in one nanoplatform when bridging an 19F compound (i.e., 4-(trifluoromethyl)benzoic acid (TFMB)-Arg-Val-Arg-Arg-Cys(StBu)-Lys-CBT) with Fe3O4.149 Aggregation of Fe3O4 produced a stable T2-weighted 1H MRI signal, while the detachment of the TFMB-ArgVal-Arg-Arg-OH residue from the nanocomplex in response to protease attenuated the paramagnetic relaxation enhancement (PRE) effect and turned on the 19F MRI signal, enabling the detection of protease activity.
Taken together, it can be considered that “structure determines function” is one of the basic principles of designing new MRI modalities and corresponding MRI CAs. In general, the design strategy of novel MRI nanocomplexes is usually inspired by the bio-application demand and distinctive insight into the characteristics of nanomaterials. Various strategies for the stimuli-responsive triggered “on/off” effect were introduced, and the “on” and “off” states reflect the changes in the structure of MRI CAs induced by the disease environment, thereby enabling the detection of the disease environment including targets, biological activity and processes. We systematically illustrated the novel MRI modes reported to date in this review and we intend to exemplify the bio-applications of these strategies, hopefully establishing the relationship that bridges the structure of MRI-based CAs with the advantages of their application. Herein, we present a simple summary on the responsive triggers and the used mechanisms as well as the composition and structure of the corresponding MRI CA in Table 1.
| Nanocomplex | MRI CAs | Trigger | Responsive mechanism | Ref. |
|---|---|---|---|---|
| NaGdF4&CaCO3-co-loaded nanospheres | T1 (Gd3+) | Endogenous pH (acidity) | Acid-activated release of Gd3+ due to the disintegration of NaGdF4 | 97 |
| Gd(III)-based probe 2 (PCP-2). | T1 (Gd3+) | Endogenous GSH | GSH-mediated self-assembly into nanoaggregates | 150 |
| P-CyFF-Gd | T1 (Gd3+) | Endogenous alkaline phosphatase (ALP) enzyme | ALP enzyme-responsive particle aggregation. | 151 |
| P-FFGd-TCO | T1 (Gd3+) | Endogenous ALP enzyme | ALP enzyme-responsive particle self-assembly | 152 |
| MSNs-PEG | T1 (Gd (DTPA)2−) | Exogenous high-intensity focused ultrasound (HIFU) | HIFU stimulation cleave PEG shedding and Gd (DTPA)2− release | 153 |
| Electrically responsive hybrid micelle (EM) | T1 (Gd3+-DTPA, USPIO) | Exogenous electric field | External electric field triggered particle disintegration | 106 |
| DNA–Mn hybrid nanoflower (DMNF) | T1 (Mn2+) | Endogenous pH (acidity) | Acid-activated release Mn2+ | 154 |
| SiO2@Au@MnO2–DOX/Apt | T1 (Mn2+) | Endogenous GSH | GSH-mediated MnO2 decomposition and Mn4+ reduction into Mn2+ | 155 |
| DOX/MnO2@PVCL NGs | T1(Mn2+) | Endogenous GSH | GSH-triggered Mn2+ release | 156 |
| Mesoporous Cu/Mn2SiO3 nanospheres (mCMSNs) | T1 (Mn2+) | Endogenous GSH | GSH-triggered mCMSNs degradation and Mn2+ release. | 157 |
| DL-Menthol (DLM)@Fe3O4 | T1 (Fe3O4) | Exogenous radiofrequency ablation | DLM-induced inertial cavitation cooperated with Fe3O4-mediatd magnetothermal transformation | 74 |
| USPION-DOX nanovesicles (USD NVs) | T1 (USPIONs) | Endogenous GSH | GSH-responsive USPIO particles disaggregation | 158 |
| IONPs | T2 (Fe3O4) | Endogenous matrix metalloproteinase (MMP) enzymes | MMP enzymes-responsive self-assembled superparamagnetic nanocluster network | 95 |
| Chlorin e6 (Ce6) & Fe3O4-co-loaded polymeric micelles (Ce6/Fe3O4-M) | T2 (Fe3O4) | Exogenous PDT-provoked singlet oxygen | Singlet oxygen shortened the transverse relaxation time (T2) | 159 |
| Cystamine dihydrochloride (Cys) connected USPIO NPs | T2 to T1 | Endogenous GSH | GSH-responsive USPIO NPs disintegrated and become monodispersed | 98 |
| USPIO NP-aggregated clusters entrapped in paramagnetic polymers | T2 to T1 | Endogenous abnormal electric discharging during epilepsy seizure | Abnormal electric discharging widens the distance between polymer and clusters, restoring the T1 MRI signal | 106 |
| Co-encapsulating paramagnetic Mn2+ (P–Mn) chelates and SPIONs | T1 and T2 | Endogenous GSH | GSH modulated the integrity of micelles and turned ‘on’ both T1 and T2 MRI signals | 107 |
| IONPs modified with HA | T1 and T2 | Endogenous pH (acidity) | Acid-activated self-assembly of monodispersed IONPs | 108 |
| Fe3O4-NO˙ NPs | T1 and T2 | Endogenous autophagy | Endogenous autophagy quenches the T1-signal of NO˙ | 123 |
| 19F-bearing nanoprobe 1-ICG nanoparticles | 19F | Endogenous reducing TME | Cascaded activated in the reducing TME and subsequently the photothermal process | 146 |
| External NIR light irradiation | ||||
| Fluorinated ionic liquids (ILs) | 19F | Endogenous pH (acidity) | Acid-activated release of trapped ILs to switch on 19F signal | 147 |
| Mn-LDH@PFPE nanoparticles | 19F&T1(Mn2+) | Endogenous pH (acidity) | Acid-activated release Mn2+ reduced the effect of paramagnetic relaxation | 47 |
5 Summary and future perspectives
