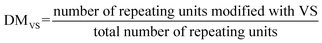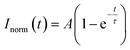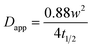Open Access Article
Open Access ArticleConstruction of multiphasic membraneless organelles towards spontaneous spatial segregation and directional flow of biochemical reactions†
Fariza
Zhorabek
 a,
Manisha Sandupama
Abesekara
a,
Jianhui
Liu
a,
Manisha Sandupama
Abesekara
a,
Jianhui
Liu
 a,
Xin
Dai
b,
Jinqing
Huang
a,
Xin
Dai
b,
Jinqing
Huang
 b and
Ying
Chau
b and
Ying
Chau
 *a
*a
aDepartment of Chemical and Biological Engineering, Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong SAR, China. E-mail: keychau@ust.hk
bDepartment of Chemistry, Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong SAR, China
First published on 13th January 2023
Abstract
Many intracellular membraneless organelles (MLOs) appear to adapt a hierarchical multicompartment organization for efficient coordination of highly complex reaction networks. Recapitulating such an internal architecture in biomimetic platforms is, therefore, an important step to facilitate the functional understanding of MLOs and to enable the design of advanced microreactors. Herein, we present a modular bottom-up approach for building synthetic multiphasic condensates using a set of engineered multivalent polymer–oligopeptide hybrids. These hybrid constructs exhibit dynamic phase separation behaviour generating membraneless droplets with a subdivided interior featuring distinct chemical and physical properties, whereby a range of functional biomolecules can be spontaneously enriched and spatially segregated. The platform also attains separated confinement of transcription and translation reactions in proximal compartments, while allowing inter-compartment communication via a directional flow of reactants. With advanced structural and functional features attained, this system can be of great value as a MLO model and as a cell-free system for multiplex chemical biosynthesis.
Introduction
In recent years, membraneless organelles (MLOs) have emerged as a new class of compartments that coordinate complex intracellular biochemistry in a spatiotemporal manner.1 They are recognized as the functional core of many cellular functions, such as stress response,2,3 regulation of gene expression4–7 and chemical signaling.8,9These dense liquid-like assemblies are formed via liquid–liquid phase separation (LLPS) of multivalent scaffold molecules.10 Scaffolds are mainly represented by intrinsically disordered proteins (IDPs) and sometimes, RNAs, where the low-complexity (LC)-, RNA-binding (RB)-domains of IDPs,11,12 and the charged backbone and aromatic bases of RNAs13 contribute to multivalent interactions. Normally, multiple IDPs coexist together with nucleic acids within one condensate, and oftentimes such heterogenous nature engenders complex internal structuration. As such, mixtures of IDPs and RNAs were found to produce core–shell architectures in stress granules,14 P-bodies15 and paraspeckles,16 multiple micro-phases in the nucleolus,17 distinct RNA-enriched cores in mitochondrial RNA-granules18 and internal vacuolization.19 Recent studies suggest that sub-structures in many cases can arise from spontaneous processes alone, exhibiting distinct morphological and biophysical characteristics20,21 and correspondingly different functional properties.22,23 For example, by spatially clustering a set of different processing factors, distinct nucleolus phases appear to sequentially host and direct multiple steps of ribosome biogenesis reactions.22–24 While more concrete functional roles of such internal architecture are yet to be revealed, the analysis of activities within other condensates25 hints at the importance of sub-compartmentalization in achieving optimal spatial and temporal regulation of highly sophisticated intracellular reactions.
Recapitulating complex architectural organization of MLOs in vitro is of high interest towards building an experimental model platform for exploring the mechanistic and functional aspects of multiphase condensates, and as next generation multicompartment microreactors. A number of artificial hierarchical membraneless structures have recently been designed using a set of synthetic polyelectrolytes,26–28 RNA and proteins/peptides,29–32 and a mixture of synthetic IDPs17,33 serving as a starting point to understand the underlying mechanism and characteristics of multiphasic phase separation, while other aspects still remain unexplored, necessitating the advancement of MLO-mimetic platforms. To date, reported multi-compartmentalization strategies have been utilized for sequential chemical transfer reactions,28 cascade enzymatic reactions,34 and spatial segregation of incompatible components.35 Nevertheless, more sophisticated bioprocessing reactions observed in living cells have rarely been demonstrated in vitro in conjunction with structurally complex membraneless systems.
In the present study, we demonstrate the construction of a novel artificial MLO system that recapitulates both the multiphasic organization and functional complexity of intracellular condensates. We use IDP-mimicking polymer–oligopeptide hybrid (IPH) constructs, as scaffold mimetics to form complex membraneless condensates in vitro. An IPH, recently reported by our group,36 is an engineered multivalent self-interacting hybrid scaffold with well-defined IDP-inspired interaction modules, which demonstrates capability to phase separate and form micro-sized liquid droplets resembling natural MLOs, with the ability to selectively enrich multiple biomolecules37 and host biochemical reactions.36 Mimicking the structural features of fused in sarcoma (FUS) protein, a prototypical IDP and a major scaffolding molecule of intracellular MLOs,38–40 the IPH consists of a flexible charge-neutral polymer chain, recapitulating the disordered and flexible random-coil like nature of IDPs, grafted with FUS-derived short oligopeptide motifs responsible for weak and reversible interactions, essential for LLPS (Fig. 1). Such a minimalist and modular design offers flexibility to finely modulate various molecular and structural parameters, allowing the engineering of multiple IPH variants with tunable intermolecular interactions. The IPH therefore appears as a promising platform for recreating the heterogeneous composition of native condensates and constructing a well-defined multi-compartment model MLO.
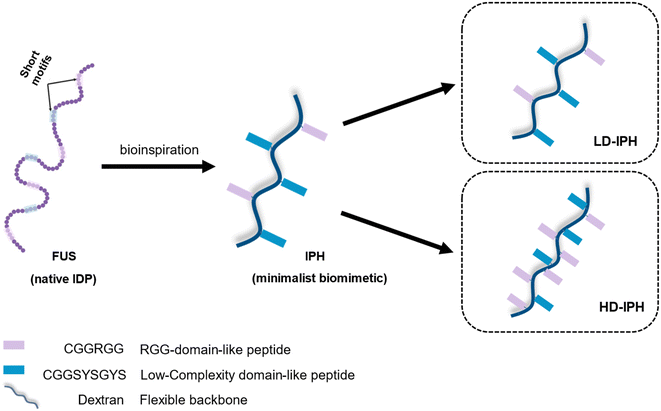 | ||
| Fig. 1 Schematic representation of the structural design and chemical composition of IDP-mimicking polymer–oligopeptide hybrid constructs. | ||
Here, using a set of different IPH constructs, in combination with RNA, we generate structurally complex synthetic membraneless organelles with two immiscible subcompartments. We find that coexisting phases display distinct chemical composition and material properties, which dictate the construct's phase-specific partitioning behaviour towards a range of molecules. We then accomplish the coupling of in vitro transcription and translation (IVTT) to demonstrate the functional aspects of the artificial multiphasic assemblies. Altogether, our findings indicate that the IPH platform provides new opportunities for spontaneous construction of hierarchical membraneless condensates in vitro with substantial mimicry of native MLOs for spatial and temporal orchestration of bioreactions.
