 Open Access Article
Open Access ArticleActivatable peptide–AIEgen conjugates for cancer imaging
Sisi
Zhou
a,
Xianbao
Sun
 *b and
Gaolin
Liang
*b and
Gaolin
Liang
 *b
*b
aJiangsu Engineering Laboratory of Smart Carbon-Rich Materials and Device, School of Chemistry and Chemical Engineering, Southeast University, Nanjing 211189, China. E-mail: xbsun@seu.edu.cn; gliang@seu.edu.cn
bState Key Laboratory of Digital Medical Engineering, School of Biological Science and Medical Engineering, Southeast University, Nanjing 211189, China
First published on 28th February 2025
Abstract
Aggregation-induced emission luminogens (AIEgens) have undergone significant development over the past decade, making substantial and profound contributions to a diverse range of research fields, prominently including cancer/disease diagnosis and therapy. Through the covalent conjugation of AIEgens with functional peptides, the resultant peptide–AIEgen conjugates possess not only the excellent biocompatibility characteristics, along with low systemic toxicity and negligible immunogenicity of peptides, but also the remarkable fluorescence properties of AIEgens. This “win–win” integration has significantly propelled the applications of peptide–AIEgen conjugates, particularly within the domain of cancer imaging. Three principal types of peptide–AIEgen conjugates, namely, tumor-targeting, tumor biomarker-responsive, and biomarker-responsive self-assembling peptide–AIEgen conjugates, are commonly devised. These conjugates confer enhanced targeting capabilities and selectivity towards tumors, thereby facilitating “smart” and precise tumor imaging with high signal-to-background ratios. In light of the crucial significance of peptide–AIEgen conjugates in tumor imaging and the recent inspiring breakthroughs that have not been encompassed in previous reviews, we present this review. We highlight the activatable peptide–AIEgen conjugates developed for tumor imaging over the past three years (from 2022 to the present). Particular attention is directed towards their design rationales, operational mechanisms, and imaging performance. Finally, prospective opportunities within this field are also reasonably deliberated.
1. Introduction
Since its discovery in 2001 by Tang and co-workers,1 aggregation-induced emission (AIE) has always been a hot topic. It has made important contributions to a multitude of research fields, such as disease diagnosis,2,3 drug screening,4 antibacterial therapy,5,6 and cancer imaging and therapy.7–9 Contrary to traditional fluorophores that suffer from aggregation-induced quenching (ACQ) of fluorescence in their aggregated states or concentrated solutions, AIE luminogens (AIEgens) are able to emit fluorescence in these cases. The mechanism underlying AIE hinges on the restriction of intramolecular motions (RIM) of AIE luminogens (AIEgens) upon aggregation.10,11 This restriction leads to the dissipation of the excited-state energy through fluorescence emission rather than the nonradiative decay pathway, thereby accounting for the unique optical properties of AIEgens. In this regard, the formation of aggregates by AIEgens is not an essential and absolute prerequisite for the activation of their fluorescence because any factor or condition that serves to impede or constrain the rotational or vibrational motions of AIEgens is likely to possess the propensity to turn on their fluorescence emission.4,12 For instance, once an AIEgen gets immobilized upon binding to certain substrates, it tends to exhibit fluorescence emission owing to the occurrence of RIM. The above AIE-governing principles enable researchers to devise fluorescent light-up AIEgens in an on-demand manner. Owing to their unique advantages, such as large Stokes shift, high signal-to-background ratios, and tailorable amenability, these light-up AIEgens have been applied to a wide range of bioimaging applications.13Among the advanced AIEgens, peptide–AIEgen conjugates, which are fabricated by conjugating AIEgens with functional peptides, have recently been intensively developed.14–18 On the one hand, peptides are nature-derived materials possessing inherent biocompatibility, biodegradability, and even bioactivity, endowing peptide–AIEgen conjugates with superior biosafety as well as low systemic toxicity and immunogenicity.19–21 On the other hand, owing to their highly programmable functions, peptides can serve as targeting motifs,22 responsive substrates,23,24 self-assembling building blocks,25 and even therapeutic agents.5,26,27 Such versatility empowers peptide–AIEgen conjugates with a wide range of biomedical applications. In particular, in the field of cancer imaging, rationally designed peptide–AIEgen conjugates confer improved targeting and selectivity toward tumors, thus enabling “smart” and precise tumor imaging with high signal-to-background ratios. Such peptide–AIEgen conjugates mainly include the following three types. First, tumor-targeting peptide–AIEgen conjugates, which are constructed by coupling AIEgens with tumor-targeting peptides or moieties, can actively target and bind to specific tumor targets and subsequently undergo RIM of AIEgens with fluorescence turned on. Second, tumor biomarker-responsive peptide–AIEgen conjugates, which covalently link AIEgens to hydrophilic peptide substrates of certain tumor biomarkers (typically, enzymes), can be specifically hydrolyzed by the corresponding biomarkers to liberate the hydrophobic AIEgens, which subsequently undergo aggregation to turn on the AIE fluorescence in biomarker-overexpressing tumor tissues. Third, biomarker-responsive self-assembling peptide–AIEgen conjugates, which are constructed by conjugating AIEgens to both biomarker-responsive peptides and self-assembling peptides, can be activated by the biomarkers and further assemble into orderly nanostructures (e.g., nanoparticles and nanofibers), which promote the closer proximity and tighter intermolecular steric interactions of AIE molecules, thereby rendering enhanced fluorescence output in tumor tissues.
In the past three years, while several excellent reviews have centered on peptide–AIEgen conjugates,28–31 no reviews dedicated to this topic from 2023 up to the present have been found. Given the significant importance of peptide–AIEgen conjugates in tumor imaging and the recent encouraging breakthroughs that have not been covered in previous reviews, we present this review. We highlight the activatable peptide–AIEgen conjugates developed for tumor imaging over the past three years (from 2022 to the present) based on the aforementioned three design strategies (Fig. 1), with particular emphasis on their design principles, operating mechanisms, and imaging performance. Finally, future opportunities in this field are also discussed. This review is anticipated to facilitate researchers in comprehensively grasping the latest progresses and prospective striving directions of peptide–AIEgen conjugates within the realm of tumor imaging and further stimulate the innovative design of more advanced peptide–AIEgen conjugates.
 | ||
| Fig. 1 Schematic illustration of design strategies for activatable peptide–AIEgen conjugates for cancer imaging. | ||
2. Binding-activated peptide–AIEgen conjugates for cancer imaging
AIEgens equipped with targeting peptides or moieties can be specifically accumulated at sites of interest through active targeting. This locally binding-induced accumulation leads to the restriction of intramolecular motion (RIM) (e.g., tetraphenylethene) or the formation of J-aggregates (e.g., bis-pyrene) of the AIEgens, thus turning their AIE fluorescence on. As such, precise and sensitive imaging of the corresponding binding targets can be realized. Notably, the majority of the targeting moieties are peptides, which offer several distinct advantages compared to other macromolecule targeting moieties: (1) high specificity – peptides can be rationally designed to exhibit highly specific affinities for certain biomarkers on tumor cells. A notable example is the RGD (arginine-glycine-aspartic acid) peptide, which has a remarkable specificity for integrin αvβ3.32 This integrin is highly expressed on the surface of tumor cells and plays a crucial role in tumor angiogenesis, cell migration, and invasion. By conjugating the RGD peptide to an AIEgen, the resultant conjugates can be accurately directed towards tumor cells. In contrast, although some antibody-based macromolecules also have targeting capabilities, their large molecular size often hinders efficient penetration into tumor tissues, thus limiting their targeting efficiency. Moreover, some other macromolecules may lack such precisely tuned binding abilities, resulting in non-specific binding and reduced imaging accuracy.33 (2) Biocompatibility and low immunogenicity – peptides are nature-derived materials with inherent biocompatibility, biodegradability, low systemic toxicity, and negligible immunogenicity. For peptide–AIEgen conjugates, these properties ensure that the conjugates can be safely used in vivo without causing significant adverse effects, enhancing their potential for clinical applications. In contrast, synthetic macromolecules (e.g., polymers) may cause immune reactions in the body due to their immunogenicity, compromising their efficacy and safety in vivo.34 (3) Versatility in function – peptides can serve multiple functions. They can act as targeting motifs, responsive substrates, self-assembling building blocks, and even therapeutic agents. For instance, in some peptide–AIEgen conjugates, the peptide segment not only targets the tumor but also participates in self-assembly processes, which can improve the stability and targeting efficiency of the conjugates. Other macromolecules may be more limited in their functional versatility. (4) Ease of modification and synthesis – solid-phase peptide synthesis (SPPS) allows for the efficient and precise synthesis of peptides with specific sequences and lengths. They can be conjugated with AIEgens in a straightforward manner, and their sequences can be adjusted to optimize binding affinity, solubility, and other properties. However, complex macromolecules such as proteins or polymers may be more challenging to modify and synthesize with the same level of precision. (5) On-demand amenability – peptides can be designed on demand to target specific molecules or pathways involved in tumor growth and progression. The programmable nature enables the development of conjugates that can specifically target unique tumor-associated antigens or biomarkers, providing a high degree of flexibility in cancer imaging applications. Other macromolecules, due to their relatively fixed structures and functions, may be difficult to design as flexibly to meet the diverse needs of different tumors. To date, various AIEgens conjugated with targeting peptides have been developed for targeted cancer imaging. Those conjugates developed in the past three years are highlighted in this section (Fig. 2). In terms of the functions of the peptide segments, these peptide–AIEgen conjugates can be roughly categorized into two types: target-binding and assembly-promoted binding peptide–AIEgen conjugates.2.1. Common AIEgens in peptide–AIEgen conjugates