In this review, we summarized the recent advances on T1 and T2 relaxation tuning with emphasis on the design strategies of nanobiotechnology and corresponding MRI CAs after a simple introduction of the basic principle of MRI. Especially, we elucidated the design principles and clarified the advantages of these MRI modes and corresponding MRI CAs in disease theranostics with an in-depth discussion on bio-applications. This review can offer distinctive insights into the design strategies of nanobiotechnology and corresponding MRI CAs based on relaxation tuning for different scientific communities. It is also believed that the presented MRI-guided therapeutic strategies in this review will significantly contribute to tackling clinical problems with the help of materials science.Although nanobiotechnology-enabled MRI modes have been deeply investigated with increasing interest, there are still obstacles preventing their further clinical application. Firstly, most procedures for the synthesis of nanocomplexes functioning as MRI CAs are uncontrollable and variable to some extent. Hence, researchers should make efforts to pursue stable and mature preparation methods that are greener, cheaper and easier to mass production. Nanobiotechnology development can furnish reliable tools and rationalize the design or synthesis of MRI CAs, where intelligently-responsive and resizable nanoparticles gradually become the mainstream trend to realize the goal of multi-functions. Secondly, although impressive progress in biomedical applications has been achieved, it is worth noting that most strategies are still in the proof-of-concept stage. The aim of most researchers is to attain disease cure in clinic, dictating that the translation from laboratory to clinic is of great significance. Thereby, it is vital to perform in-depth studies on the biodistribution, biocompatibility and interaction of MRI CAs with biological systems before their translation to clinical applications. Meanwhile, preclinical and clinical research should be urgently accelerated. Thirdly, during the past decade, the utilization of water-soluble fluoropolymers in biomedical applications has grown rapidly and 19F MRI has been developed as a convenient diagnostic technique. Nevertheless, few fluoride-based MRI CAs have been systematically investigated in comparison to 1H MRI CAs. When designing fluorinated nanocompounds for biological applications, the important safety concern should be considered, where chemically-stable fluorine-containing molecules without releasing polyfluoroalkyl substances in the environment and human body are preferable, and thus require meticulous exploration. Also, the imaging sensitivity needs to be elevated, where the introduction of paramagnetic units with a relaxation enhancement effect in the course of fluoride-based CA design is considered to further improve the imaging sensitivity. Finally, more attention and efforts are needed for the exploration of refractory diseases besides cancer.
Abbreviations
| MRI | Magnetic resonance imaging |
| CAs | Contrast agents |
| SNR | Signal-to-noise ratio |
| SPIONs | Superparamagnetic iron oxide nanoparticles |
| GQD | Graphene quantum dots |
| PGQD | Paramagnetic GQD |
| PEG | Polyethylene glycol |
| DOX | Doxorubicin |
| Ce6 | Chlorin e6 |
| UMFNPs | Ultrasmall manganese ferrite nanoparticles |
| HSA | Human serum albumin |
| FBS | Fetal bovine serum |
| BSA | Bovine serum albumin |
| IONPs | Iron oxide nanoparticles |
| MMP | Metalloproteinase |
| LDH | Layered double hydroxide |
| USPIO NPs | Ultrasmall superparamagnetic iron oxide nanoparticles |
| HA | Hyaluronic acid |
| MRET | Magnetic resonance tuning |
| TMRET | Two-way MERT |
| NIR | Near-infrared |
| NMR | Nuclear magnetic resonance |
| PRE | Paramagnetic relaxation enhancement |
| ALP | Alkaline phosphatase |
| HIFU | High-intensity focused ultrasound |
| GSH | Glutathione |
| CT | Computed tomography |
| PAI | Photoacoustic imaging |
| PET | Positron emission tomography |
| NIR-PL | Near-infrared persistent luminescence |
| PDT | Photodynamic therapy |
| SDT | Sonodynamic therapy |
| CDT | Chemodynamic therapy |
| PSs | Photosensitizers |
| CAT | Catalase |
| UCNPs | Upconversion nanoparticles |
| MHT | Magnetic hyperthermia therapy |
| AMF | Alternating magnetic field |
| RFA | Radiofrequency ablation |
| DLM | DL-Menthol |
| PTT | Photothermal therapy |
| P–Mn | Paramagnetic Mn2+ |
| EDTA | Ethylene diamine tetraacetic acid |
| RBC | Red blood cells |
| CO | Carbon monoxide |
| NO | Nitric oxide |
| H2S | Hydrogen sulfide |
| TME | Tumor microenvironment |
| RNS | Reactive nitrogen species |
| ROS | Reactive oxygen species |
| DCs | Dendritic cells |
| NSCs | Neural stem cells |
| MDSCs | Myeloid-derived suppressor cells |
| GFP | Green fluorescence protein |
| CEST | Chemical exchange saturation transfer |
| CBT | 2-Cyanobenzothiazole |
| TFMB | 4-(Trifluoromethyl) benzoic acid |
| CSRT | Coordination-substitution-based relaxation tuning |
| FLI | Fluorescence imaging |
| PFCs | Perfluorocarbons |
| ITI | Infrared thermal imaging |
| NPs | Nanoparticles |
| ASO | Antisense oligonucleotides |
| ATO | Arsenic trioxide |
| MMP | Matrix metalloproteinase |
| Olsa | Olsalazine |
| CSRT | Coordination-substitution-based relaxation tuning |
| UCI | Ultrasound contrast imaging |
Authors’ contributions
K. Zhang and T. Wang conceived the theme, organized the structure and collect the references and materials. X. Zhang, Y. Xu, Y. Xu and Y. Zhang apply the copyright. T. Wang and K. Zhang wrote the paper, and K. Zhang revised manuscript and re-organized the figures.Conflicts of interest
The authors declare no competing interests.Acknowledgements
This work was supported by Outstanding Youth Fund of National Natural Science Foundation of China (No. 82022033), Shanghai Rising-Star Program (Grant No. 19QA1406800), Shanghai Talent Development Fund (Grant No. 2019040), the Fundamental Research Funds for the Central Universities (22120210561) and Shanghai Young Top-Notch Talent, National Natural Science Foundation of China (Grant No. 82171942) and Science and Technology Commission of Shanghai Municipality (Grant No. 20ZR1443400).References
- J. Kim, Y. Piao and T. Hyeon, Chem. Soc. Rev., 2009, 38, 372–390 RSC.