Results and discussion
Internal organization of heterogeneous condensates
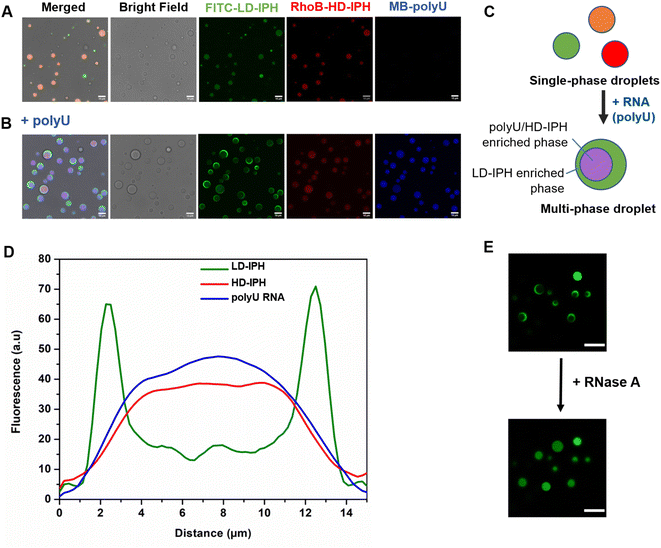
Previous in vitro observations and molecular simulations suggest that the interplay of multiple factors gives rise to an internal hierarchy of MLOs. The immiscibility of components can potentially be induced by the imbalance of intermolecular interactions within the network of competing binding partners, associated with differences in the charge density and valency,26,27 mixture stoichiometry,30 rate of formation/dissociation of complexes,41 and presence of substrate specificity,15 while the relative rank of organization of two or more immiscible phases is dictated by the interfacial tension between phases.21With the IPH-derived platform,36 we utilized the difference in the valency of scaffold molecules and the presence of a common binding partner for competitive interactions. The IPH is composed of vinyl-sulfone (VS) functionalized dextran (Dex-VS) and a set of cysteine-terminated ‘sticker’-oligopeptides: CGGSYSGYS and CGGRGG, where peptides are grafted on the dextran backbone via a click reaction between the sulfhydryl group of cysteine residues and the VS groups of Dex.38–40 Such a design enables easy modulation of the construct's valency by changing the density of ‘sticker’-sequences on the polymer chain, that is, by altering the degree of modification (DM) of the construct. Variability in interaction parameters was explored in two IPH variants with a DM of 38% and 92% (Fig. 1). Hereafter, the first variant with a low density of interacting “sticker”-oligopeptides is referred to as LD-IPH, while the latter high-density hybrid as HD-IPH.
In the biological context, the majority of multiphasic MLOs contain RNA molecules as central scaffold components that initiate and modulate their phase separation. Moreover, serving as a common binding element of multiple scaffold proteins, under different molecular stoichiometry and interaction scenarios, RNA is known to induce the immiscibility of components, mediating both the formation of subcompartments and their organization within MLOs.30–32,42 To recapitulate this aspect of MLO composition, synthetic homopolymeric RNA, polyU was included as a model RNA component in our artificial construct and as a common binding component for LD- and HD-IPHs. Here, both of them are expected to interact with the model RNA primarily through CGGRGG-stickers, via long-range electrostatic attraction between positively charged Arg-residues and the negatively charged phosphate backbone of RNA, as well as short-range cation–π interaction and π–π stacking between arginine's guanidinium group and uracil rings.43 CGGSYSGYS-stickers can also engage in π–π stacking with RNA via aromatic residues Tyr. Thus, HD- and LD-IPHs would appear as competitive binding partners of polyU RNA in this mixing scenario.
In the absence of RNA, a mixture of LD- and HD-IPH constructs engenders homogeneously mixed droplets containing both IPHs, in addition to droplets containing only LD-IPH or HD-IPH coexisting in a solution (Fig. 2A). The addition of polyU into the LD/HD mixture induced the reorganization of the droplet components and the formation of biphasic droplets with a partial engulfment morphology (Fig. 2B and C). PolyU preferentially associates with HD-IPH, forming an inner compartment, while displacing LD-IPH to the surface, as demonstrated by fluorescence intensity profiles across these heterogeneous condensates as shown in Fig. 2D. More specifically, LD-IPH was concentrated primarily in the outer layer of the condensate with a partitioning coefficient, K1, of 20 ± 1.83, while showing only slight localization in the inner layer (K2 = 4.7 ± 0.98). Here, K is defined as the ratio of fluorescence intensity within the corresponding layer of the condensate and the external dilute phase. Both polyU and HD-IPH were 2-times more enriched in the inner core part compared to the outer layer (Table 1). Such patterning was driven by competing IPH-RNA interactions and difference in surface tension, following the rules governing intra-droplet organization in biological condensates. HD-IPH, owing to the higher availability of Tyr and Arg side chains, can more effectively compete for available polyU, forming multiple modes of interactions via ion pairing with RNA's phosphate backbone and aromatic interactions with nucleobases. A fully saturated HD-IPH/polyU complex demonstrates more dense and more hydrophobic assemblies, both parameters associated with a higher surface tension,17,27 forcing it to be buried in the inner phase. Concurrently, LD-IPH with lower valency was positioned between a dilute water phase and inner HD-IPH/polyU complex, due to its interaction potential with both inner phase components, and to minimize the interfacial free energy of this multiphase condensate, consistent with the simulation studies of multivalent protein species.44
| Outer condensed phasea (K1) | Inner condensed phasea (K2) | |
|---|---|---|
| a Denotes partitioning coefficients calculated by dividing the mean fluorescence intensities of labelled components inside either the outer (Io) or the inner (Ii) compartment of the condensed phase by the mean intensities in the external dilute phase (Idil), as K1 = Io/Idil and K2 = Ii/Idil. | ||
| LD-IPH | 20 ± 1.83 | 4.71 ± 0.98 |
| HD-IPH | 8.5 ± 1.13 | 20.7 ± 5.8 |
| polyU RNA | 8.57 ± 1.32 | 17.1 ± 1.36 |
This pattern was observed for different mixing protocols, both when (1) LD-IPH was premixed with polyU at a charge-matching ratio, with the subsequent addition of HD-IPH (Fig. S1A†), and (2) HD-IPH was complexed with polyU at a charge-matching ratio, followed by the addition of LD-IPH (Fig. S1B†). Notably, instances with more than one inner phase appearance within one droplet could also be observed; however these locally nucleated subcompartments eventually coalescence reaching an equilibrium biphasic droplet state (ESI Video 1†). Droplets with their equilibrium morphology can be preserved for over 24 h (Fig. S2†).