In the realm of peptide–AIEgen conjugates, several AIEgens play crucial roles, including tetraphenylethene (TPE), quinoline-malononitrile (QM), PyTPA, and bis-pyrene (BP). Tetraphenylethene (TPE) is one of the most well-known archetypes of AIEgens.35 It consists of a central olefin bond flanked by four peripheral phenyl rings, presenting a highly twisted propeller-like structure. In dilute solutions, the phenyl rings of TPE can freely rotate or twist. When excited by light, this leads to non-radiative decay, resulting in an initial fluorescence-off state. However, in its aggregated state, the rotation and twisting of the benzene rings are restricted. The highly twisted structure of TPE prohibits π–π aggregation, thus circumventing aggregation-caused quenching (ACQ). Quinoline-malononitrile (QM) is another important AIEgen. It is derived from the traditional dicyanomethylene-4H-pyran chromophore by replacing the oxygen in the scaffold.36 Structurally, it combines a quinoline ring with a malononitrile group, forming a core structure that serves as the foundation for its unique optical properties. In a dilute solution, QM molecules exist in a dispersed state. The molecular conformation allows for relatively free movement of its components. When excited by light, the energy absorbed by QM can be dissipated through non-radiative processes, resulting in a weak or negligible fluorescence signal in the dilute solution state. When QM molecules aggregate, the conformation twist induced by the N-ethyl group blocks π–π stacking between aromatic rings (face-to-face stacking), suppressing non-radiative decay. As a result, the excited-state energy is predominantly released through radiative decay, leading to a substantial enhancement in fluorescence intensity. QM emits yellow fluorescence, with its maximal emission at around 590 nm. PyTPA is an AIEgen that links a strong electron donating group (diphenylamino) to the AIE scaffold (α-cyanostilbene). It is nearly non-fluorescent in aqueous media owing to the twisted intramolecular charge transfer (TICT) effect and free intramolecular motions, while emitting red fluorescence centred at 620–650 nm upon aggregation.37,38 Bis-pyrene (BP) has large π-conjugated structures. Different from TPE, which follows the restriction of intramolecular motion (RIM) mechanism, BP forms J-aggregates to emit excimer fluorescence (at around 520 nm). These properties enable it to be used in peptide–AIEgen conjugates for specific imaging purposes.392.2. Target-binding peptide–AIEgen conjugates
Peptides with specific affinities for certain biomarkers of tumor cells are frequently employed to construct target-binding peptide–AIEgen conjugates. For example, short phase-separating peptides (PSPs) can bind with RNA to form dynamic droplet-like assemblies in response to environmental cues in cellular membrane-less organelles and nuclear bodies, playing vital roles in various biological events, such as the regulation of RNA metabolism and gene expression in tumor cells. However, traditional fluorophores employed for the labelling of PSPs suffered from the ACQ effect, thus showing limitations in the visualization of intracellular RNA. To address this issue, Song and co-workers linked a prototypical AIEgen tetraphenylethylene (TPE) to an RNA-binding PSP (RRASL)2, affording a peptide–AIEgen conjugate TPE-(RRASL)2 (compound 1 in Fig. 2).40 This conjugate could bind to anionic polyU RNA to form coacervates through liquid–liquid phase separation (LLPS) in a positive charge-dependent mechanism (Fig. 3a), exhibiting an RNA dose-dependent fluorescence profile (Fig. 3b). After incubation of TPE-(RRASL)2 with a human gallbladder cancer cell line SGC-996, the fluorescence of TPE could be observed (Fig. 3c), which was colocalized with the fluorescence of a standard RNA dye SYTO 13 (Fig. 3d and e), as indicated by a high Pearson correlation coefficient of ∼0.95. This indicated that TPE-(RRASL)2 could be used for intracellular RNA imaging in living cells. Notably, it was found that the LLPS process was promoted by the conjugation of TPE on peptide, which improved the sensitivity of the probe. This work would motivate the design of droplet-based biosensors without sacrificing the dispersibility of endogenous probes under physiological conditions. Moreover, given that abnormal RNA metabolism and gene expression are hallmarks of tumor cells, the ability of TPE-(RRASL)2 to image intracellular RNA can help reveal the molecular mechanisms of tumorigenesis and metastasis at the RNA level, which is crucial for tumor diagnosis.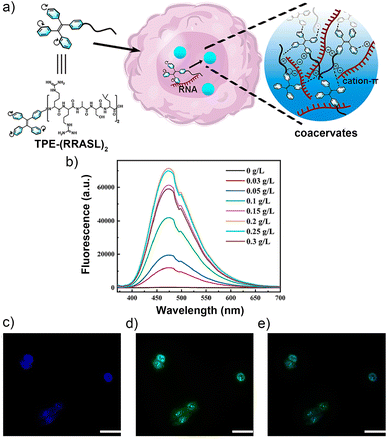 | ||
| Fig. 3 Target-binding peptide–AIEgen conjugate TPE-(RRASL)2 for RNA imaging in SGC-996 tumor cells. (a) Schematic illustration of TPE-(RRASL)2/polyU RNA coacervates inside cells. (b) Fluorescence spectrometer analysis showing an increase in the photoluminescence intensity of peptide solution with increasing polyU concentration. (c) TPE-(RRASL)2 penetrates through SGC-996 cells and translocates into the cell nucleus. Concentration of TPE-(RRASL)2 is 0.12 mM. (d) Fluorescence microscopy images of SGC-996 cells, stained using a standard RNA dye, SYTO 13. (e) TPE-(RRASL)2 and SYTO 13 are colocalized in SGC-996 cells. Scale bars, 50 μm. Reproduced from ref. 40 with permission from American Chemical Society, Copyright 2023. | ||
The aggregation of β-structure-rich proteins is a pathological hallmark of many diseases, such as Alzheimer's disease and cancers (e.g., medullary thyroid carcinoma). Zheng and colleagues proposed a peptide–AIEgen conjugate G7-TBA (compound 2 in Fig. 2) by linking a highly hydrophilic amyloid peptide GNNQQNY to an AIEgen 4-(1,2,2-triphenylvinyl)benzoic acid (TBA, with TPE as the AIE core).41 This conjugate could bind to amyloid aggregates of three different amyloid proteins (Aβ, hIAPP, and hCT), which turned on the AIE fluorescence. Moreover, it could also serve as an amyloid modulator that accelerated amyloid fibrillization and modulated amyloid-induced cell toxicity towards human SH-SY5Y neuroblastoma cells and rat insulinoma RIN-m5F tumor cells. Notably, the peptide GNNQQNY bound to amyloid aggregation via the formation of steric β-zippers, exceeding the binding affinity between commercial ThT and amyloid aggregates. Moreover, it should be noted that amyloid aggregation is not only related to neurodegenerative diseases but also bears significance in certain cancers, such as medullary thyroid carcinoma. The conjugate G7-TBA can potentially be used to detect amyloid-related pathological changes in these cancer types, providing new insights into tumor-associated protein misfolding events. This work would inspire researchers to design efficient peptide-based theranostic agents for combating neurodegenerative diseases and cancers.
Autophagy plays a pivotal role in the development of a wide variety of diseases, such as cancers. To visualize autophagy inside cells, Wen and co-workers designed a peptide–AIEgen conjugate LKT (compound 3 in Fig. 2), which consisted of an LC3-interacting peptide motif (DDDWTHL), a triple-G peptide linker (GGG), a cell-penetrating poly-K peptide motif (KKKKKKKKKK), and an AIEgen (TPE).42 With this design, LKT was able to target intracellular LC3 proteins inside HeLa cells without the need for transfection, and non-fluorescent LKT could be activated to exhibit strong AIE fluorescence upon binding with the LC3 protein. Given that cytoplasmic LC3 protein on the autophagosome membrane is the hallmark of autophagy, LKT was able to isolate tumor cell subpopulations differing in autophagy levels.
More recently, Li and co-workers designed a peptide–AIEgen conjugate TPE-Pep1 (compound 4 in Fig. 2) by linking TPE to a peptide GSRKR that could bind with low molecular weight heparin (LMWH).43 They further co-assembled this conjugate with gold nanoclusters that were modified with another LMWH-targeting peptide (RKRLQVQLSIRT), affording hybrid assemblies. These assemblies enabled ultrasensitive detection of LMWH at the ppb level through ratiometric fluorescence imaging, which was superior to the gold standard assay. It should be noted that abnormal blood coagulation is a common phenomenon in cancer patients. Tumor cells can trigger a hypercoagulable state, and heparin, especially LMWH, is often used in cancer patients to prevent thrombosis.44 The peptide-based organic–inorganic hybrid self-assembly system provides a highly sensitive and visual method for LMWH detection with a detection limit of 0.0004 IU mL−1. This system demonstrates its potential to detect low-abundance biomarkers in tumor imaging.