- J. P. Liu, E. Fullerton, O. Gutfleisch and D. J. Sellmyer, Nanoscale Magnetic Materials and Applications, Springer, New York, NY, 2009, vol. 1 Search PubMed.
- B. Blasiak, F. C. J. M. van Veggel and B. Tomanek, J. Nanomater., 2013, 1–12 Search PubMed.
- Y. A. Pirogov, Phys. Procedia, 2016, 82, 3–7 CrossRef CAS.
- J. Wahsner, E. M. Gale, A. Rodriguez-Rodriguez and P. Caravan, Chem. Rev., 2019, 119, 957–1057 CrossRef CAS PubMed.
- M. Legacz, K. Roepke, M. Giersig and U. Pison, Adv. Nanopart., 2014, 3, 41–53 CrossRef CAS.
- Y.-K. Peng, S. C. E. Tsang and P.-T. Chou, Mater. Today, 2016, 19, 336–348 CrossRef CAS.
- V. P. Grover, J. M. Tognarelli, M. M. Crossey, I. J. Cox, S. D. Taylor-Robinson and M. J. McPhail, J. Clin. Exp. Hepatol., 2015, 5, 246–255 CrossRef.
- I. Tirotta, V. Dichiarante, C. Pigliacelli, G. Cavallo, G. Terraneo, F. B. Bombelli, P. Metrangolo and G. Resnati, Chem. Rev., 2015, 115, 1106–1129 CrossRef CAS PubMed.
- H. Matsushita, S. Mizukami, F. Sugihara, Y. Nakanishi, Y. Yoshioka and K. Kikuchi, Angew. Chem., Int. Ed., 2014, 53, 1008–1011 CrossRef CAS PubMed.
- R. Kumar, W. S. Shin, K. Sunwoo, W. Y. Kim, S. Koo, S. Bhuniya and J. S. Kim, Chem. Soc. Rev., 2015, 44, 6670–6683 RSC.
- L. Jamgotchian, S. Vaillant, E. Selingue, A. Doerflinger, A. Belime, M. Vandamme, G. Pinna, W. L. Ling, E. Gravel, S. Meriaux and E. Doris, Nanoscale, 2021, 13, 2373–2377 RSC.
- J. Kim, N. Lee and T. Hyeon, Philos. Trans. R. Soc., A, 2017, 375, 20170022 CrossRef PubMed.
- J. K. Oh and J. M. Park, Prog. Polym. Sci., 2011, 36, 168–189 CrossRef CAS.
- K. Ulbrich, K. Hola, V. Subr, A. Bakandritsos, J. Tucek and R. Zboril, Chem. Rev., 2016, 116, 5338–5431 CrossRef CAS PubMed.
- Y. Xiao and J. Du, J. Mater. Chem. B, 2020, 8, 354–367 RSC.
- Y. Lin, K. Zhang, R. Zhang, Z. She, R. Tan, Y. Fan and X. Li, J. Mater. Chem. B, 2020, 8, 5973–5991 RSC.
- D. Weishaupt, V. D. Kochli, B. Marincek and E. E. Kim, J. Nucl. Med., 2007, 48, 1910 CrossRef.
- S. Mastrogiacomo, W. Dou, J. A. Jansen and X. F. Walboomers, Mol. Imaging Biol., 2019, 21, 1003–1019 CrossRef CAS PubMed.
- Z. R. Stephen, F. M. Kievit and M. Zhang, Mater. Today, 2011, 14, 330–338 CrossRef CAS.
- L. G. Hanson, Concepts Magn. Reson., Part A, 2008, 32A, 329–340 CrossRef.
- Y. D. Xiao, R. Paudel, J. Liu, C. Ma, Z. S. Zhang and S. K. Zhou, Int. J. Mol. Med., 2016, 38, 1319–1326 CrossRef CAS PubMed.
- D. Pan, A. H. Schmieder, S. A. Wickline and G. M. Lanza, Tetrahedron, 2011, 67, 8431–8444 CrossRef CAS.
- N. A. Keasberry, M. Banobre-Lopez, C. Wood, G. J. Stasiuk, J. Gallo and N. J. Long, Nanoscale, 2015, 7, 16119–16128 RSC.
- B. H. Kim, N. Lee, H. Kim, K. An, Y. I. Park, Y. Choi, K. Shin, Y. Lee, S. G. Kwon, H. B. Na, J. G. Park, T. Y. Ahn, Y. W. Kim, W. K. Moon, S. H. Choi and T. Hyeon, J. Am. Chem. Soc., 2011, 133, 12624–12631 CrossRef CAS PubMed.
- R. Chen, A. Canales and P. Anikeeva, Nat. Rev. Mater., 2017, 2, 16093 CrossRef CAS PubMed.
- Y. Hu, S. Mignani, J. P. Majoral, M. Shen and X. Shi, Chem. Soc. Rev., 2018, 47, 1874–1900 RSC.
- V. Gulani, F. Calamante, F. G. Shellock, E. Kanal and S. B. Reeder, Lancet Neurol., 2017, 16, 564–570 CrossRef.
- T. J. Fraum, D. R. Ludwig, M. R. Bashir and K. J. Fowler, J. Magn. Reson. Imaging, 2017, 46, 338–353 CrossRef PubMed.
- Y. Yang, S. Chen, H. Li, Y. Yuan, Z. Zhang, J. Xie, D. W. Hwang, A. Zhang, M. Liu and X. Zhou, Nano Lett., 2019, 19, 441–448 CrossRef CAS.
- Y. Li, H. Dong, Q. Tao, C. Ye, M. Yu, J. Li, H. Zhou, S. Yang, G. Ding and X. Xie, Biomaterials, 2020, 250, 120056 CrossRef CAS PubMed.
- J. G. Penfield and R. F. Reilly, Jr., Nat. Clin. Pract. Nephrol., 2007, 3, 654–668 Search PubMed.
- L. Henderson, O. Neumann, C. Kaffes, R. Zhang, V. Marangoni, M. K. Ravoori, V. Kundra, J. Bankson, P. Nordlander and N. J. Halas, ACS Nano, 2018, 12, 8214–8223 CrossRef CAS PubMed.