The removal of polyU with the addition of RNase A caused the droplets to change back to a uniformly mixed state (Fig. 2E), signifying the importance of RNA molecules in driving spatial organization inside LD-IPH/HD-IPH droplets. In repeated experiments with other single-stranded RNAs (ssRNAs) of different compositions and lengths, the same internal droplet rearrangement of LD- and HD-IPH took place (Fig. S3†), hinting towards the generic nature of RNA-associated driving forces for internal compartmentalization of condensates.
Multiphasic organelles display distinct biophysical properties
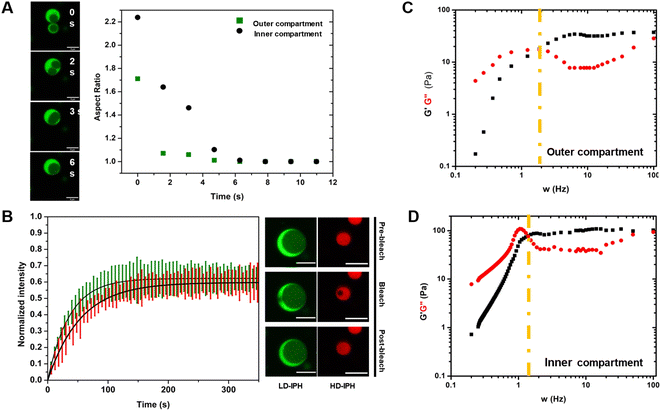
Reconstructed multiphasic droplets possess liquid-like characteristics, as both outer and inner compartments undergo complete fusion and adapt spherical shapes, indicated by a final aspect ratio (major axis/minor axis of coalescing droplets) of 1.0 (Fig. 3A). However, the fusion of the LD-IPH-enriched outer phase occurs faster than the fusion of the inner compartment implying different bulk material properties of each phase. To further investigate their material characteristics, we separately evaluated the linear viscoelastic properties of each domain, using the passive microrheology technique with optical tweezers.45 Two phases were shown to exhibit the properties of complex Maxwell fluids (Fig. 3C and D), reminiscent of other in vitro condensate systems,45,46 each with a distinct terminal relaxation time (τR) corresponding to the average rearrangement time of the interacting network. For the HD-IPH/polyU enriched core, the characteristic τR was 1.13 ± 0.88 s compared to 0.50 ± 0.12 s for the LD-IPH enriched outer domain, signifying that the inner compartment displays elastic-like (G′) behaviour over a longer timescale than the shell part. Consistently, the characteristic viscosity (η) of the inner phase was relatively higher than that of the outer phase, with an estimated η of 23.3 ± 12.1 Pa s and 11.8 ± 5.12 Pa s, respectively. The dissimilar viscosity of subcompartments is a hallmark of native multiphase condensates,20,25,47 for which the values range from 0.7 to 103 Pa s.In addition, the two phases exhibited different diffusivities of its component (Fig. 3B). The outer phase showed faster molecular dynamics with a characteristic recovery half-time, t1/2, of 26.21 ± 6.71 s, while the inner HD-IPH/polyU-rich phase exhibited relatively stalled dynamics (t1/2 = 38.15 ± 5.28 s). Overall, combined with the fusion characteristics and frequency-dependent viscoelastic response, the artificial multiphase MLO reconstruction with LD-IPH/HD-IPH/polyU recapitulates the inhomogeneous dynamics of stress granules25 and L-bodies,20 with a dense core and more dynamic shell.
Partitioning behavior of multiphase organelles
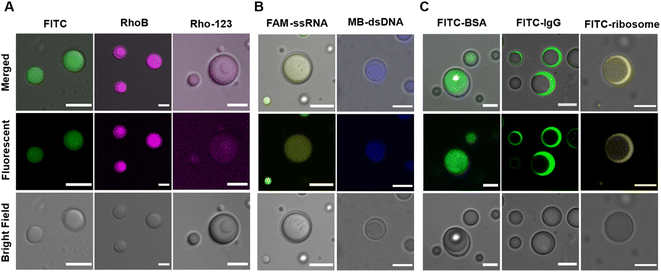
Besides scaffold molecules, there are also a substantial portion of “client” molecules that are dispensable for MLO assembly but enriched within them via partitioning (or sequestration).48,49 Multicompartment MLOs additionally demonstrate the ability to internally sort and segregate their ‘clients’,25,26,32 dictated by the different chemical compositions and physical characteristics of distinct internal phases.A mixture of LD-, HD-IPH and polyU RNA was shown to engender spatially segregated phases with relatively different chemical and physical characteristics, which could introduce a difference in their corresponding solute partitioning behaviour. Using a set of intrinsically fluorescent and fluorescently labelled molecules, the sequestration behaviour of our multiphasic droplet model was investigated (Fig. 4 and S4†). Both anionic fluorescent isothiocyanate (FITC) and zwitterionic rhodamine B (RhoB) demonstrated sequestration into the droplet, with a slight increase in fluorescent signals in the inner compartment (K2 = 20.7) relative to the outer shell (K1 = 14.5), while a cationic dye (Rho123) showed no preferential enrichment in neither of the phases and was equally distributed both outside and inside of droplets (Fig. 4A). The partitioning of FITC and RhoB can be driven by non-specific electrostatic and aromatic interactions, supported by positively charged arginine residues and aromatic-rich peptide modules that are present on both LD-IPH and HD-IPH, with the IPH with higher valency (HD-IPH) being more competitive. Both ssRNA and dsDNA showed much stronger relative preferential accumulation to the inner phase (Fig. 4B and S4†), compared to FITC which can be attributed to additional base-pairing interactions provided by polyU RNA along with a higher density of binding elements on HD-IPH.