2.3. Assembly-promoted binding of peptide–AIEgen conjugates
In recent years, efforts have also been made to conjugate AIEgens with peptides that have both targeting and self-assembly functions.27,45 These peptide–AIEgen conjugates are able to pre-form ordered assemblies under physiological conditions. The ordered peptide assemblies can protect the peptide–AIEgen conjugates from being degraded under physiological conditions, improve their circulation time, and enhance their binding affinity to the target of interest on tumors.45 Moreover, these peptide–AIEgen conjugates, capable of in situ morphological transformation or self-assembly, enable selective enrichment of AIEgens at the tumor sites for improving the signal-to-background ratio.46 Additionally, targeted imaging combined with therapy allows for enhanced imaging of tumors, as well as therapeutic efficacy.47,48 For example, Wang and co-workers designed a peptide–AIEgen conjugate TAP (compound 5 in Fig. 2), which linked a TPE moiety to a self-assembling peptide segment (FFVLK), followed by a peptide sequence (KYNETWRSED) that was able to target the programmed death-1 ligand (PD-L1).49TAP first self-assembled into nanofibers in the aqueous solution, leading to the enrichment and ordered arrangement of TPE and KYNETWRSED moieties, which provided enhanced AIE fluorescence (excitation: 405 nm; emission: 520–580 nm) and binding affinity to PD-L1, respectively. Upon specific binding to PD-L1 that was overexpressed on the surface of tumor cells, TAP nanofibers were accumulated in the tumor tissues. Confocal imaging clearly showed TAP's preference for MC38 (high PD-L1) tumor cells over NIH/3T3 (low PD-L1) cells, validating its targeting ability at the cellular level. Moreover, in BALB/c nude mice with MC38 tumors, TAP nanofibers accumulated significantly in tumors, with high fluorescence signals, which were significantly stronger than those of their non-targeting counterpart (TN@TTP). Additionally, ex vivo imaging further supported this tumor selectivity. Notably, the in vivo fluorescence imaging experiments showed that the ratio of fluorescence signal intensity at the tumor tissue to non-tumor normal tissue for the TAP nanofiber group reached a maximum of 4.9 at 8 h. This indicated the significant fluorescence enhancement of TAP nanofibers at the tumor site compared to normal tissues. Collectively, TAP enabled enhanced and real-time tumor imaging in vivo, as well as photothermal immunotherapy.Bispyrene (BP) represents another typical AIEgen, yet it exhibits a distinct AIE mechanism. Recently, Yang and co-workers proposed a peptide-BP conjugate M1 (compound 6 in Fig. 2) for imaging early gastric cancer.32M1 consisted of a BP moiety, a self-assembling peptide sequence FFVLK for forming β-sheet nanofibers, a short polyethylene glycol moiety for improving biocompatibility, and a RGD motif for targeting integrin αvβ3 on tumor cell surfaces (Fig. 4a). Owing to the strong intermolecular π–π conjugate stacking of BP, M1 assembled into nanoparticles in aqueous solutions. With improved binding affinity, M1 nanoparticles could efficiently target tumor cells and accumulate on cell surfaces. Notably, upon this binding, the original hydrophilic–hydrophobic balance of M1 was disrupted, leading to a morphological nanoparticle-to-nanofiber transformation on tumor cell surfaces. Additionally, M1 showed high tumor selectivity. It bound preferentially to αvβ3-overexpressing cells, such as HUVECs and U87, and accumulated significantly in tumors in mouse xenograft and rabbit gastric cancer models, highlighting its selectivity for tumor-associated angiogenesis. Moreover, the formed nanofibers rendered efficient retention and long-term imaging (up to 96 h). In mouse in vivo imaging, at 96 h, the fluorescence intensity in tumor tissues treated with Cy7-labeled M1 was 85.4-fold higher than that in the PBS group and 28.8-fold higher than that of free Cy7, indicating significant fluorescence enhancement. Given that the absorption of BP (310–420 nm) overlapped with the wavelength of blue laser endoscopy (BLE) (410–450 nm), M1 was applied to visualize early gastric cancer in a rabbit model by the endoscopic technique. It highlighted tumors with yellow-green fluorescence under BLE (Fig. 4b), which showed high clinical significance. Notably, the fluorescence emission shift of BP is crucial for differentiating aggregates from basal signals in early gastric cancer diagnosis. It not only validates the self-assembly and targeting ability of M1 but also enhances the sensitivity and specificity of the imaging method. More recently, Jokerst and colleagues designed a peptide–AIEgen conjugate (ACE)LAGGGGFFPra(PyTPE)PASG(PEI), namely, (PAS)1F2 (compound 7 in Fig. 2).25 The peptide segments FF and PAS were used for self-assembly, while the positively charged low-molecular-weight branched polyethylenimine (PEI) acted as a proton sponge, which could target the membrane of subcellular organelles (e.g., lysosomes and mitochondria) through electrostatic interactions. (PAS)1F2 could form nanoassemblies with dense surface charges (Fig. 4c), which conferred enhanced targeting ability (Fig. 4d), as well as toxicity toward lysosomes and mitochondria, triggering immunogenic cell death (ICD) of cancer cells and subsequently activating immune cells. With this conjugate, imaging and triggering lysosome-regulated immunogenic cancer cell death were achieved. Structurally, (PAS)1F2 lacks specific targeting moieties for cancer cells, as indicated by comparable cytotoxicity on non-cancer cells (HEK 293 t and Vero E6) and cancer cells (HeLa and SKOV-3). This implies that (PAS)1F2 may not exhibit high tissue selectivity in vivo. However, (PAS)1F2 contains a terminal LAG segment, which is a post-cleavage fragment of PLG*LAG, a cleavable substrate of matrix metalloproteinases-2 (MMP-2). This fragment allows for future functionalization with MMP-2 cleavable cargos (e.g., imaging agents or drugs), which may endow (PAS)1F2 with tumor-specific tissue selectivity.
 | ||
| Fig. 4 Binding and assembly of peptide–AIEgen conjugates. (a) The molecular structure and constituent units (fluorescent unit, hydrogen-bonding unit, target unit, and hydrophilic unit) of M1, and fibrillogenesis of M1 NPs induced by the binding of ligand-receptor mechanism. (b) The endoscopic images of gastric tissue in three endoscopic modes (WLI, LCI, and BLI) before and after administration. Reproduced from ref. 32 used under the terms of the Creative Commons CC BY license. (c) Scheme showing the formation of the proton sponge nano-assembly from peptide–AIEgen conjugate (PAS)1F2. The assembly enhances the aggregation-induced fluorescence, charge density, and cell cytotoxicity. (d) Confocal images and analysis of Mander's colocalization coefficients showing the colocalization of (PAS)1F2 with (i) lysosomes and (ii) mitochondria. Reproduced from ref. 25 with permission from John Wiley and Sons, Copyright 2024. | ||
3. Enzyme-cleavable peptide–AIEgen conjugates
For a stimuli-cleavable peptide–AIEgen conjugate, the peptide functions as the specific substrate of the stimulus of interest (typically, an enzyme), and it should be a hydrophilic sequence. This endows the peptide–AIEgen conjugate with high water-solubility, promoting its molecular dissociation and thereby rendering a non-emissive initial state. This can be alternatively achieved by conjugation of hydrophilic moieties (e.g., polyethylene glycol) to the peptide–AIEgen conjugate.50,51 Once cleaved by the stimulus, the hydrophobic AIEgen is liberated and subsequently undergoes aggregation, emitting fluorescence. In this section, we highlight those innovative stimuli-cleavable peptide–AIEgen conjugates proposed in the past three years, which are classified and discussed according to the type of AIEgens.3.1. Enzyme-cleavable peptide–TPE conjugates
Owing to its easy synthesis, availability, and modification, TPE has been frequently conjugated to peptides to fabricate stimulus-activatable peptide–AIEgen conjugates for a broad range of applications.52–55 For instance, Zhang and colleagues linked TPE to a caspase-3-activatable peptide Asp-Glu-Val-Asp (DEVD), an amphiphilic segment H9-PEG8, and a αvβ3 integrin-targeting peptide Arg-Gly-Asp (RGD), affording a peptide–AIEgen conjugate TPR.55TPR could self-assemble into micelles, which allowed for targeted delivery and caspase-3-controlled release of drug doxorubicin (DOX), enabling apoptosis caspase-3 monitoring during tumor chemotherapy. Caspase-6 is another apoptosis executioner caspase. To reveal the role of caspase-6 in apoptosis, in 2023, Groborz and co-workers also developed a peptide–TPE conjugate Ac-EVEID-KTPE (compound 8 in Fig. 5), which was designed by conjugating TPE with a caspase-6-cleavable peptide Ac-EVEID.56 Notably, Ac-EVEID-KTPE exhibited high selectivity towards caspase-6 with a higher signal-to-noise ratio than commercial coumarin-based probes. In the same year, Zhong and colleagues proposed a dual enzyme-activated peptide–TPE conjugate TPE-1p (compound 9 in Fig. 5).57 Upon the sequential activation by alkaline phosphatase (ALP) and chymotrypsin-like enzyme (ChT-L), both of which are biomarkers of tumors, TPE-1p was converted to TPE-Y, followed by self-assembly into nanofibers with strong AIE fluorescence signals (Fig. 6a and b). Linear relationships were noted between fluorescence intensity at 480 nm of TPE-1p and the concentration of either ALP or ChT-L, with limits of detection being 0.031 U mL−1 for ALP and 1.2 U mL−1 for ChT-L, respectively. These results demonstrated the quantitative relationship between enzymatic transformation and fluorescence intensity. Notably, this dual stimulus-triggered activation strategy provided enhanced tumor selectivity, which is crucial for precise tumor imaging.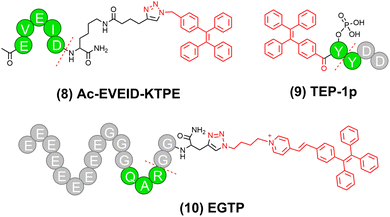 | ||
| Fig. 5 Representative stimuli-activatable peptide-TPE conjugates developed in the past three years. Dotted lines indicated the cleavage sites. | ||
 | ||
| Fig. 6 (a) Chemical structure of TPE-1p and the proposed enzymatic transformation process by ALP and ChT-L; TPE-1p cannot be recognized and activated by ChT-L without ALP-catalyzed dephosphorylation of the peptide. (b) Schematic illustration of TPE-1p for monitoring ChT-L activity of proteasomes in cancer cells. Reproduced from ref. 57 with permission from American Chemical Society, Copyright 2023. (c) Molecular structure of EGTP and its usage for TMPRSS2-responsive imaging and accurate inhibitor screening: (i) negatively charged amphiphilic EGTP cannot cross the cell membrane of cells with low TMPRSS2 expression; (ii) EGTP can be cleaved by cells with high expression of TMPRSS2; (iii) surface potential of the PyTPE segment changes after removal of the negatived polyglutamic acid; (iv) and can be internalized into the cells; (v) high concentrations of the PyTPE segment can aggregate with enhanced fluorescence; (vi) no fluorescence is observed once TMPRSS2 is suppressed by its inhibitors. Reproduced from ref. 63 with permission from American Chemical Society, Copyright 2023. | ||
Researchers have also employed TPE derivatives with red-shifted emission wavelengths to fabricate stimuli-activatable peptide-AIE conjugates, which provided improved tissue penetration depths.58–61 For instance, Song and colleagues conjugated a DEVD peptide to tetraphenylethene pyridinium (PyTPE), which emitted bright yellow fluorescence centered around 600 nm upon aggregation.62 They linked this conjugate to sulfhydryl PE-coupled nanogapped gold nanoparticles (AuNNPs), realizing precise apoptosis imaging and radiotherapy of tumors. Later, in 2023, Jokerst and colleagues linked PyTPE to a peptide sequence containing a polyglutamic acid peptide (nine glutamic acids), a triglycine spacer, and a transmembrane protease serine 2 (TMPRSS2)-cleavable peptide (QAR) (Fig. 6c).63 The obtained peptide–PyTPE conjugate EGTP (compound 10 in Fig. 5) was able to form highly negatively charged and amphiphilic nanoparticles. Upon cleavage by TMPRSS2 on the tumor cell membrane, these nanoparticles underwent surface potential changes and subsequent selective internalization by TMPRSS2-overexpressing tumor cells with AIE fluorescence turned on inside the cells. The fluorescence intensity of EGTP at 590 nm increased over time in the presence of TMPRSS2, while this increase was blocked by the TMPRSS2 inhibitor camostat. Moreover, the fluorescence intensity of EGTP increased by 22.61-fold and 9.96-fold with increasing concentrations of both EGTP and TMPRSS2. This significant fluorescence enhancement indicated that the aggregation of PyTPE after enzymatic cleavage had a substantial impact on fluorescence intensity.