- W. Xie, Z. Guo, Z. Cao, Q. Gao, D. Wang, C. Boyer, M. Kavallaris, X. Sun, X. Wang, L. Zhao and Z. Gu, ACS Biomater. Sci. Eng., 2019, 5, 2555–2562 CrossRef CAS PubMed.
- T. Sun, Y. Liu, C. Zhou, L. Zhang, X. Kang, S. Xiao, M. Du, Z. Xu, Y. Liu, G. Liu, M. Gong and D. Zhang, Nanoscale, 2021, 13, 7638–7647 RSC.
- P. Zhang, Y. Hou, J. Zeng, Y. Li, Z. Wang, R. Zhu, T. Ma and M. Gao, Angew. Chem., Int. Ed., 2019, 58, 11088–11096 CrossRef CAS PubMed.
- L. Palagi, E. Di Gregorio, D. Costanzo, R. Stefania, C. Cavallotti, M. Capozza, S. Aime and E. Gianolio, J. Am. Chem. Soc., 2021, 143, 14178–14188 CrossRef CAS PubMed.
- B. Chen, Z. Guo, C. Guo, Y. Mao, Z. Qin, D. Ye, F. Zang, Z. Lou, Z. Zhang, M. Li, Y. Liu, M. Ji, J. Sun and N. Gu, Nanoscale, 2020, 12, 5521–5532 RSC.
- R. Jin, B. Lin, D. Li and H. Ai, Curr. Opin. Pharmacol., 2014, 18, 18–27 CrossRef CAS PubMed.
- Z. Zhou, L. Yang, J. Gao and X. Chen, Adv. Mater., 2019, 31, 1804567 CrossRef PubMed.
- C. Du, J. Wang, X. Liu, H. Li, D. Geng, L. Yu, Y. Chen and J. Zhang, Biomaterials, 2020, 230, 119581 CrossRef CAS PubMed.
- N. Zhang, Y. Wang, C. Zhang, Y. Fan, D. Li, X. Cao, J. Xia, X. Shi and R. Guo, Theranostics, 2020, 10, 2791–2802 CrossRef CAS PubMed.
- M. Ma, H. Zhu, J. Ling, S. Gong, Y. Zhang, Y. Xia and Z. Tang, ACS Nano, 2020, 14, 4036–4044 CrossRef CAS PubMed.
- X. Xue, R. Bo, H. Qu, B. Jia, W. Xiao, Y. Yuan, N. Vapniarsky, A. Lindstrom, H. Wu, D. Zhang, L. Li, M. Ricci, Z. Ma, Z. Zhu, T. Y. Lin, A. Y. Louie and Y. Li, Biomaterials, 2020, 257, 120234 CrossRef CAS PubMed.
- C. Zhang, K. Yan, C. Fu, H. Peng, C. J. Hawker and A. K. Whittaker, Chem. Rev., 2022, 122, 167–208 CrossRef CAS PubMed.
- J. Lv and Y. Cheng, Chem. Soc. Rev., 2021, 50, 5435–5467 RSC.
- C. Zhang, L. Li, F. Y. Han, X. Yu, X. Tan, C. Fu, Z. P. Xu and A. K. Whittaker, Small, 2019, 15, 1902309 CrossRef PubMed.
- K. Zhang, H. Chen, P. Li, X. Bo, X. Li, Z. Zeng and H. Xu, ACS Appl. Mater. Interfaces, 2015, 7, 18590–18599 CrossRef CAS PubMed.
- Z. Wang, J. Liu, T. Li, J. Liu and B. Wang, J. Mater. Chem. B, 2014, 2, 4748–4753 RSC.
- Y. Yong, L. Zhou, S. Zhang, L. Yan, Z. Gu, G. Zhang and Y. Zhao, NPG Asia Mater., 2016, 8, e273 CrossRef CAS.
- Z. Chen, Y. Peng, Y. Li, X. Xie, X. Wei, G. Yang, H. Zhang, N. Li, T. Li, X. Qin, S. Li, C. Wu, F. You, H. Yang and Y. Liu, ACS Nano, 2021, 15, 16683–16696 CrossRef CAS PubMed.
- L. Xia, X. Meng, L. Wen, N. Zhou, T. Liu, X. Xu, F. Wang, Z. Cheng, Z. Yang and H. Zhu, Small, 2021, 17, 2100378 CrossRef CAS PubMed.
- X. Han, Y. Xu, Y. Li, X. Zhao, Y. Zhang, H. Min, Y. Qi, G. J. Anderson, L. You, Y. Zhao and G. Nie, ACS Nano, 2019, 13, 4379–4391 CrossRef CAS PubMed.
- S. M. Janib, A. S. Moses and J. A. MacKay, Adv. Drug Delivery Rev., 2010, 62, 1052–1063 CrossRef CAS PubMed.
- R. Zou, J. Li, T. Yang, Y. Zhang, J. Jiao, K. L. Wong and J. Wang, Theranostics, 2021, 11, 8448–8463 CrossRef CAS PubMed.
- Z. Zeng, C. Fang, Y. Zhang, C.-X. Chen, Y.-F. Zhang and K. Zhang, Front. Bioeng. Biotechnol., 2021, 9, 784602 CrossRef PubMed.
- L. Lei, S. Cai, Y. Zhang, L. Yang, J. Deng, H. Mei, X. Zhang, K. Zhang, B. He and J. Cao, Adv. Funct. Mater., 2022, 32, 2103394 CrossRef CAS.
- H. Mei, X. Zhang, S. Cai, X. Zhang, Y. Zhang, Z. Guo, W. Shi, R. Chu, K. Zhang, J. Cao and B. He, Nano Today, 2021, 41, 101305 CrossRef CAS.
- R. An, X. Cheng, S. Wei, Y. Hu, Y. Sun, Z. Huang, H. Y. Chen and D. Ye, Angew. Chem., Int. Ed., 2020, 59, 20636–20644 CrossRef CAS PubMed.
- Y. Wang, Y. Li, Z. Zhang, L. Wang, D. Wang and B. Z. Tang, Adv. Mater., 2021, 33, 2103748 CrossRef CAS PubMed.