Interestingly, spatial organization of negatively charged biological macromolecules based on size was observed (Fig. 4C). FITC-BSA (Mw = 65 kDa) was preferentially localized in the inner phase, owing to increased interacting moieties on HD-IPH (for electrostatic attraction and/or aromatic interactions), whereas FITC-IgG (150 kDa) and FITC-ribosome (∼2.5 MDa) were primarily partitioned in the outer phase. This phenomenon is reminiscent of the properties of native condensates exhibiting size-selective filtering of solutes50 due to differential mesh sizes. Based on this, we speculate that the denser core consisting of HD-IPH/polyU displays a relatively smaller characteristic mesh size compared to the less viscous outer phase, signifying the possibility of size-dependent sorting of solutes within the multiphasic droplet. These results support that the artificial multiphasic MLO platform enables differential partitioning of ‘client’-molecules, like native complex MLOs.
Towards spatial organization of complex reactions
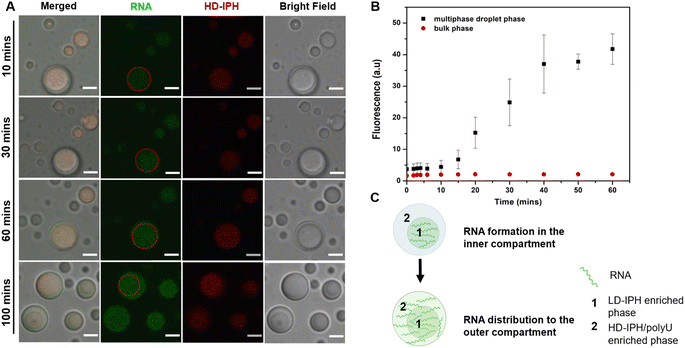
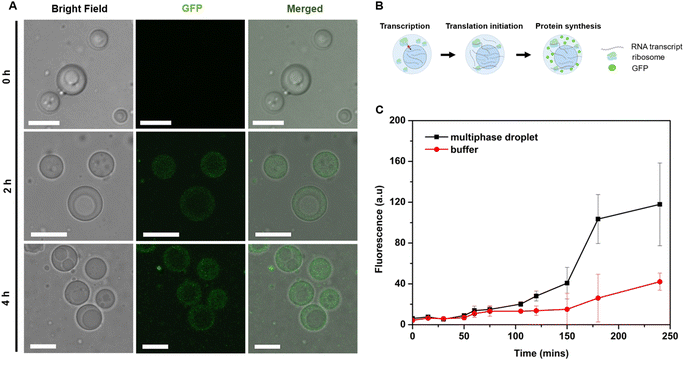
Phase-specific sequestration of biomolecules allows biological condensates to realize multiple processes in a confined spatial location and to direct the flow of reactions, allowing incremental accumulation and release of reactants and products from one sub-compartment to the other.17,23 By taking advantage of the complex architecture of the engineered multiphase MLO-platform and its differential partitioning behaviour (Fig. 4), we demonstrate here that the mimetic system can recapitulate the functional complexity of its native counterpart by coupling transcription and translation in proximal compartments within the same droplet.First, to demonstrate the confinement of in vitro transcription, IVTx – a mixture containing T7 polymerase, a set of NTPs, reaction buffer and dsDNA template were added to the biphasic droplet. To visualize the generation of RNA, the Broccoli aptamer sequence was used, which upon binding to a DFHBI-1T small molecule probe can fluoresce from the aptamer-activated Broccoli-DFHBI-1T complex. Fig. 5A and B show that in the first 60 min the fluorescence intensity of Broccoli-DFHBI-1T 1T locally increased within the inner phase due to RNA synthesis, with low fluorescence in the surrounding dilute phase. To verify that the signal concentration inside the droplet is not due to the partitioning of RNA molecules synthesized outside the droplet, we centrifuged a mixture of droplets and IVTx components and recorded the signals in the supernatant, where no fluorescence enhancement was observed (Fig. S5†), confirming that IVTx machinery is compartmentalized into the droplets. From microscope observations, the initial fluorescent signals were traced from the inner subcompartment, hinting towards phase-specific confinement of the reaction. To verify that, we have tested the partitioning characteristics of FITC-labelled T7 polymerase, which demonstrated distinct accumulation in the inner compartment (Fig. S6†). All these results, together with the previous observation of dsDNA preferential partitioning into the core of the multiphase droplets (Fig. 4B), support that the IVTx reaction occurs in the droplet, more specifically in its inner phase.
Further observations showed that starting from 60 min, the RNA produced in the inner compartment slowly moved to the outer phase and distributed equally in both phases (Fig. 5C), suggesting the saturation of binding sites in the inner phase. We hypothesize that this is particularly significant for the initiation of the translation step, which is expected to occur in the outer shell part, where the ribosome is preferentially localized (Fig. 4C).
To confirm this, full in vitro transcription and translation (IVTT) machinery was incorporated into the multiphasic droplet along with plasmid DNA encoding positively charged GFP (+36). The first noticeable signs of GFP could be traced after 2 h under a confocal fluorescence microscope (Fig. 6A), followed by rapid fluorescent signal progression that took place exclusively in the outer compartment (Fig. 6C). Given that the ribosome is preferentially sequestered in the outer phase (Fig. 4C), these results suggest that in vitro translation takes place in the shell part of the multiphase condensate, coupled to the in vitro transcription localized in the inner core-phase (Fig. 5).
To quantify the yield of GFP, the fluorescence intensities were correlated with protein concentrations. Generally, the protein production capacity in the presence of droplets was relatively lower than in the standard buffer without droplets, with an approximate yield of 6.5 ng μL−1 for the droplet-containing sample, and 10 ng μL−1 for the buffer without droplets after 4 h of reaction. The lower yield of the multiphase IPH-droplet system might be related to the high viscosity of both phases (23.3 Pa.s for the inner phase and 11.8 Pa.s in the outer phase), where, upon addition of full IVTT machinery, all components within the droplets might experience substantially stalled diffusion. For example, the mobility (Dapp) of LD-IPH and HD-IPH was reduced nearly threefold in the presence of IVTT machinery (Fig. S7†), suggesting the effect on diffusion both in the inner and outer compartments.