3.2. Enzyme-cleavable peptide–QM conjugates
Quinoline-malononitrile (QM) is another AIEgen that emits yellow fluorescence (maximal emission at around 590 nm). QM has also been employed in designing peptide–AIEgen probes for bioimaging applications.64,65In 2021, Zhu and colleagues designed a cathepsin E-cleavable peptide–QM conjugate QM-HSP-CPP for early diagnosis of pancreatic cancer (PC).23QM-HSP-CPP consisted of a fluorophore QM, a hydrophobic CTSE-cleavable peptide HSP (Ala-Gly-Phe-Ser-Leu-Pro-Ala-Lys-Arg), and a hydrophilic cell penetrating peptide CPP (eight repeated Arg). Compared with the control probe QM-HSP that did not contain the CPP moiety, QM-HSP-CPP was well dispersed in aqueous solutions with low background fluorescence and exhibited significantly improved cancer cell penetration. Upon specific cleavage by CTSE, QM-HSP-CPP was activated to emit AIE fluorescence (600 nm), showing a high signal ratio (9.3 fold) between tumor and paratumor tissue in intraoperative human PC sections, whereas the immunofluorescence (IF) staining exhibited an insignificant difference (1.2 fold). In 2022, the same group proposed an Atg4B-activated peptide–QM conjugate QM-GFTN (compound 11 in Fig. 7) by conjugating QM to a hydrophilic peptide Gly-Phe-Thr-Asn (GFTN).66 Upon cleavage by Atg4B, a biomarker for autophagy, QM-GFTN was converted to QM with its AIE fluorescence turned on (Fig. 8a). QM-GFTN was able to selectively respond to Atg4B and visualize autophagy in living cells and mouse tissues. In solution, Atg4B-treated QM-GFTN exhibited a 6.75-fold higher fluorescence intensity than the intrinsic fluorescence of the probe at 90 min. Moreover, in HeLa, PC12, and SW1990 cells treated with rapamycin (an autophagy inducer that increases Atg4B expression), QM-GFTN exhibited a 42.3-fold enhanced fluorescence intensity compared to PBS-treated cells, respectively. These results indicated that Atg4B-mediated cleavage of QM-GFTN led to a significant increase in fluorescence intensity. Notably, QM-GFTN exhibited much enhanced “turn-on” fluorescence signals than the commercial fluorescent probe autophagic vacuole dye dansylcadaverine (MDC) when visualizing autophagy in human tumor tissue slices (Fig. 8b, c). Later, in 2023, Wang and colleagues designed a dual-enzyme-responsive peptide–QM conjugate QM-IEPD-DEVD-Cys(StBu)-Lys-CBT (compound 12 in Fig. 7) and linked it to manganese–zinc (Mn–Zn) ferrite magnetic nanoparticles (MZF-MNPs), affording a nanoprobe QM(GP)–MZF(CP) for fluorescence and MR dual-modal imaging of tumor response to immunotherapy.67 During immunotherapy, granzyme B produced by cytotoxic T cells and apoptosis-related caspase-3 together cleaved the peptide of QM-IEPD-DEVD-Cys(StBu)-Lys-CBT, releasing QM with its AIE fluorescence turned on.
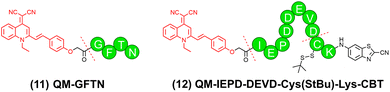 | ||
| Fig. 7 Representative stimuli-activatable peptide–QM conjugates developed in the past three years. Dotted lines indicated the cleavage sites. | ||
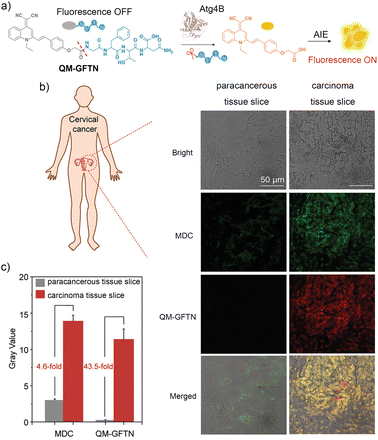 | ||
| Fig. 8 Atg4B-activatable peptide–QM conjugate QM-GFTN for autophagy monitoring. (a) Illustration of QM-GFTN for Atg4B activity study. Once responding to autophagy-related protease Atg4B, the specific cleavage of the GFTN peptide by Atg4B induces the hydrophobic AIE residues to aggregate rapidly with significantly intense fluorescence emission. (b) Fluorescence imaging of the human tumor tissue slices stained with QM-GFTN (10 μM) and MDC (10 μM) and (c) the corresponding fluorescence intensity. QM-GFTN: λex = 405 nm, λem = 650–750 nm; MDC: λex = 405 nm, λem = 450–550 nm; Scale bar = 50 μm, and the error bars represent S.D. with n = 3. Reproduced from ref. 66 with permission from Wiley-VCH GmbH, Copyright 2021. | ||
3.3. Enzyme-cleavable peptide–PyTPA conjugates
In recent years, the Lou and Xia groups have made significant efforts to design peptide–PyTPA conjugates for biomedical applications.68–70 For example, considering that the aggregation degree of AIEgens in aqueous media was difficult to control, Lou's group conjugated PyTPA to a peptide RRRRRRGGPLGLAGPra to afford a weakly fluorescent peptide–PyTPA conjugate MP for quantitative detection of matrix metalloproteinase-2 (MMP-2).71 Upon cleavage by MMP-2, the hydrophobic PyTPA moiety was released from MP, absorbed by negatively charged nanoparticles (NPs), and subsequently underwent the self-assembly of NPs to photonic crystals (PCs) on slippery lubricant-infused porous substrates (SLIPS). Through this two-step enrichment, the aggregation degree of PyTPA was largely enhanced, which provided much enhancement of fluorescence, enabling ultrasensitive detection of MMP-2 secreted by tumor cells directly. Later, in 2022, Lou and colleagues designed a Cathepsin B-cleavable peptide–PyTPA conjugate GCP (compound 13 in Fig. 9) by linking PyTPA to a caged GO203 peptide via a peptide substrate of Cathepsin B.69GCP could assemble with miR-140 into GCP/miR-140 nanoparticles, which were able to sense cellular Cathepsin B, as well as realized combined photodynamic therapy (PDT) and photodynamic therapy (PDT) of tumors. More recently, in 2023, Lou and colleagues proposed a peptide–PyTPA conjugate DMFA (compound 14 in Fig. 9) by linking PyTPA to a peptide sequence containing a positively charged amphiphilic α-helix-forming peptide GRFKRFRKKFKKLFKKLSPVIPLLHL, an MMP-2 cleavable peptide PLGLAG, and a β-sheet-forming peptide KLVFFGG (Fig. 10a).72DMFA could self-assemble into nanoparticles in aqueous solution. Upon capture by the tumor cell membrane, DMFA could be cleaved by the pericellular overexpressed MMP-2 into two parts, namely, the α-helix-forming peptide and the PyTPA-conjugated β-sheet-forming peptide. The latter peptide further assembled into nanofibers (Fig. 10a). This nanoparticle-to-nanofiber transformation on the cell membrane was tracked by AIE fluorescence, which reflected MMP-2 levels (Fig. 10b). Meanwhile, this morphological change further contribute to inhibiting the growth and metastasis of tumor cells by decreasing Na+/K+-ATPase activity.72 In the same year, Lou and co-workers developed a reactive oxygen species (ROS)-responsive peptide–PyTPA conjugate TPAMCF (compound 15 in Fig. 9).73 Owing to the phosphatidylserine-targeting capacity of its peptide segment (i.e., CLIKKPF), TPAMCF-formed nanoparticles could target apoptotic foam cells and further respond to the locally upregulated ROS. This led to the release of PyTPA, which exhibited “turn-on” AIE fluorescence in the lipid droplets inside the foam cells.73 Recently, a PyTPA derivative was also employed to construct a peptide–AIEgen conjugate APD (compound 16 in Fig. 9), which could be activated by caspase-3 during tumor cell apoptosis. The addition of caspase-3 to APD rendered a sustained fluorescence light-up around 700 nm, and the intensity at 120 min was about 100-fold enhancement compared to APD alone. This indicated a significant increase in fluorescence intensity due to the enzymatic transformation. With APD, chemo-therapeutic feedback monitoring using fluorescence imaging was realized.74 | ||
| Fig. 9 Representative stimuli-activatable peptide–PyTPA conjugates developed in the past three years. Dotted lines indicated the cleavage sites. | ||
 | ||
| Fig. 10 (a) Chemical structure and illustration of matrix metalloproteinase-2 (MMP-2)-activatable peptide–PyTPA conjugate DMFA. (b) CLSM images and intensity of HeLa cells, MCF-7 cells, and HeLa + ARP100 incubated with DMFA, DFA, and LFA for 4 h. The linear regions across the HeLa cells, MCF-7 cells, and HeLa cells + ARP100 are marked by red arrows, blue arrows, and purple arrows, respectively. Reproduced from ref. 72. Used under the terms of the Creative Commons CC BY license. | ||
4. Stimuli-activatable self-assembling peptide–AIEgen conjugates