- H. Fan, G. Yan, Z. Zhao, X. Hu, W. Zhang, H. Liu, X. Fu, T. Fu, X. B. Zhang and W. Tan, Angew. Chem., Int. Ed., 2016, 55, 5477–5482 CrossRef CAS PubMed.
- T. Zhang, Z. Jiang, L. Chen, C. Pan, S. Sun, C. Liu, Z. Li, W. Ren, A. Wu and P. Huang, Nano Res., 2020, 13, 273–281 CrossRef CAS.
- J. Zhu, T. Xiao, J. Zhang, H. Che, Y. Shi, X. Shi and J. C. M. van Hest, ACS Nano, 2020, 14, 11225–11237 CrossRef CAS PubMed.
- J. Xu, R. Shi, G. Chen, S. Dong, P. Yang, Z. Zhang, N. Niu, S. Gai, F. He, Y. Fu and J. Lin, ACS Nano, 2020, 14, 9613–9625 CrossRef CAS PubMed.
- N. Wang, Q. Zeng, R. Zhang, D. Xing and T. Zhang, Theranostics, 2021, 11, 2334–2348 CrossRef CAS PubMed.
- I. M. Obaidat, B. Issa and Y. Haik, Nanomaterials, 2015, 5, 63–89 CrossRef PubMed.
- M.-H. Chan, M.-R. Hsieh, R.-S. Liu, D.-H. Wei and M. Hsiao, Chem. Mater., 2019, 32, 697–708 CrossRef.
- T. Kim, E. Momin, J. Choi, K. Yuan, H. Zaidi, J. Kim, M. Park, N. Lee, M. T. McMahon, A. Quinones-Hinojosa, J. W. Bulte, T. Hyeon and A. A. Gilad, J. Am. Chem. Soc., 2011, 133, 2955–2961 CrossRef CAS PubMed.
- Z. Li, P. W. Yi, Q. Sun, H. Lei, H. Li Zhao, Z. H. Zhu, S. C. Smith, M. B. Lan and G. Q. M. Lu, Adv. Funct. Mater., 2012, 22, 2387–2393 CrossRef CAS.
- C. Bai, Z. Jia, L. Song, W. Zhang, Y. Chen, F. Zang, M. Ma, N. Gu and Y. Zhang, Adv. Funct. Mater., 2018, 28, 1802281 CrossRef.
- J. Shen, T. W. Rees, Z. Zhou, S. Yang, L. Ji and H. Chao, Biomaterials, 2020, 251, 120079 CrossRef CAS PubMed.
- E. Ximendes, R. Marin, Y. Shen, D. Ruiz, D. Gomez-Cerezo, P. Rodriguez-Sevilla, J. Lifante, P. X. Viveros-Mendez, F. Gamez, D. Garcia-Soriano, G. Salas, C. Zalbidea, A. Espinosa, A. Benayas, N. Garcia-Carrillo, L. Cusso, M. Desco, F. J. Teran, B. H. Juarez and D. Jaque, Adv. Mater., 2021, 33, 2100077 CrossRef CAS PubMed.
- C. Yan, D. Liu, L. An, Y. Wang, Q. Tian, J. Lin and S. Yang, Anal. Chem., 2020, 92, 8254–8261 CrossRef CAS PubMed.
- Y. Fang, H.-Y. Li, H.-H. Yin, S.-H. Xu, W.-W. Ren, S.-S. Ding, W.-Z. Tang, L.-H. Xiang, R. Wu, X. Guan and K. Zhang, ACS Appl. Mater. Interfaces, 2019, 11, 11251–11261 CrossRef CAS PubMed.
- K. Zhang, H. Y. Li, J. Y. Lang, X. T. Li, W. W. Yue, Y. F. Yin, D. Du, Y. Fang, H. Wu, Y. X. Zhao and C. Xu, Adv. Funct. Mater., 2019, 29, 1905124 CrossRef CAS.
- K. Zhang, H. Chen, F. Li, Q. Wang, S. Zheng, H. Xu, M. Ma, X. Jia, Y. Chen, J. Mou, X. Wang and J. Shi, Biomaterials, 2014, 35, 5875–5885 CrossRef CAS PubMed.
- Y. Zhang, L. Guo, F. Kong, L. Duan, H. Li, C. Fang and K. Zhang, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2021, 13, e1733 Search PubMed.
- G. J. Stasiuk, H. Smith, M. Wylezinska-Arridge, J. L. Tremoleda, W. Trigg, S. K. Luthra, V. Morisson Iveson, F. N. Gavins and N. J. Long, Chem. Commun., 2013, 49, 564–566 RSC.
- T. Ma, Y. Hou, J. Zeng, C. Liu, P. Zhang, L. Jing, D. Shangguan and M. Gao, J. Am. Chem. Soc., 2018, 140, 211–218 CrossRef CAS PubMed.
- K. Umezawa, M. Yoshida, M. Kamiya, T. Yamasoba and Y. Urano, Nat. Chem., 2017, 9, 279–286 CrossRef CAS PubMed.
- J. Jo, C. H. Lee, R. Kopelman and X. Wang, Nat. Commun., 2017, 8, 471 CrossRef PubMed.
- Z. Geng, F. Chen, X. Wang, L. Wang, Y. Pang and J. Liu, Biomaterials, 2021, 275, 120897 CrossRef CAS PubMed.
- S. Anbu, S. H. L. Hoffmann, F. Carniato, L. Kenning, T. W. Price, T. J. Prior, M. Botta, A. F. Martins and G. J. Stasiuk, Angew. Chem., Int. Ed., 2021, 60, 10736–10744 CrossRef CAS PubMed.
- B. Duan, D. Wang, H. Wu, P. Xu, P. Jiang, G. Xia, Z. Liu, H. Wang, Z. Guo and Q. Chen, ACS Biomater. Sci. Eng., 2018, 4, 3047–3054 CrossRef CAS PubMed.
- Y. Zhang, Y. Yin, W. Zhang, H. Li, T. Wang, H. Yin, L. Sun, C. Su, K. Zhang and H. Xu, J. Nanobiotechnol., 2021, 19, 161 CrossRef CAS PubMed.