The majority of the droplets maintained the dual-phase arrangement in the protein synthesis process for ∼4 h (Fig. 6A), after which the first signs of aggregates started to appear. At the 24 h time-mark, there were only a few droplets remained with fluorescent proteins entrapped in the outer phase (Fig. S8†). The formation of aggregates might be caused by increased crowding within the condensed phase driven by the continuous translation of a target protein. High crowding conditions contribute to the reduction of free volume, thus increasing the intermolecular interactions and producing aggregates of IPH-chains with proteins, accompanied by the disintegration of droplets. A similar phenomenon was observed in the gene expression system entrapped in coacervates.51
Via the demonstration of two-stage coupled reactions for protein expression, it is evident that the spatially complex LD-IPH/HD-IPH/polyU droplet was able to attain the spatial segregation of components and compartment-specific reaction confinement, without restricting communication between two separate reactions, and rather engendering the directional flow of reactants. Despite the lack of enhancement of the reaction yield, the results show that the sub-compartmentalized artificial MLO is a functional system, with two distinct phases capable of spatiotemporal arrangement and coupling of complex biochemical reactions. Such functional features shown by the minimalist IPH-derived artificial platform resonates with the recent observations of functionalities of more complex natural and engineered multiphasic MLOs.22,23 Moreover, the inner and outer phases of the artificial MLO, exhibiting different internal densities, also recapitulate the actual cellular settings, where transcription takes place in a crowded and viscous nucleoplasm, while translation occurs in a relatively less crowded and less dense cytoplasm. By further adjusting the material states of each phase to more closely mimic the optimal cellular conditions for these two processes, we expect that there is room to improve the coupled two stage cell-free protein expression offered by the biphasic IPH-based platform.52
Conclusions
We have demonstrated that IDP-mimicking polymer–oligopeptide hybrids (IPHs) present a new and simple approach for spontaneous formation of multiphasic membraneless assemblies. The obtained droplets with two coexisting compartments display characteristics analogous to native spatially complex MLOs, including liquid-like behaviour, differential dynamics of components and viscoelastic properties within each phase, and phase-specific segregation of ‘client’-molecules. Additionally, this synthetic platform could accommodate a set of complex biochemical reactions with functional features of reaction confinement, enhancement, and directional flow. We believe that IPH-derived multiphasic constructs can serve as a modular platform for a wide range of applications, in particular, as cell-free bioreactors with MLO-mimicking features.Experimental
Synthesis of IPHs
The degree of modification (DM) of the oligopeptide–polymer construct is defined as the ratio of the number of dextran repeating units conjugated to “sticker”-oligopeptides to the total number of repeating units in the polymer, essentially determining the density of interacting “sticker”- units per dextran -“spacer” chain, and thus the overall valency of the IPH construct.To conjugate cysteine-terminated ‘sticker’-oligopeptides, CGGRGG and CGGSYSGYS, dextran is functionalized with thiol-reactive vinyl-sulfone (VS) groups. Thus, to afford an IPH with desired DMstickers, dextran must contain sufficient VS groups:
For the synthesis of HD-IPH (DMstickers = 92%), Dex-VS with similar DMVS = 92% was synthesized. 30 mg of dextran (40 kDa) was dissolved in water at 10 mg mL−1 concentration. 5 M NaOH was used to adjust the solution pH to 12.3 and divinyl sulfone was quickly added to the solution at 1.2× molar excess to the hydroxyl groups of dextran. The reaction was held under constant stirring for 1440 s and terminated by adjusting the pH of the solution to mildly acidic (pH = 3.0) using 5 M HCl. The final aqueous solution was transferred to dialysis tubing (MWCO 3.5 kDa, Spectra/Por Dialysis Membrane, Spectrum Labs, USA) and dialyzed against ultra-pure water for 5 days with 2 times water replacement daily. Purified Dex-VS solution was further lyophilized and stored at −20 °C for further use. DMVS was quantified from 1H NMR by comparing the integrals of the VS 1H peak to that of the dextran 1H1 peak.
The synthesis of HD-IPH follows thiol-Michael addition chemistry between the thiol groups of cysteine and VS groups. The reaction was held in NaCl aqueous solution (3 M), to prevent potential phase separation of oligopeptide–polymer hybrids during the synthesis. N2 pre-treated NaCl solution was used to first dissolve two peptides, with an additional 10 min N2 flow to prevent potential disulfide bond formation. Then 40 kDa Dex-VS, Dm = 92%, was added at 2 mg mL−1 and dissolved under rigorous stirring, followed by acidity adjustment to pH 6.0 to initiate the reaction. The total molarity of peptides added was at 1.5× excess to VS of Dex-VS. The N2 flow was maintained for another 10 min, and then the reagents were left to react for 24 h under continuous stirring. The materials were then dialyzed (MWCO 3.5 kDa) against ultrapure water for 5 days with two times water replacement daily. After dialysis, HD-IPH solution was lyophilized and stored at −20° for further use. The DM of peptides was quantified by comparing the integrations of peptide characteristic peaks with the integration of the 1H1 peak of dextran.
LD-IPHs and FITC labelled LD-IPHs were synthesized, purified and characterized following a previously reported method.36
Preparation of heterogenous droplets
Fluorophore-labelled LD-IPH and HD-IPH were mixed with their corresponding unlabelled versions at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio and were used for the subsequent assemblies of droplets. For LD-/HD-IPH droplets, both construct mixtures were added to a physiology-mimicking buffer (150 mM NaCl, 10 mM HEPES, and 10 wt% PEG 8000 crowding agent) followed by vortexing for 20 s and incubation for 15 min. For mixtures involving polyU, several protocols were evaluated:
1 ratio and were used for the subsequent assemblies of droplets. For LD-/HD-IPH droplets, both construct mixtures were added to a physiology-mimicking buffer (150 mM NaCl, 10 mM HEPES, and 10 wt% PEG 8000 crowding agent) followed by vortexing for 20 s and incubation for 15 min. For mixtures involving polyU, several protocols were evaluated:
(1) To the pre-formed and equilibrated LD-/HD-IPH droplets, polyU RNA is added at a specific charge ratio (N/P) to a final concentration of LD-IPH and HD-IPH of 6 μM. The mixture is vigorously vortexed for 20 s and kept on ice for further equilibration.
(2) Pre-formed HD-IPH droplets are mixed with polyU at a specific N/P ratio and incubated on ice for 15 min, following by addition of 6 μM LD-IPH. The mixture is vigorously vortexed for 20 s and kept on ice for further equilibration. The final concentration of HD-IPH is 6 μM.
(3) Pre-formed LD-IPH droplets are mixed with polyU at a specific N/P ratio and incubated on ice for 15 min, following by addition of 6 μM HD-IPH. The mixture is vigorously vortexed for 20 s and kept on ice for further equilibration. The final concentration of IPH is 6 μM.
For all mixtures, methylene blue stain (20 μM) was added after final incubation to visualize RNA.