Stimuli-activatable peptide–AIEgen conjugates enable selective imaging of the stimuli of interest, which relies on the aggregation of AIEgens after their activation.75 By integrating short self-assembling peptide sequences into the peptide scaffold, efforts aim to realize the ordered self-assembly of the activated AIEgens, which would promote the regular arrangement of AIE molecules. Compared with their randomly aggregated counterparts, orderly arranged AIEgen aggregates have closer proximity and tighter intermolecular steric interactions, which provide more strengthened restriction to the intramolecular motions of AIEgens, thus rendering enhanced turn-on AIE fluorescence.76,77 This strategy has demonstrated its effectiveness in fabricating highly sensitive AIE probes.The pioneering exploration was made by Ding and co-workers, who designed a TPE-based peptide–AIEgen conjugate TPE-GFFYK(DVEDEE-Ac).78 This conjugate could be specifically cleaved by caspase-3 to yield TPE-GFFYK, which was able to form nanofibrous network structures with orderly arranged TPE molecules, rendering higher sensitivity in sensing caspase-3 in tumor cells. Interestingly, they made a pertinent “butterflies-in-a-box” metaphor to understand this concept, namely, when many butterflies were put into a box, they would have more difficulty in flapping their wings if they were arranged in good order compared to if they were randomly arranged.78 Later, the same group devised another peptide–TPE conjugate TPE-Py-pYK(TPP)pY by linking a TPE derivative TPE-Py to a self-assembling peptide pYK(TPP)pY, which contained a triphenylphosphonium-modified lysine flanked by two phosphotyrosines.79TPE-Py-pYK(TPP)pY could be activated by ALP to form nanoassemblies within tumor tissues, which turned on the AIE fluorescence and produced singlet oxygen to cause lysosomal membrane permeabilization (LMP), realizing enhanced imaging and immunotherapy (via LMP-invoked immunogenic cell death) of tumors.
Notably, the characteristic highly twisted molecular conformation of the TPE moiety tends to hinder the assembly of peptide–TPE conjugates, thereby lowering the fluorescence efficiency. To address this issue, Yang and colleagues recently optimized the structural parameters when designing a self-assembled peptide–TPE conjugate F3 (compound 17 in Fig. 11) for sensing alkaline phosphatase (ALP), which is a typical biomarker of tumors.80 As shown in Fig. 12a, they revealed that: (1) a glycine linker between the self-assembled peptide sequence and the TPE moiety was not necessitated, because it would hinder the restriction of peptide assembly on the intramolecular rotation of TPE, thus inhibiting the fluorescence output; (2) increasing the number of phenylalanine residues to three would improve the self-assembly and luminescence ability of the peptide–TPE conjugate; (3) increasing the number of phosphorylated tyrosine residues to two would well balance the hydrophobicity and amphiphilicity of the conjugate, as well as provide good responsiveness to ALP. These findings provided insightful guidance to the future design of self-assembling peptide–AIEgen conjugates.
 | ||
| Fig. 11 Representative stimuli-activatable self-assembling peptide–PyTPA conjugates developed in the past three years. Dotted lines indicated the cleavage sites. | ||
 | ||
| Fig. 12 (a) Molecular structures of the AIEgen-peptides and the correlation between the structure and the ability to detect ALP. Reproduced from ref. 80 with permission from American Chemical Society, Copyright 2022. (b) Schematic illustration of the design strategy of probes FA and BFA with single- or dual-quenching effects, respectively, which could be lighted up with strong red fluorescence intensity by furin. Reproduced from ref. 86 with permission from the Royal Society of Chemistry, Copyright 2023. (c) Schematic illustration of tandem targeting and dual aggregation of Ala-Biotin-QMT for accurate and enhanced near-infrared fluorescence imaging of tumors. Reproduced from ref. 94 with permission from American Chemical Society, Copyright 2024. | ||
Recently, bis-pyrene (BP) was also employed to construct self-assembling peptide–AIEgen conjugates. In 2023, Ren and colleagues devised PAHN-1 (compound 18 in Fig. 11) by conjugating the BP moiety to a multi-functional peptide containing a self-assembling and mitochondrial-targeting peptide CFFFVLKLAKLAK, a caspase-3-cleavable peptide DEVD, and a penetrating peptide AKRGARSTA for targeting neuropilin-1 (NRP-1)-expressing solid tumors.81 This caspase-3-activatable self-assembling peptide–BP conjugate could self-assemble into nanoparticles, which showed extraordinary homing and penetration ability toward NRP-1 receptor-positive 4T1 breast cancer cells. After cellular uptake, PAHN-1 nanoparticles could further target the mitochondria and upregulate the expression of caspase-3 through mitochondrial dysfunction. The upregulated caspase-3, in turn, cleaved PAHN-1 to yield the BP-CFFFVLKLAKLAKDEVD residue with decreased hydrophilicity, which rendered tighter aggregation of BP molecules.
Peptide–AIEgen conjugates with positively charged peptides might non-specifically bind to ambient proteins through electrostatic interactions, which restrict their intramolecular motions, thus giving rise to non-negligible “false-positive” AIE fluorescence signals. To suppress the background signal of an imaging probe, a widely used strategy is to modify the probe with a quencher moiety, which renders a low background based on the FRET mechanism.82–85 For instance, Hao and co-workers modified a dark quencher BHQ3 to RVRRGFF-AIE (FA), affording a self-assembling peptide–AIEgen conjugate BFA (compound 19 in Fig. 11) for imaging the furin activity in tumors.86 Compared with its BHQ3-free counterpart (i.e., FA), BFA showed significantly enhanced anti-interference ability owing to the dual “quenching” effect (Fig. 12b). Quantitative analysis showed that in MDA-MB-468 cells, when coupled with a negatively charged polymer γ-PGA for charge shielding, BFA exhibited 5.3-fold and 3.1-fold higher fluorescence intensities than those in HEK 293T and LoVo cells, respectively. This indicated that PGA–BFA could be used to differentiate furin-overexpressing cancer cells from normal cells.
The peptide assembly mechanism for the above conjugates is mainly based on the well-known orderly packing of dipeptide Phe–Phe (FF) or its derivatives.87–89 Alternatively, the click reaction between 2-cyanobenzothiazole (CBT) and cysteine (Cys), which is also able to trigger the assembly of peptide,90,91 has also been employed to construct self-assembling peptide–AIEgen conjugates. Briefly, by covalently flanking a peptide–AIEgen scaffold by the CBT and Cys moieties, the resulting conjugate would undergo the CBT-Cys click reaction to form cyclic dimers, which further aggregate into nanostructures through π–π stacking. Notably, this process enables dual aggregation of the loaded AIEgen (first aggregation into dimers and second aggregation into nanostructures), which confers high AIE fluorescence output. Moreover, by caging the amino and/or the sulfhydryl groups of the terminal Cys with the specific substrates of stimuli of interest, self-assembly of the peptide–AIEgen conjugates would be controlled in an on-demand manner. This type of stimuli-activatable self-assembling peptide–AIEgen conjugate was pioneered by our group. In 2017, we designed a peptide–TPE conjugate Ac-Arg-Val-Arg-Arg-Cys(StBu)-Lys(TPE)-CBT.92 It could be specifically cleaved by furin to yield Cys(StBu)-Lys(TPE)-CBT, which further underwent the CBT-Cys click reaction under tumor-characteristic reduction conditions and subsequent assembly, rendering 3.4-fold enhanced AIE fluorescence signals over its single AIE counterpart in furin-overexpressing tumor cells.