- X. Qian, X. Han, L. Yu, T. Xu and Y. Chen, Adv. Funct. Mater., 2019, 30, 1907066 CrossRef.
- V. S. Lelyveld, E. Brustad, F. H. Arnold and A. Jasanoff, J. Am. Chem. Soc., 2011, 133, 649–651 CrossRef CAS PubMed.
- M. Z. Koylu, S. Asubay and A. Yilmaz, Molecules, 2009, 14, 1537–1545 CrossRef CAS PubMed.
- Y. Li, X. Zhao, X. Liu, K. Cheng, X. Han, Y. Zhang, H. Min, G. Liu, J. Xu, J. Shi, H. Qin, H. Fan, L. Ren and G. Nie, Adv. Mater., 2020, 32, 1906799 CrossRef CAS PubMed.
- P. Mi, D. Kokuryo, H. Cabral, H. Wu, Y. Terada, T. Saga, I. Aoki, N. Nishiyama and K. Kataoka, Nat. Nanotechnol., 2016, 11, 724–730 CrossRef CAS PubMed.
- T. Hyeon, Chem. Commun., 2003, 927–934, 10.1039/b207789b.
- N. Tran and T. J. Webster, J. Mater. Chem., 2010, 20, 8760–8767 RSC.
- C. Kaittanis, S. A. Naser and J. M. Perez, Nano Lett., 2006, 7, 380–383 CrossRef PubMed.
- Q. Wang, Z. Liang, F. Li, J. Lee, L. E. Low and D. Ling, Exploration, 2021, 1, 20210009 CrossRef.
- J. Gallo, N. Kamaly, I. Lavdas, E. Stevens, Q. D. Nguyen, M. Wylezinska-Arridge, E. O. Aboagye and N. J. Long, Angew. Chem., Int. Ed., 2014, 53, 9550–9554 CrossRef CAS PubMed.
- A. Shen, X. Meng, X. Gao, X. Xu, C. Shao, Z. Tang, Y. Liu, W. Bu and P. Wang, Adv. Funct. Mater., 2019, 29, 1803832 CrossRef.
- Z. Yi, Z. Luo, N. D. Barth, X. Meng, H. Liu, W. Bu, A. All, M. Vendrell and X. Liu, Adv. Mater., 2019, 31, 1901851 CrossRef PubMed.
- D. Ma, M. Shi, X. Li, J. Zhang, Y. Fan, K. Sun, T. Jiang, C. Peng and X. Shi, Bioconjugate Chem., 2020, 31, 352–359 CrossRef CAS PubMed.
- X. Zhu, J. Li, P. Peng, N. Hosseini Nassab and B. R. Smith, Nano Lett., 2019, 19, 6725–6733 CrossRef CAS PubMed.
- Z. Zhao, X. Wang, Z. Zhang, H. Zhang, H. Liu, X. Zhu, H. Li, X. Chi, Z. Yin and J. Gao, ACS Nano, 2015, 9, 2749–2759 CrossRef CAS PubMed.
- S. Ren, J. Yang, L. Ma, X. Li, W. Wu, C. Liu, J. He and L. Miao, ACS Appl. Mater. Interfaces, 2018, 10, 31947–31958 CrossRef CAS PubMed.
- F. Li, D. Zhi, Y. Luo, J. Zhang, X. Nan, Y. Zhang, W. Zhou, B. Qiu, L. Wen and G. Liang, Nanoscale, 2016, 8, 12826–12833 RSC.
- Z. Zhou, D. Huang, J. Bao, Q. Chen, G. Liu, Z. Chen, X. Chen and J. Gao, Adv. Mater., 2012, 24, 6223–6228 CrossRef CAS PubMed.
- D. W. Yoo, J.-H. Lee, T.-H. Shin and A. J. Cheon, Acc. Chem. Res., 2011, 44, 863–874 CrossRef CAS PubMed.
- J. S. Choi, S. Kim, D. Yoo, T. H. Shin, H. Kim, M. D. Gomes, S. H. Kim, A. Pines and J. Cheon, Nat. Mater., 2017, 16, 537–542 CrossRef CAS PubMed.
- C. Wang, W. Sun, J. Zhang, J. Zhang, Q. Guo, X. Zhou, D. Fan, H. Liu, M. Qi, X. Gao, H. Xu, Z. Gao, M. Tian, H. Zhang, J. Wang, Z. Wei, N. J. Long, Y. Mao and C. Li, Nat. Biomed. Eng., 2021, 5, 278–289 CrossRef CAS PubMed.
- Z. Wang, X. Xue, H. Lu, Y. He, Z. Lu, Z. Chen, Y. Yuan, N. Tang, C. A. Dreyer, L. Quigley, N. Curro, K. S. Lam, J. H. Walton, T. Y. Lin, A. Y. Louie, D. A. Gilbert, K. Liu, K. W. Ferrara and Y. Li, Nat. Nanotechnol., 2020, 15, 482–490 CrossRef CAS PubMed.
- C. Pan, J. Lin, J. Zheng, C. Liu, B. Yuan, O. U. Akakuru, M. Zubair Iqbal, Q. Fang, J. Hu, J. Chen, J. Lin, Q. Dai, X. Guo, Z. Li, T. Zhang, C. Xu, X. Ma, T. Chen, A. Wu and Y. Jin, Nanoscale, 2021, 13, 6461–6474 RSC.
- K. M. Ward, A. H. Aletras and R. S. Balaban, J. Magn. Reson., 2000, 143, 79–87 CrossRef CAS PubMed.
- Y. Yuan, J. Zhang, X. Qi, S. Li, G. Liu, S. Siddhanta, I. Barman, X. Song, M. T. McMahon and J. W. M. Bulte, Nat. Mater., 2019, 18, 1376–1383 CrossRef CAS PubMed.
- Z. Ren, S. Sun, R. Sun, G. Cui, L. Hong, B. Rao, A. Li, Z. Yu, Q. Kan and Z. Mao, Adv. Mater., 2020, 32, 1906024 CrossRef CAS PubMed.