Imaging experiments
The multiphase droplet formation and fusion kinetics were visualized by confocal fluorescence microscopy. Pre-formed and equilibrated droplet-samples were deposited on a surface-treated confocal dish and left for 5–10 min to settle. To differentiate the localization of individual components within droplet constructs, mixtures with a FITC-labelled IPH and RhoB-labelled HD were viewed with two fluorescent channels in line-sequential mode, using a 488 nm laser (0.4%) for FITC-IPH in the emission range 498–520 nm and 552 nm (1%) for RhoB-HD, using 562–630 nm emission. RNA localization was viewed with methylene blue staining (10 μM) using a 638 nm laser at 0.3% intensity and 663–780 nm emission settings.Fluorescence recovery after photobleaching
The circular region of interest (ROI) with a diameter of 2 μm was defined in the corresponding phase in fluorescence-labelled multiphase droplets. Photobleaching was performed on 488 nm and 552 nm laser lines for inner and outer phases, respectively. The images and fluorescence intensity of the ROI were recorded every 2 s for about 10 minutes using NIS-Elements software. The percentage recovery of fluorescence intensity over time was normalized using a reference ROI in an unbleached droplet.Normalized data were then fitted to a single exponential equation in MATLAB:
t1/2 = τ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
Using the radius of the photobleached region (w), the apparent diffusion coefficient (Dapp) was approximated using the Axelrod model equation:53
Microrheology with static optical tweezers (mSOT)
The measurement of the frequency-dependent viscoelastic moduli of multiphase droplet constructs was conducted on an optical tweezers setup (LUMICKS, m-Trap) with a 60× objective.First, IPH-droplets were prepared in filtered physiology-mimicking buffer containing 1 μm polystyrene beads (0.0005%), followed by 15 min incubation.
A homemade chamber on the coverslip was prepared using Gene Frame (Thermo Fisher) arranged into two chambers separated by tiny incision, as a path for droplets to move from one chamber to the other. The upper chamber was loaded with droplet solution and the bottom chamber was filled with 10 μL of the buffer. An optical trap was first positioned on the top chamber to trap beads embedded into the droplet (with a size of 8–10 μm) and to move the droplet sample to the bottom chamber to prevent any interference from other droplets during bead trajectory tracking. The temperature of the stage was kept constant at 22 °C, and to minimize the potential local temperature change from the laser (35% intensity).
The trap-constrained motion of the bead inside the droplet was tracked using a bright field camera. The raw data in the form trajectory of the bead in X–Y coordinates over time were extracted and used for subsequent analysis using a MATLAB script built using reported data analysis steps.45,54
Droplet-mediated in vitro transcription
Multiphase droplets were formed in the buffer containing a 3 wt% PEG 8000 crowding agent, 150 mM NaCl, 10 mM HEPES by sequential addition of HD-IPH, polyU and LD-IPH followed by 20 s vortexing and incubation on ice for 15 min. To the pre-formed droplets, the components of the MEGAscript kit (Thermo Fisher), more specifically 4 NTPs (2 mM), T7 polymerase (1 U μL−1), DFHBI-1T (25 μM) and 1× reaction buffer, were added, and the solution was placed on ice for 15 min to allow equilibration, protected from light. Then, pre-incubated in vitro transcription (IVTx) assembly with multiphase droplets was deposited on a confocal dish and the reaction was initiated with the addition of 50 nM template DNA (sequence shown below). Reaction progression was monitored under a laser scanning microscope (Leica SP8) using a 100× oil objective with a 488 nm argon laser, at 22 °C.Broccoli aptamer template: sense strand: 5′-TAA TAC GAC TCA CTA TAG GAG ACG GTC GGG TCC AGA TAT TCG TAT CTG TCG AGT AGA GTG TGG GCT C-3′
Antisense (template) strand: 5′-TAA TAC GAC TCA CTA TAG GAG ACG GTC GGG TCC AGA TAT TCG TAT CTG TCG AGT AGA GTG TGG GCT C-3′
To check whether IVTx could occur outside the droplets in the dispersed phase, solution with droplets incubated with the IVTx kit was centrifuged at 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g at 25 °C for 15 min to remove the condensed (droplet) phase. Then, plasmid DNA was added to the collected supernatant, and the solution was transferred to a 384-well plate to trace any fluorescence change on a microplate reader (FlexStation 3, Molecular Devices). The results were compared to the buffer containing full IVTx components and plasmid DNA.
000×g at 25 °C for 15 min to remove the condensed (droplet) phase. Then, plasmid DNA was added to the collected supernatant, and the solution was transferred to a 384-well plate to trace any fluorescence change on a microplate reader (FlexStation 3, Molecular Devices). The results were compared to the buffer containing full IVTx components and plasmid DNA.
Droplet-mediated cell-free protein expression
Protein expression was performed using a PURExpress reconstituted E. coli protein expression system (New England BioLabs) consisting of two solution mixtures (A and B). Solution A is composed of small molecules such amino acids and rNTPs, and Solution B includes ribosomes, T7 RNA polymerase, translation factors, energy regeneration enzymes and aminoacyl-tRNA synthetases.To perform transcription/translation reactions, the first inner compartment, composed of the HD-IPH construct and polyU, was formed, keeping the N/P ratio of components at 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. To the pre-formed HD-IPH/polyU droplets, 6 μL of Solution B was added, followed by gentle mixing by using a pipette and 5 min incubation on ice. Following that, LD-IPH was added to the solution at 12 μM concentration. The mixture was vortexed for 20 s and equilibrated on ice for 15 min, followed by addition of 8 μL of Solution A. The reaction was initiated with the addition of plasmid DNA of positively charged GFP (+36) (posGFP). For all reactions, a buffer containing a 3 wt% PEG 8000 crowding agent, 150 mM NaCl and 10 mM HEPES was used.
1. To the pre-formed HD-IPH/polyU droplets, 6 μL of Solution B was added, followed by gentle mixing by using a pipette and 5 min incubation on ice. Following that, LD-IPH was added to the solution at 12 μM concentration. The mixture was vortexed for 20 s and equilibrated on ice for 15 min, followed by addition of 8 μL of Solution A. The reaction was initiated with the addition of plasmid DNA of positively charged GFP (+36) (posGFP). For all reactions, a buffer containing a 3 wt% PEG 8000 crowding agent, 150 mM NaCl and 10 mM HEPES was used.