Very recently, in 2024, we employed an NIR AIEgen (i.e., QMT93) to construct another stimuli-activatable self-assembling peptide–AIEgen conjugate β-tBu-Ala-Cys(StBu)-Lys(Biotin)-Pra(QMT)-CBT (Ala-Biotin-QMT) (compound 20 in Fig. 11).94 It consisted of a β-tBu-Ala moiety serving as the substrate of leucine aminopeptidase (LAP, a tumor biomarker), a caged Cys residue for subsequent click reaction with the terminal CBT moiety, a lysine residue modified with a biotin group for tumor targeting, and a pra residue modified with the AIEgen QMT (Fig. 12c). Notably, Ala-Biotin-QMT was capable of dual targeting (i.e., active targeting via biotin-receptor binding, and passive targeting via LAP-mediated hydrolyzation of the conjugate) and dual aggregation (i.e., dimer formation and consequent aggregation), rendering 4.8-fold and 7.9-fold higher NIR AIE fluorescence signals over the control groups in HepG2 cells and HepG2 tumor-bearing mice, respectively. As such, simultaneous enhancement of imaging precision and tumor sensitivity was achieved. With a similar strategy, we also devised a caspase-1-activatable self-assembling peptide–AIEgen conjugate Ac-Trp-Glu-His-Asp-Cys(StBu)-Pra(QMT)-CBT (QMT-CBT) for sensitive imaging of Alzheimer's disease.95 These studies are inspiring for tailoring highly accurate and sensitive self-assembling peptide–AIEgen conjugates for the diagnosis of cancers and other diseases.
5. Summary and outlook
Activatable peptide–AIEgen conjugates combine the functionality of peptides and aggregation-induced emission properties of AIEgens, exhibiting outstanding advantages such as superior biosafety, sensitivity and selectivity. Therefore, they have been widely developed and applied in the biomedical field, particularly in tumor imaging in recent years. The design strategies mainly include the following three types: First, peptide–AIEgen conjugates that couple AIEgens with tumor-targeting peptides or moieties can actively target and bind to specific tumor targets through specific recognition or electrostatic interactions. This binding process leads to the restriction of the intramolecular motions of AIEgens, thus lighting up the fluorescence in the tumor area. Second, for those AIEgens conjugated with hydrophilic substrate peptides of certain tumor biomarkers, the hydrophobic AIEgens will be released upon the cleavage by the corresponding biomarkers, followed by aggregation, which turns on the AIE fluorescence in biomarker-overexpressing tumor tissues. Third, those AIEgens conjugated with both biomarker-responsive peptides and self-assembling peptides can not only be activated by the corresponding biomarkers but also further stack into orderly assemblies (e.g., nanoparticles and nanofibers), which promote the denser packing of AIE molecules, thereby rendering enhanced fluorescence output in tumor tissues. In this review, we focus on these three strategies and highlight the activatable peptide–AIEgen conjugates developed for tumor imaging in the past three years (since 2022), with emphasis on their design principles, operating mechanisms and imaging performance. Future efforts in this field might focus on addressing the following challenges.First, near-infrared peptide–AIEgen conjugates: currently, most peptide–AIEgen conjugates use AIEgens that are excited by visible light, which severely limits their ability to visualize deep-seated tumor tissues. Therefore, using NIR light-excitable AIEgens to construct peptide–AIEgen conjugates would be conducive to their applications in vivo and is highly encouraged.96–98 For instance, with the assistance of two-photon fluorescence imaging technique, a γ-glutamyltranspeptidase-responsive AIEgen can be excited with a NIR-II laser, which renders significantly enhanced tissue penetration and signal-to-background ratios.99
Second, ratiometric peptide–AIEgen conjugates: ratiometric fluorescence techniques employ two sets of fluorescence signals, which vary differently with respect to the concentration of target analytes, resulting in a ratiometric signal that is independent of both probe concentration and environmental fluctuations.100 This allows for quantification of analytes over a broad concentration range. Although various innovative ratiometric AIEgens have been devised,101,102 peptide–AIEgen conjugates that enable ratiometric imaging of tumors remain scarce.
Third, multi-locked peptide–AIEgen conjugates: in the intricate milieu of biological systems, the conventionally used single stimulus-activatable (i.e., single-locked) peptide–AIEgen conjugates are prone to “false positive” fluorescence signals within healthy tissues, and they may be inadequate for precisely indicating the occurrence of tumors.103 As a consequence, dual or multi-locked peptide–AIEgen conjugates are highly desired. By employing dual or more biomarkers as “keys” to turn on their fluorescence, multi-locked peptide–AIEgen conjugates enable the concurrent detection of distinct tumor biomarkers, thereby rendering substantially augmented imaging accuracy for tumor imaging.104
Fourth, more imaging applications during cancer management: with current or more advanced peptide–AIEgen conjugates, efforts are also encouraged to be paid to other imaging fields during cancer management that remain seldomly explored with current conjugates, such as immune response, bacterial infection, imaging-guided therapy, and drug screening.
Overall, activatable peptide–AIEgen conjugates have opened new opportunities in many research fields (e.g., chemistry, chemical biology, medicine, materials science, etc). We hope this review could provide a timely reference for in-depth research and further development in this field and inspire relevant researchers to explore the application potential as well as innovative directions of activatable peptide–AIEgen conjugates in the realm of cancer imaging and other related fields.
Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Author contributions
All the authors conceived the outline of the manuscript. S. Z. and X. S. wrote the original draft of the manuscript. G. L. revised and supervised the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant 22304024, 22234002, and 22204019), National Key Research and Development Program of China (Grant 2023YFF0724100), and Natural Science Foundation of Jiangsu Province (Grant BK20232007).References
- J. D. Luo, Z. L. Xie, J. W. Y. Lam, L. Cheng, H. Y. Chen, C. F. Qiu, H. S. Kwok, X. W. Zhan, Y. Q. Liu, D. B. Zhu and B. Z. Tang, Chem. Commun., 2001, 1740–1741, 10.1039/B105159h.
- H. Li, H. Kim, J. Han, V. Nguyen, X. J. Peng and J. Yoon, Aggregate, 2021, 2, e51 CrossRef.
- Y. Duo, G. Luo, W. Zhang, R. Wang, G. G. Xiao, Z. Li, X. Li, M. Chen, J. Yoon and B. Z. Tang, Chem. Soc. Rev., 2023, 52, 1024–1067 RSC.
- J. You, Y. Ma, H. Song, J. Hou and E. Zhao, Trac. Trends Anal. Chem., 2024, 180, 117886 CrossRef.
- J. Y. Cai, M. Zhang, J. Q. Peng, Y. Q. Wei, W. C. Zhu, K. Z. Guo, M. Gao, H. Wang, H. M. Wang and L. Wang, Adv. Mater., 2024, 36, 2400531 CrossRef PubMed.
- M. M. S. Lee, E. Y. Yu, J. H. C. Chau, J. W. Y. Lam, R. T. K. Kwok and B. Z. Tang, Adv. Mater., 2024, 2407707, DOI:10.1002/adma.202407707.
- C. Chen, X. Y. Zhang, Z. Y. Gao, G. X. Feng and D. Ding, Nat. Protoc., 2024, 19, 2408–2434 CrossRef PubMed.
- H. Wang, D. Jiao, D. Feng, Q. Liu, Y. Huang, J. Hou, D. Ding and W. Zhang, Adv. Mater., 2024, 36, e2311733 CrossRef.
- D. Mao and B. Liu, Matter, 2021, 4, 350–376 CrossRef.
- Z. Zhao, H. K. Zhang, J. W. Y. Lam and B. Z. Tang, Angew. Chem., Int. Ed., 2020, 59, 9888–9907 CrossRef.
- H. Ou, S. Dai, R. Liu and D. Ding, Sci. China Chem., 2019, 62, 929–932 CrossRef.
- J. Miao, M. Miao, Y. Jiang, M. Zhao, Q. Li, Y. Zhang, Y. An, K. Y. Pu and Q. Q. Miao, Angew. Chem., Int. Ed., 2022, 62, e202216351 CrossRef.
- G. L. Niu, R. Y. Zhang, X. J. Shi, H. Park, S. Xie, R. T. K. Kwok, J. W. Y. Lam and B. Z. Tang, Trac. Trends Anal. Chem., 2020, 123, 115769 CrossRef CAS.
- W. X. Jiang, C. Cheng, X. L. Qiu, L. Chen, X. Guo, Y. L. Luo, J. X. Wang, J. R. Wang, Z. Y. Xie, P. Li, Z. G. Wang, H. T. Ran, Z. Y. Zhou and J. L. Ren, Adv. Sci., 2023, 10, 2204989 CrossRef CAS.
- W. Zhou, L. Chen, H. Li, M. Wu, M. Liang, Q. Liu, W. Wu, X. Jiang and X. Zhen, ACS Nano, 2024, 18, 19771–19782 CAS.
- J. Yang, J. Wei, F. Luo, J. Dai, J. J. Hu, X. Lou and F. Xia, Top. Curr. Chem., 2020, 378, 47 CrossRef CAS PubMed.
- F. Xia, J. Wu, X. Wu, Q. Hu, J. Dai and X. Lou, Acc. Chem. Res., 2019, 52, 3064–3074 CrossRef CAS PubMed.
- P. Bao, C. Li, H. Ou, S. Ji, Y. Chen, J. Gao, X. Yue, J. Shen and D. Ding, Biomater. Sci., 2021, 9, 437–442 RSC.