- Z. Sha, S. Yang, L. Fu, M. Geng, J. Gu, X. Liu, S. Li, X. Zhou and C. He, Nanoscale, 2021, 13, 5077–5093 RSC.
- P. Zhang, J. Zeng, Y. Li, C. Yang, J. Meng, Y. Hou and M. Gao, Angew. Chem., Int. Ed., 2021, 60, 8130–8138 CrossRef CAS PubMed.
- E. M. R. Lake, X. Ge, X. Shen, P. Herman, F. Hyder, J. A. Cardin, M. J. Higley, D. Scheinost, X. Papademetris, M. C. Crair and R. T. Constable, Nat. Methods, 2020, 17, 1262–1271 CrossRef CAS PubMed.
- M. Yang, Y. Zhang, C. Fang, L. Song, Y. Wang, L. Lu, R. Yang, Z. Bu, X. Liang, K. Zhang and Q. Fu, Adv. Mater., 2022, 34, 2109522 CrossRef CAS PubMed.
- L. Zheng, S. Liu, X. Cheng, Z. Qin, Z. Lu, K. Zhang and J. Zhao, Adv. Sci., 2019, 6, 1900099 CrossRef PubMed.
- K. Zhang, Y. Fang, Y. He, H. Yin, X. Guan, Y. Pu, B. Zhou, W. Yue, W. Ren, D. Du, H. Li, C. Liu, L. Sun, Y. Chen and H. Xu, Nat. Commun., 2019, 10, 5380 CrossRef PubMed.
- H. Cao, L. Duan, Y. Zhang, J. Cao and K. Zhang, Signal Transduction Targeted Ther., 2021, 6, 426 CrossRef CAS PubMed.
- X. Guan, H.-H. Yin, X.-H. Xu, G. Xu, Y. Zhang, B.-G. Zhou, W.-W. Yue, C. Liu, L.-P. Sun, H.-X. Xu and K. Zhang, Adv. Funct. Mater., 2020, 30, 2000326 CrossRef CAS.
- T. Luo, D. Wang, L. Liu, Y. Zhang, C. Han, Y. Xie, Y. Liu, J. Liang, G. Qiu, H. Li, D. Su, J. Liu and K. Zhang, Adv. Sci., 2021, 8, 2101065 CrossRef CAS PubMed.
- C. He and D. J. Klionsky, Annu. Rev. Genet., 2009, 43, 67–93 CrossRef CAS PubMed.
- L. Robke, L. Laraia, M. A. Carnero Corrales, G. Konstantinidis, M. Muroi, A. Richters, M. Winzker, T. Engbring, S. Tomassi, N. Watanabe, H. Osada, D. Rauh, H. Waldmann, Y. W. Wu and J. Engel, Angew. Chem., Int. Ed., 2017, 56, 8153–8157 CrossRef CAS PubMed.
- C. Zhang, J. Ren, J. He, Y. Ding, D. Huo and Y. Hu, Biomaterials, 2018, 179, 186–198 CrossRef CAS PubMed.
- K. Zhang, Y. Cheng, W. Ren, L. Sun, C. Liu, D. Wang, L. Guo, H. Xu and Y. Zhao, Adv. Sci., 2018, 5, 1800021 CrossRef PubMed.
- J. Chen, H. Luo, Y. Liu, W. Zhang, H. Li, T. Luo, K. Zhang, Y. Zhao and J. Liu, ACS Nano, 2017, 11, 12849–12862 CrossRef CAS PubMed.
- E. D. Gregorio, G. Ferrauto, E. Gianolio, S. Lanzardo, C. Carrera, F. Fedeli and S. Aime, ACS Nano, 2015, 9, 8239–8248 CrossRef PubMed.
- A. Barandov, B. B. Bartelle, C. G. Williamson, E. S. Loucks, S. J. Lippard and A. Jasanoff, Nat. Commun., 2019, 10, 897 CrossRef PubMed.
- B. Lin, L. Lu, Y. Wang, Q. Zhang, Z. Wang, G. Cheng, X. Duan, F. Zhang, M. Xie, H. Le, X. Shuai and J. Shen, Nano Lett., 2021, 21, 806–815 CrossRef CAS PubMed.
- Y. Liu, F. Yang, C. Yuan, M. Li, T. Wang, B. Chen, J. Jin, P. Zhao, J. Tong, S. Luo and N. Gu, ACS Nano, 2017, 11, 1509–1519 CrossRef CAS PubMed.
- X. Jia, Y. Zhang, Y. Zou, Y. Wang, D. Niu, Q. He, Z. Huang, W. Zhu, H. Tian, J. Shi and Y. Li, Adv. Mater., 2018, 30, 1704490 CrossRef PubMed.
- P. Zhao, Z. Jin, Q. Chen, T. Yang, D. Chen, J. Meng, X. Lu, Z. Gu and Q. He, Nat. Commun., 2018, 9, 4241 CrossRef PubMed.
- J. Wu, Z. Meng, A. A. Exner, X. Cai, X. Xie, B. Hu, Y. Chen and Y. Zheng, Biomaterials, 2021, 276, 121001 CrossRef CAS PubMed.
- C. Xie, D. Cen, Z. Ren, Y. Wang, Y. Wu, X. Li, G. Han and X. Cai, Adv. Sci., 2020, 7, 1903512 CrossRef CAS PubMed.
- W. J. Rogers, C. H. Meyer and C. M. Kramer, Nat. Clin. Pract. Cardiovasc. Med., 2006, 3, 554–562 CrossRef CAS PubMed.
- J. Zhang, M. Yang, X. Fan, M. Zhu, Y. Yin, H. Li, J. Chen, S. Qin, H. Zhang, K. Zhang and F. Yu, J. Nanobiotechnol., 2022, 20, 103 CrossRef CAS PubMed.
- Y. Yin, X. Jiang, L. Sun, H. Li, C. Su, Y. Zhang, G. Xu, X. Li, C. Zhao, Y. Chen, H. Xu and K. Zhang, Nano Today, 2021, 36, 101009 CrossRef CAS.