The in vitro translation in combination with transcription was monitored using a plasmid DNA construct of posGFP. Similarly, after depositing the IVTT/IPH mixture on a pre-treated confocal dish, the reaction was initiated with the addition of 300 ng of plasmid DNA solution. The reaction was monitored at different time points over 24 h under a laser scanning microscope (Nikon C2) using a 63× oil objective with a 488 nm laser, at 22 °C.
Quantification of the protein yield
To quantify the yield of the end-product, the fluorescence intensities were approximated to protein concentrations using a calibration curve derived from confocal fluorescence imaging. For the case with condensates, a standard curve was generated based on the fluorescence intensities of GFP trapped in IPH droplets at different concentrations, while for a non-droplet sample separate calibration was made. Imaging was performed under a Nikon C2 microscope with the same acquisition settings used for in vitro translation monitoring.Data availability
All datasets supporting this article are provided within the paper and its ESI file. An additional video with multiphase droplets is uploaded as the ESI. The MATLAB code for optical tweezers-based microrheology analysis has been deposited in GitHub at https://github.com/fafreeze/bioinspire.git.Author contributions
Conceptualization: Z. F., J. L., and Y. C.; methodology: Z. F., M. S. A., and J. L.; investigation: Z. F., M. S. A., and X. D.; visualization: Z. F. and Y. C.; supervision: J. L., Y. C., and J. H.; writing—original draft: Z. F. and Y. C.; writing—review & editing: Z. F., M. S. A., J. L., X. D., J. H., and Y. C.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge the financial support by the Hong Kong Research Grant Council (GRF 16102520) and Hong Kong PhD Fellowship Scheme for M.S.A.References
- S. Boeynaems, S. Alberti, N. L. Fawzi, T. Mittag, M. Polymenidou, F. Rousseau, J. Schymkowitz, J. Shorter, B. Wolozin, L. Van Den Bosch, P. Tompa and M. Fuxreiter, Trends Cell Biol., 2018, 28, 420–435 CrossRef CAS PubMed.
- W. van Leeuwen and C. Rabouille, Traffic, 2019, 20, 623–638 CAS.
- D. Marijan, R. Tse, K. Elliott, S. Chandhok, M. Luo, E. Lacroix and T. E. Audas, FEBS Lett., 2019, 593, 3162–3172 CrossRef CAS PubMed.
- E. M. Langdon and A. S. Gladfelter, Annu. Rev. Microbiol., 2018, 72, 255–271 CrossRef CAS PubMed.
- J. Sheu-Gruttadauria and I. J. MacRae, Cell, 2018, 173, 946–957 CrossRef CAS PubMed .e16.
- K. Shrinivas, B. R. Sabari, E. L. Coffey, I. A. Klein, A. Boija, A. V. Zamudio, J. Schuijers, N. M. Hannett, P. A. Sharp, R. A. Young and A. K. Chakraborty, Mol. Cell, 2019, 75, 549–561 CrossRef CAS PubMed.
- D. Mateju, B. Eichenberger, F. Voigt, J. Eglinger, G. Roth and J. A. Chao, Cell, 2020, 183, 1801–1812 CrossRef CAS PubMed.
- X. Su, J. A. Ditlev, E. Hui, W. Xing, S. Banjade, J. Okrut, D. S. King, J. Taunton, M. K. Rosen and R. D. Vale, Science, 2016, 352, 595–599 CrossRef CAS PubMed.
- X. Chen, X. Wu, H. Wu and M. Zhang, Nat. Neurosci., 2020, 23, 301–310 CrossRef CAS PubMed.
- Y. Shin, J. Berry, N. Pannucci, M. P. Haataja, J. E. Toettcher and C. P. Brangwynne, Cell, 2017, 168, 159–171 CrossRef CAS PubMed.
- E. Gomes and J. Shorter, J. Biol. Chem., 2019, 294, 7115–7127 CrossRef CAS PubMed.
- C. P. Brangwynne, P. Tompa and R. V. Pappu, Nat. Phys., 2015, 11, 899–904 Search PubMed.
- K. Rhine, V. Vidaurre and S. Myong, Annu. Rev. Biophys., 2020, 49, 247–265 Search PubMed.
- J. R. Wheeler, T. Matheny, S. Jain, R. Abrisch and R. Parker, Elife, 2016, 5, e18413 Search PubMed.
- D. W. Sanders, N. Kedersha, D. S. W. Lee, A. R. Strom, V. Drake, J. A. Riback, D. Bracha, J. M. Eeftens, A. Iwanicki, A. Wang, M. T. Wei, G. Whitney, S. M. Lyons, P. Anderson, W. M. Jacobs, P. Ivanov and C. P. Brangwynne, Cell, 2020, 181, 306–324 CrossRef CAS PubMed.
- J. A. West, M. Mito, S. Kurosaka, T. Takumi, C. Tanegashima, T. Chujo, K. Yanaka, R. E. Kingston, T. Hirose, C. Bond, A. Fox and S. Nakagawa, J. Cell Biol., 2016, 214, 817–830 CrossRef CAS PubMed.
- M. Feric, N. Vaidya, T. S. Harmon, D. M. Mitrea, L. Zhu, T. M. Richardson, R. W. Kriwacki, R. V. Pappu and C. P. Brangwynne, Cell, 2016, 165, 1686–1697 CrossRef CAS PubMed.
- T. Rey, S. Zaganelli, E. Cuillery, E. Vartholomaiou, M. Croisier, J. C. Martinou and S. Manley, Nat. Cell Biol., 2020, 22, 1180–1186 CrossRef CAS PubMed.
- H. B. Schmidt and R. Rohatgi, Cell Rep., 2016, 16, 1228–1236 CrossRef CAS PubMed.
- C. R. Neil, S. P. Jeschonek, S. E. Cabral, L. C. O'Connell, E. A. Powrie, J. P. Otis, T. R. Wood and K. L. Mowry, Mol. Biol. Cell, 2021, 32, ar37 CrossRef CAS PubMed.
- M. Feric, N. Vaidya, T. S. Harmon, R. W. Kriwacki, R. V Pappu, C. P. Brangwynne, M. Feric, N. Vaidya, T. S. Harmon, D. M. Mitrea, L. Zhu, T. M. Richardson, R. W. Kriwacki, R. V Pappu and C. P. Brangwynne, Cell, 2016, 165, 1686–1697 CrossRef CAS PubMed.
- L. Zhu, T. M. Richardson, L. Wacheul, M. T. Wei, M. Feric, G. Whitney, D. L. J. Lafontaine and C. P. Brangwynne, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 17330–17335 CrossRef CAS PubMed.
- R.-W. Yao, G. Xu, Y. Wang, L. Shan, P.-F. Luan, Y. Wang, M. Wu, L.-Z. Yang, Y.-H. Xing, L. Yang and L.-L. Chen, Mol. Cell, 2019, 76, 767–783 CrossRef CAS PubMed.
- D. L. J. Lafontaine, J. A. Riback, R. Bascetin and C. P. Brangwynne, Nat. Rev. Mol. Cell Biol., 2021, 22, 165–182 CrossRef CAS PubMed.
- C. M. Fare, A. Villani, L. E. Drake and J. Shorter, Open Biol., 2022, 11, 210137 CrossRef.
- G. A. Mountain and C. D. Keating, Biomacromolecules, 2020, 21, 630–640 CrossRef CAS PubMed.
- T. Lu and E. Spruijt, J. Am. Chem. Soc., 2020, 142, 2905–2914 CrossRef CAS PubMed.
- N. G. Moreau, N. Martin, P. Gobbo, T. Y. D. Tang and S. Mann, Chem. Commun., 2020, 56, 12717–12720 RSC.
- R. S. Fisher and S. Elbaum-Garfinkle, Nat. Commun., 2020, 11, 4628 CrossRef CAS PubMed.
- T. Kaur, M. Raju, I. Alshareedah, R. B. Davis, D. A. Potoyan and P. R. Banerjee, Nat. Commun., 2021, 12, 872 CrossRef CAS PubMed.
- P. R. Banerjee, A. N. Milin, M. M. Moosa, P. L. Onuchic and A. A. Deniz, Angew. Chem., 2017, 129, 11512–11517 CrossRef.
- S. Boeynaems, A. S. Holehouse, V. Weinhardt, D. Kovacs, J. Van Lindt and C. Larabell, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 7889–7898 CrossRef CAS.
- Y. Jo and Y. Jung, Chem. Sci., 2020, 11, 1269–1275 RSC.
- Y. Chen, M. Yuan, Y. Zhang, S. Liu, X. Yang, K. Wang and J. Liu, Chem. Sci., 2020, 11, 8617–8625 RSC.
- A. F. Mason, N. A. Yewdall, P. L. W. Welzen, J. Shao, M. Van Stevendaal, J. C. M. V. Hest, D. S. Williams and L. K. E. A. Abdelmohsen, ACS Cent. Sci., 2019, 5, 1360–1365 CrossRef CAS PubMed.
- J. Liu, F. Zhorabek, X. Dai, J. Huang and Y. Chau, ACS Cent. Sci., 2022, 8, 493–500 CrossRef CAS PubMed.
- J. Liu, F. Zhorabek, T. Zhang, J. W. Y. Lam, B. Z. Tang and Y. Chau, Small, 2022, 18, 2201721 CrossRef CAS PubMed.
- C. He, S. Zheng, Y. Luo and B. Wang, Theranostics, 2018, 8, 237–255 CrossRef CAS PubMed.
- B. S. Schuster, G. L. Dignon, W. S. Tang, F. M. Kelley, A. K. Ranganath, C. N. Jahnke, A. G. Simpkins, R. M. Regy, D. A. Hammer, M. C. Good and J. Mittal, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 11421–11431 CrossRef CAS PubMed.
- F. Luo, X. Gui, H. Zhou, J. Gu, Y. Li, X. Liu, M. Zhao, D. Li, X. Li and C. Liu, Nat. Struct. Mol. Biol., 2018, 25, 341–346 CrossRef CAS PubMed.
- K. Gasior, J. Zhao, G. McLaughlin, M. G. Forest, A. S. Gladfelter and J. Newby, Phys. Rev. E, 2019, 99, 1–11 CrossRef PubMed.
- E. M. Langdon, Y. Qiu, A. Ghanbari Niaki, G. A. McLaughlin, C. A. Weidmann, T. M. Gerbich, J. A. Smith, J. M. Crutchley, C. M. Termini, K. M. Weeks, S. Myong and A. S. Gladfelter, Science, 2018, 360, 922–927 CrossRef CAS PubMed.
- I. Alshareedah, T. Kaur, J. Ngo, H. Seppala, L. A. D. Kounatse, W. Wang, M. M. Moosa and P. R. Banerjee, J. Am. Chem. Soc., 2019, 141, 14593–14602 CrossRef CAS PubMed.
- I. Sanchez-Burgos, J. R. Espinosa, J. A. Joseph and R. Collepardo-Guevara, Biomolecules, 2021, 11, 1–19 CrossRef PubMed.
- I. Alshareedah, M. M. Moosa, M. Pham, D. A. Potoyan and P. R. Banerjee, Nat. Commun., 2021, 12, 6620 CrossRef CAS.
- L. Jawerth, E. Fischer-Friedrich, S. Saha, J. Wang, T. Franzmann, X. Zhang, J. Sachweh, M. Ruer, M. Ijavi, S. Saha, J. Mahamid, A. A. Hyman and F. Jülicher, Science, 2020, 370, 1317–1323 CrossRef CAS PubMed.
- C. P. Brangwynne, T. J. Mitchison and A. A. Hyman, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 4334–4339 CrossRef CAS PubMed.
- S. F. Banani, A. M. Rice, W. B. Peeples, Y. Lin, S. Jain, R. Parker and M. K. Rosen, Cell, 2016, 166, 651–663 CrossRef CAS PubMed.
- D. Song, Y. Jo, J. M. Choi and Y. Jung, Nat. Commun., 2020, 11, 1–13 CrossRef.
- M.-T. Wei, S. Elbaum-Garfinkle, A. S. Holehouse, C. C.-H. Chen, M. Feric, C. B. Arnold, R. D. Priestley, R. V Pappu and C. P. Brangwynne, Nat. Chem., 2017, 9, 1118–1125 CrossRef CAS PubMed.
- T. Y. D. Tang, D. Van Swaay, A. DeMello, J. L. Ross Anderson and S. Mann, Chem. Commun., 2015, 51, 11429–11432 RSC.
- X. Ge, D. Luo and J. Xu, PLoS One, 2011, 6, e28707 CrossRef CAS.
- D. Axelrod, D. E. Koppel, J. Schlessinger, E. Elson and W. W. Webb, Biophys. J., 1976, 16, 1055–1069 CrossRef CAS PubMed.
- M. Tassieri, G. M. Gibson, R. M. L. Evans, A. M. Yao, R. Warren, M. J. Padgett and J. M. Cooper, Phys. Rev. E, 2010, 81, 1–5 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2sc05438h |
| This journal is © The Royal Society of Chemistry 2023 |