- S. Y. Qin, J. Q. Feng, Y. J. Cheng, W. L. Liu, A. Q. Zhang, L. Wang, H. Wang and X. Z. Zhang, Coord. Chem. Rev., 2024, 502, 215600 CrossRef CAS.
- L. L. Li, Z. Y. Qiao, L. Wang and H. Wang, Adv. Mater., 2018, 31, 1804971 CrossRef PubMed.
- J. Dai, X. Dong, R. Liu, B. Chen, X. Dong, Q. Wang, J. J. Hu, F. Xia and X. Lou, Biomaterials, 2022, 285, 121528 CrossRef CAS PubMed.
- H. Li, H. Kim, C. Zhang, S. Zeng, Q. Chen, L. Jia, J. Wang, X. Peng and J. Yoon, Coord. Chem. Rev., 2022, 473, 214818 Search PubMed.
- Z. Zhu, Q. Wang, X. Chen, Q. Wang, C. Yan, X. Zhao, W. Zhao and W. H. Zhu, Adv. Mater., 2022, 34, e2107444 Search PubMed.
- G. Qi, F. Hu, L. Shi, M. Wu and B. Liu, Angew. Chem., Int. Ed., 2019, 58, 16229–16235 Search PubMed.
- T. Y. He, J. Wen, W. J. Wang, Z. L. Hu, C. X. Ling, Z. C. Zhao, Y. Cheng, Y. C. Chang, M. Xu, Z. C. Jin, L. Amer, L. Sasi, L. Fu, N. F. Steinmetz, T. M. Rana, P. Wu and J. V. Jokerst, Adv. Mater., 2024, 36, 2307679 Search PubMed.
- D. Fan, X. Liu, Y. Ren, Z. Luo, Y. Li, J. Dong, S. V. Wegner, F. Chen and W. Zeng, Acta Pharm. Sin. B, 2024, 14, 1759–1771 CrossRef CAS PubMed.
- X. L. Sha, G. T. Lv, Q. H. Chen, X. Cui, L. Wang and X. Cui, J. Mater. Chem. B, 2024, 12, 3676–3685 RSC.
- J. Ouyang, L. H. Sun, F. Zeng and S. Z. Wu, Coord. Chem. Rev., 2022, 458, 214438 CrossRef CAS.
- N. Y. Zhang, X. J. Hu, H. W. An, J. X. Liang and H. Wang, Biomaterials, 2022, 287, 121655 CrossRef CAS.
- B. Chen, H. Yuan, W. Zhang, J. Hu, X. Lou and F. Xia, Biosensors, 2022, 12, 667 CrossRef CAS PubMed.
- B.-L. Song, X.-H. Zhang, Z.-Y. Qiao and H. Wang, CCS Chem., 2022, 4, 437–455 Search PubMed.
- Q. Luo, C. Fan, W. Ying, X. Peng, Y. Hu, Z. Luan, S. Ye, C. Gong, Y. Huang, Y. Xiao, Y. Chen, M. Xing, L. Wang and S. Yang, Adv. Sci., 2023, 10, e2203918 CrossRef.
- L. L. Tundisi, J. A. Ataide, J. S. R. Costa, D. D. Coelho, R. B. Liszbinski, A. M. Lopes, L. Oliveira-Nascimento, M. B. de Jesus, A. F. Jozala, C. Ehrhardt and P. G. Mazzola, Colloids Surf. B Biointerfaces, 2023, 222, 113043 CrossRef CAS.
- Z. F. Yuan, P. McMullen, S. Luozhong, P. Sarker, C. J. Tang, T. Wei and S. Y. Jiang, Chem. Sci., 2023, 14, 2033–2039 RSC.
- D. D. La, S. V. Bhosale, L. A. Jones and S. V. Bhosale, ACS Appl. Mater. Interfaces, 2018, 10, 12189–12216 Search PubMed.
- Z. Q. Guo, C. X. Yan and W. H. Zhu, Angew. Chem., Int. Ed., 2020, 59, 9812–9825 Search PubMed.
- J. Dai, Y. Cheng, J. Wu, Q. Wang, W. Wang, J. Yang, Z. Zhao, X. Lou, F. Xia, S. Wang and B. Z. Tang, ACS Nano, 2020, 14, 14698–14714 CrossRef CAS PubMed.
- C. Y. Yu, H. Xu, S. Ji, R. T. Kwok, J. W. Lam, X. Li, S. Krishnan, D. Ding and B. Z. Tang, Adv. Mater., 2017, 29, 1606167 CrossRef.
- P.-P. He, X.-D. Li, L. Wang and H. Wang, Acc. Chem. Res., 2019, 52, 367–378 CrossRef CAS.
- S. Yang, H. Yu, X. Xu, T. Yang, Y. Wei, R. Zan, X. Zhang, Q. Ma, H. C. Shum and Y. Song, ACS Nano, 2023, 17, 8195–8203 CrossRef CAS PubMed.
- Y. Tang, D. Zhang, X. Gong and J. Zheng, Adv. Funct. Mater., 2022, 32, 2208022 CrossRef CAS.
- W. Zhang, P. Wei, L. Liu, T. Ding, Y. Yang, P. Jin, L. Zhang, Z. Zhao, M. Wang, B. Hu, X. Jin, Z. Xu, H. Zhang, Y. Song, L. Wang, S. Zhong, J. Chen, Z. Yang, Z. Chen, Y. Wu, Z. Ye, Y. Xu, Y. Zhang and L. P. Wen, Autophagy, 2023, 19, 3062–3078 CrossRef CAS.
- F. Fan, X. M. Chen, J. Lin, M. Lin, L. Li, Y. Gu, Y. Chai, H. Zhang, X. Chen and Q. Li, Adv. Funct. Mater., 2023, 34, 2314832 CrossRef.
- N. T. Connell, G. A. Abel and J. M. Connors, Thromb. Res., 2017, 150, 53–58 Search PubMed.
- L. Zhang, D. Jing, N. Jiang, T. Rojalin, C. M. Baehr, D. Zhang, W. Xiao, Y. Wu, Z. Cong, J. J. Li, Y. Li, L. Wang and K. S. Lam, Nat. Nanotechnol., 2020, 15, 145–153 Search PubMed.
- F. Zhang, Y. He, X. Jin, S. Xiang, H. T. Feng and L. L. Li, J. Phys. Chem. B, 2021, 125, 10224–10231 CrossRef.
- T. Wang, Z. Gao, Y. Zhang, Y. Hong, Y. Tang, K. Shan, X. Kong, Z. Wang, Y. Shi and D. Ding, J. Control. Release, 2022, 351, 272–283 CrossRef PubMed.
- Y. Qian, W. Wang, Z. Wang, X. Jia, Q. Han, I. Rostami, Y. Wang and Z. Hu, ACS Appl. Mater. Interfaces, 2018, 10, 7871–7881 CrossRef.
- L. Fu, J. Zhang, C. Wu, W. Wang, D. Wang, Z. Hu and Z. Wang, Nano Res., 2022, 15, 7286–7294 CrossRef.
- C. Zhang, L. H. Liu, W. X. Qiu, Y. H. Zhang, W. Song, L. Zhang, S. B. Wang and X. Z. Zhang, Small, 2018, 14, e1703321 CrossRef PubMed.
- M. Yuan, X. Fang, Y. Wu, Y. J. Xu, H. J. Feng, J. Mu, Z. X. Chen, Y. H. Lin, Q. R. Fu, W. Du, H. H. Yang and J. B. Song, Anal. Chem., 2022, 94, 5204–5211 CrossRef PubMed.
- J. Li, H. Gao, R. Liu, C. Chen, S. Zeng, Q. Liu and D. Ding, Sci. China Chem., 2020, 63, 1428–1434 CrossRef.
- Y. Deng, C. Fu, A. Xu, R. He, W. Lu and M. Liu, Spectrochim. Acta Part A Mol. Spectrosc., 2024, 316, 124334 CrossRef.
- K. Han, W. Y. Zhang, Z. Y. Ma, S. B. Wang, L. M. Xu, J. Liu, X. Z. Zhang and H. Y. Han, ACS Appl. Mater. Interfaces, 2017, 9, 16043–16053 CrossRef PubMed.
- W. Qin, Y. Wu, Y. Hu, Y. Dong, T. Hao and C. Zhang, ACS Appl. Bio Mater., 2021, 4, 1038–1044 CrossRef.
- K. M. Groborz, M. Kalinka, J. Grzymska, S. Kolt, S. J. Snipas and M. Poreba, Chem. Sci., 2023, 14, 2289–2302 RSC.
- Q. Jiao, Y. Zheng, S. Pei, X. Luo, X. Wu, K. Xu and W. Zhong, Anal. Chem., 2023, 95, 9097–9106 CrossRef PubMed.
- Y. Cheng, F. Huang, X. Min, P. Gao, T. Zhang, X. Li, B. Liu, Y. Hong, X. Lou and F. Xia, Anal. Chem., 2016, 88, 8913–8919 CrossRef PubMed.
- Y. Cheng, C. L. Sun, R. Liu, J. L. Yang, J. Dai, T. Y. Zhai, X. D. Lou and F. Xia, Angew. Chem., Int. Ed., 2019, 58, 5049–5053 CrossRef PubMed.
- Y. Cheng, A. E. Clark, J. Zhou, T. He, Y. Li, R. M. Borum, M. N. Creyer, M. Xu, Z. Jin, J. Zhou, W. Yim, Z. Wu, P. Fajtova, A. J. O'Donoghue, A. F. Carlin and J. V. Jokerst, ACS Nano, 2022, 16, 12305–12317 CrossRef PubMed.
- Z. Gao, H. Gao, D. Zheng, T. Xu, Y. Chen, C. Liang, L. Wang, D. Ding and Z. Yang, Sci. China Chem., 2020, 63, 398–403 CrossRef.
- M. Yuan, X. Fang, Y. Wu, Y. Xu, H. Feng, J. Mu, Z. Chen, Y. Lin, Q. Fu, W. Du, H. Yang and J. Song, Anal. Chem., 2022, 94, 5204–5211 CrossRef PubMed.
- Y. Cheng, A. E. Clark, W. Yim, R. M. Borum, Y. C. Chang, Z. Jin, T. He, A. F. Carlin and J. V. Jokerst, Anal. Chem., 2023, 95, 3789–3798 CrossRef PubMed.
- Z. Zhu, Q. Wang, H. Liao, M. Liu, Z. Liu, Y. Zhang and W. H. Zhu, Natl. Sci. Rev., 2021, 8, nwaa198 CrossRef PubMed.
- K. Gu, W. Qiu, Z. Guo, C. Yan, S. Zhu, D. Yao, P. Shi, H. Tian and W. H. Zhu, Chem. Sci., 2019, 10, 398–405 RSC.
- Y. Lyu, X. Chen, Q. Wang, Q. Li, Q. Wang, X. Li, Z. Zhu, C. Yan, X. Zhao and W. H. Zhu, Adv. Funct. Mater., 2021, 32, 2108571 Search PubMed.
- M. Li, J. Tang, C. Lin, A. Shen, X. Ma, J. Wu, X. Gao and P. Wang, Adv. Healthcare Mater., 2023, 12, e2300602 Search PubMed.
- J. L. Yang, J. Dai, Q. Wang, Y. Cheng, J. J. Guo, Z. J. Zhao, Y. N. Hong, X. D. Lou and F. Xia, Angew. Chem., Int. Ed., 2020, 59, 20405–20410 Search PubMed.
- J. Dai, J. J. Hu, X. Q. Dong, B. Chen, X. Y. Dong, R. Liu, F. Xia and X. D. Lou, Angew. Chem., Int. Ed., 2022, 61, e202117798 Search PubMed.
- H. Jia, D. Ding, J. Hu, J. Dai, J. Yang, G. Li, X. Lou and F. Xia, Adv. Mater., 2021, 33, e2104615 Search PubMed.
- F. Wu, Y. Huang, X. Yang, J. J. Hu, X. Lou, F. Xia, Y. Song and L. Jiang, Anal. Chem., 2021, 93, 16257–16263 Search PubMed.
- W. Zhang, J. J. Hu, R. Liu, J. Dai, L. Yuan, Y. Liu, B. Chen, M. Gong, F. Xia and X. Lou, Adv. Sci., 2023, 10, e2207228 Search PubMed.
- J. Liu, Z. He, Y. Zhong, L. Zhu, M. Yan, N. Mou, K. Qu, X. Qin, G. Wang, K. Zhang, W. Yang and W. Wu, ACS Appl. Mater. Interfaces, 2023, 15, 47381–47393 CrossRef PubMed.
- D. Yao, Y. Wang, K. Bian, B. Zhang and D. Wang, Biomaterials, 2023, 292, 121920 CrossRef PubMed.
- S. Lu, X. Guo, F. Zhang, X. Li, M. Zou and L.-L. Li, Chin. Chem. Lett., 2021, 32, 1947–1952 CrossRef CAS.
- X. Zhang, C. Ren, F. Hu, Y. Gao, Z. Wang, H. Li, J. Liu, B. Liu and C. Yang, Anal. Chem., 2020, 92, 5185–5190 CrossRef CAS PubMed.
- Y. Zhao, X. Zhang, Z. Li, S. Huo, K. Zhang, J. Gao, H. Wang and X. J. Liang, Adv. Mater., 2017, 29, 1601128 CrossRef.
- A. Han, H. Wang, R. T. Kwok, S. Ji, J. Li, D. Kong, B. Z. Tang, B. Liu, Z. Yang and D. Ding, Anal. Chem., 2016, 88, 3872–3878 CrossRef CAS PubMed.
- S. L. Ji, J. Li, X. C. Duan, J. T. Zhang, Y. F. Zhang, M. Q. Song, S. G. Li, H. L. Chen and D. Ding, Angew. Chem., Int. Ed., 2021, 60, 26994–27004 CrossRef CAS PubMed.
- L. Zhang, Y. Li, G. Mu, L. Yang, C. Ren, Z. Wang, Q. Guo, J. Liu and C. Yang, Anal. Chem., 2022, 94, 2236–2243 CrossRef CAS PubMed.
- W. Jiang, C. Cheng, X. Qiu, L. Chen, X. Guo, Y. Luo, J. Wang, J. Wang, Z. Xie, P. Li, Z. Wang, H. Ran, Z. Zhou and J. Ren, Adv. Sci., 2023, 10, e2204989 CrossRef PubMed.
- Y. Hu, J. Yu, M. Xu and K. Pu, J. Am. Chem. Soc., 2024, 146, 12656–12663 CrossRef CAS PubMed.
- Z. Hai, J. Wu, D. Saimi, Y. Ni, R. Zhou and G. Liang, Anal. Chem., 2018, 90, 1520–1524 CrossRef CAS.
- T. T. Wang, Q. C. Wei, Z. T. Zhang, M. T. Lin, J. J. Chen, Y. Zhou, N. N. Guo, X. C. Zhong, W. H. Xu, Z. X. Liu, M. Han and J. Q. Gao, Biomater. Sci., 2020, 8, 118–124 RSC.
- Y. Yuan, R. Zhang, X. Cheng, S. Xu and B. Liu, Chem. Sci., 2016, 7, 4245–4250 RSC.
- S. Ji, S. Li, H. Gao, J. Wang, K. Wang, W. Nan, H. Chen and Y. Hao, Biomater. Sci., 2023, 11, 2221–2229 RSC.
- Y. Q. Wang, J. H. Weng, X. D. Wen, Y. X. Hu and D. J. Ye, Biomater. Sci., 2021, 9, 406–421 RSC.
- Y. Z. Zhong, J. Zhan, G. H. Xu, Y. M. Chen, Q. Qin, X. Liao, S. D. Ma, Z. M. Yang and Y. B. Cai, Angew. Chem., Int. Ed., 2021, 60, 8121–8129 CrossRef.
- H. He, X. Lin, J. Guo, J. Wang and B. Xu, ACS Nano, 2020, 14, 6947–6955 CrossRef.
- G. L. Liang, H. J. Ren and J. H. Rao, Nat. Chem., 2010, 2, 239 CrossRef.
- Z. X. Chen, M. Chen, K. X. Zhou and J. H. Rao, Angew. Chem., Int. Ed., 2020, 59, 7864–7870 CrossRef.
- X. Liu and G. Liang, Chem. Commun., 2017, 53, 1037–1040 RSC.
- L. Xu, Y. Deng, H. Gao, Y. Yao, X. Liu, W. Zhan, G. Liang and X. Sun, Nanoscale, 2024, 16, 11538–11541 Search PubMed.
- Y. Deng, L. Xu, X. Liu, Q. Jiang, X. Sun, W. Zhan and G. Liang, J. Am. Chem. Soc., 2024, 146, 25462–25466 Search PubMed.
- L. Xu, H. Gao, W. Zhan, Y. Deng, X. Liu, Q. Jiang, X. Sun, J. J. Xu and G. Liang, J. Am. Chem. Soc., 2023, 145, 27748–27756 Search PubMed.
- W. H. Xu, D. Wang and B. Z. Tang, Angew. Chem., Int. Ed., 2021, 60, 7476–7487 CrossRef.
- C. Yan, J. Dai, Y. Yao, W. Fu, H. Tian, W. H. Zhu and Z. Guo, Nat. Protoc., 2023, 18, 1316–1336 CrossRef.
- Q. S. Zhang, P. Yu, Y. Fan, C. X. Sun, H. S. He, X. Liu, L. F. Lu, M. Y. Zhao, H. X. Zhang and F. Zhang, Angew. Chem., Int. Ed., 2021, 60, 3967–3973 CrossRef.
- H. C. Shen, L. D. Du, C. H. Xu, B. Z. Wang, Q. Q. Zhou, R. Q. Ye, R. T. K. Kwok, J. W. Y. Lam, G. C. Xing, J. W. Sun, T. M. Liu and B. Z. Tang, ACS Nano, 2024, 18, 20268–20282 Search PubMed.
- L. Y. Wu, W. H. Zeng, L. D. Feng, Y. X. Hu, Y. D. Sun, Y. X. Yan, H. Y. Chen and D. J. Ye, Sci. China Chem., 2020, 63, 741–750 CrossRef.
- X. Chen, Q. Yang, X. Lv, Y. Xiong, B. Zhong Tang and X. Huang, Coord. Chem. Rev., 2024, 516, 215970 CrossRef.
- K. Han, S. B. Wang, Q. Lei, J. Y. Zhu and X. Z. Zhang, ACS Nano, 2015, 9, 10268–10277 CrossRef PubMed.
- P. H. Cheng and K. Y. Pu, Chem. Soc. Rev., 2024, 53, 10171–10188 RSC.
- Y. Yuan, C. J. Zhang, R. T. K. Kwok, D. Mao, B. Z. Tang and B. Liu, Chem. Sci., 2017, 8, 2723–2728 RSC.
| This journal is © The Royal Society of Chemistry 2025 |