- H. Wu, H. Li, Y. Liu, J. Liang, Q. Liu, Z. Xu, Z. Chen, X. Zhang, K. Zhang and C. Xu, Bioact. Mater., 2022, 13, 223–238 CrossRef CAS PubMed.
- L.-J. Luo, X.-M. Liu, X. Zhang, J. Liu, Y. Gao, T.-Y. Sun and L.-L. Li, Nano Lett., 2022, 22, 1694–1702 CrossRef CAS PubMed.
- A. J. Grippin, B. Wummer, T. Wildes, K. Dyson, V. Trivedi, C. Yang, M. Sebastian, H. R. Mendez-Gomez, S. Padala, M. Grubb, M. Fillingim, A. Monsalve, E. J. Sayour, J. Dobson and D. A. Mitchell, ACS Nano, 2019, 13, 13884–13898 CrossRef CAS PubMed.
- M. L. Senders, A. E. Meerwaldt, M. M. T. van Leent, B. L. Sanchez-Gaytan, J. C. van de Voort, Y. C. Toner, A. Maier, E. D. Klein, N. A. T. Sullivan, A. M. Sofias, H. Groenen, C. Faries, R. S. Oosterwijk, E. M. van Leeuwen, F. Fay, E. Chepurko, T. Reiner, R. Duivenvoorden, L. Zangi, R. M. Dijkhuizen, S. Hak, F. K. Swirski, M. Nahrendorf, C. Perez-Medina, A. J. P. Teunissen, Z. A. Fayad, C. Calcagno, G. J. Strijkers and W. J. M. Mulder, Nat. Nanotechnol., 2020, 15, 398–405 CrossRef CAS PubMed.
- S. Lim, H. Y. Yoon, H. J. Jang, S. Song, W. Kim, J. Park, K. E. Lee, S. Jeon, S. Lee, D. K. Lim, B. S. Kim, D. E. Kim and K. Kim, ACS Nano, 2019, 13, 10991–11007 CrossRef CAS PubMed.
- Y. Zhang, H. Zhang, B. Li, H. Zhang, B. Tan and Z. Deng, Nano Res., 2018, 11, 1625–1641 CrossRef CAS.
- J. Liu, J. Xu, J. Zhou, Y. Zhang, D. Guo and Z. Wang, Int. J. Nanomed., 2017, 12, 1113–1126 CrossRef CAS PubMed.
- C. Verry, S. Dufort, B. Lemasson, S. Grand, J. Pietras, I. Troprès, Y. Crémillieux, F. Lux, S. Mériaux, B. Larrat, J. Balosso, G. L. Duc, E. L. Barbier and O. Tillement, Sci. Adv., 2020, 6, eaay5279 CrossRef CAS PubMed.
- H. Lin, X. Tang, A. Li and J. Gao, Adv. Mater., 2021, 33, 2005657 CrossRef CAS PubMed.
- X. Tang, X. Gong, A. Li, H. Lin, C. Peng, X. Zhang, X. Chen and J. Gao, Nano Lett., 2020, 20, 363–371 CrossRef CAS PubMed.
- X. Zhu, X. Tang, H. Lin, S. Shi, H. Xiong, Q. Zhou, A. Li, Q. Wang, X. Chen and J. Gao, Chem, 2020, 6, 1134–1148 CAS.
- X. Tang, X. Gong, J. Ming, D. Chen, H. Lin and J. Gao, Anal. Chem., 2020, 92, 16293–16300 CrossRef CAS PubMed.
- Z. Ding, H. Sun, S. Ge, Y. Cai, Y. Yuan, Z. Hai, T. Tao, J. Hu, B. Hu, J. Wang and G. Liang, Adv. Funct. Mater., 2019, 29, 1903860 CrossRef CAS.
- H. Li, D. Luo, C. Yuan, X. Wang, J. Wang, J. P. Basilion and T. J. Meade, J. Am. Chem. Soc., 2021, 143, 17097–17108 CrossRef CAS PubMed.
- R. Yan, Y. Hu, F. Liu, S. Wei, D. Fang, A. J. Shuhendler, H. Liu, H. Y. Chen and D. Ye, J. Am. Chem. Soc., 2019, 141, 10331–10341 CrossRef CAS PubMed.
- Y. Hu, J. Zhang, Y. Miao, X. Wen, J. Wang, Y. Sun, Y. Chen, J. Lin, L. Qiu, K. Guo, H. Y. Chen and D. Ye, Angew. Chem., Int. Ed., 2021, 60, 18082–18093 CrossRef CAS PubMed.
- C. A. Cheng, W. Chen, L. Zhang, H. H. Wu and J. I. Zink, J. Am. Chem. Soc., 2019, 141, 17670–17684 CrossRef CAS PubMed.
- H. Zhao, J. Lv, F. Li, Z. Zhang, C. Zhang, Z. Gu and D. Yang, Biomaterials, 2021, 268, 120591 CrossRef CAS PubMed.
- T. T. Zhang, C. H. Xu, W. Zhao, Y. Gu, X. L. Li, J. J. Xu and H. Y. Chen, Chem. Sci., 2018, 9, 6749–6757 RSC.
- F. Xu, J. Zhu, L. Lin, C. Zhang, W. Sun, Y. Fan, F. Yin, J. C. M. van Hest, H. Wang, L. Du and X. Shi, Theranostics, 2020, 10, 4349–4358 CrossRef CAS PubMed.
- C. Liu, D. Wang, S. Zhang, Y. Cheng, F. Yang, Y. Xing, T. Xu, H. Dong and X. Zhang, ACS Nano, 2019, 13, 4267–4277 CrossRef CAS PubMed.
- D. Liu, Z. Zhou, X. Wang, H. Deng, L. Sun, H. Lin, F. Kang, Y. Zhang, Z. Wang, W. Yang, L. Rao, K. Yang, G. Yu, J. Du, Z. Shen and X. Chen, Biomaterials, 2020, 244, 119979 CrossRef CAS PubMed.
- K. Deng, B. Wu, C. X. Wang, Q. Wang, H. Yu, J. M. Li, K. H. Li, H. Y. Zhao and S. W. Huang, Adv. Healthcare Mater., 2020, 9, 2000533 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |



