Open Access Article
Open Access ArticleHaralick's texture analysis to predict cellular proliferation on randomly oriented electrospun nanomaterials†
Nora
Bloise
 ab,
Lorenzo
Fassina
ab,
Lorenzo
Fassina
 c,
Maria Letizia
Focarete
d,
Nadia
Lotti
c,
Maria Letizia
Focarete
d,
Nadia
Lotti
 ef and
Livia
Visai
ef and
Livia
Visai
 *ab
*ab
aDepartment of Molecular Medicine, Centre for Health Technologies (CHT), INSTM UdR of Pavia, University of Pavia, 27100 Pavia, Italy. E-mail: livia.visai@unipv.it
bMedicina Clinica-Specialistica, UOR5 Laboratorio di Nanotecnologie, ICS Maugeri, IRCCS, 27100 Pavia, Italy
cDepartment of Electrical, Computer and Biomedical Engineering, Centre for Health Technologies (CHT), University of Pavia, 27100 Pavia, Italy
dDepartment of Chemistry “Giacomo Ciamician”, INSTM UdR of Bologna, University of Bologna, 40126 Bologna, Italy
eCivil, Chemical, Environmental and Materials Engineering Department, University of Bologna, 40131 Bologna, Italy
fInterdepartmental Center for Industrial Research on Advanced Applications in Mechanical Engineering and Materials Technology, CIRI-MAM, University of Bologna, 40131 Bologna, Italy
First published on 16th February 2022
Abstract
Using a computer vision approach we have extracted the Haralick's texture features of randomly oriented electrospun nanomaterials in order to predict the proliferative behavior of cells which were subsequently seeded onto the nanosurfaces.
Introduction
New advances in cell biology, regenerative medicine and tissue engineering have been made possible by our understanding of how the extracellular matrix (ECM) or the biomaterial surfaces can control specific cell behaviors such as adhesion, growth, differentiation and migration. Work in this area, an interdisciplinary research field called nanobiotechnology (NBT), clearly shows that living cells have an extraordinary and intrinsic capacity to sense, integrate and respond to environmental signals at the micro- and nanoscale. Polymer surfaces, for example, with specific chemical modifications and with micro/nano topographic designs are able to affect the above mentioned cell behaviors and so determine whether a material has preliminary cytocompatibility and is useful for biological and medical purposes.1,2 In addition to chemical modification of the surface, topography characteristics such as roughness and surface nanoscopic pores can also impact cellular behavior.3–5 Topography can be evaluated via a computer vision approach where an imaging technique (e.g., scanning electron microscopy (SEM)) is followed by a texture analysis of the obtained images and in this field, in the past few years, classic 1st-order texture analysis of the image gray-levels (e.g., their mean, variance, skewness, and kurtosis) has evolved into a more sophisticated 2nd-order method. This approach also considers the relations between two or more pixels at a time, illuminating patterns and objects in an image as well as its topography.6 A successful way to perform a texture analysis of an image is via the so-called Haralick's features;7 these features are calculated from the gray-level co-occurrence matrix (GLCM), which is defined over an image and reveals the distribution of co-occurring pixel grayscale values. The GLCM and its Haralick features are used in many fields of texture analysis, for example in land-use and forest-type classification,8 pollen detection,9 fabric defect recognition,10 and plant leaf classification.11 Usage in a medical context includes skin texture,12 magnetic resonance imaging (MRI) images of the liver,13 X-ray mammography,14 breast cancer,15 brain cancer,16 tumor phenotype,17 and tumor classification.18 In the present work, from a tissue engineering viewpoint, we have extracted the Haralick's textural features of unseeded randomly oriented electrospun nanomaterials in order to predict the proliferative behavior of cells which were subsequently seeded onto the nanosurfaces. In other words, we have studied nanodetails of the biomaterial surface via a computer vision approach to assess the biological effects of the biomaterial nanoscale topography.Results
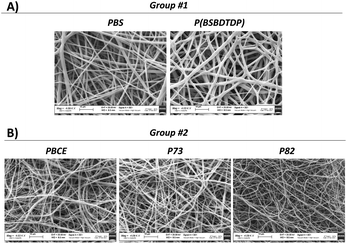
Fig. 1 shows the SEM pictures of both scaffold groups [group #1 and group #2] used for texture analysis. Both groups contain electrospun scaffolds made of uniform, bead-free, and randomly arranged fibres.In Table 1, two Haralick's features of the scaffolds are shown. In group #1, the comparison between the two materials was significant for both features (p < 0.05). In group #2, for contrast feature, the significant comparisons (p < 0.05) were PBCE vs. P73 and P73 vs. P82, whereas, for the variance feature, the significant comparison was between P73 and P82 (p < 0.05).
| Group #1 | Group #2 | ||||
|---|---|---|---|---|---|
| PBS | P(BSBDTDP) | PBCE | P73 | P82 | |
| Contrast | 0.0674 ± 0.0058 | 0.0848 ± 0.0080 | 0.0614 ± 0.0092 | 0.0805 ± 0.0126 | 0.0675 ± 0.0182 |
| Variance | 3.4383 ± 0.2259 | 4.3883 ± 0.4602 | 3.3133 ± 0.3389 | 3.8867 ± 0.6601 | 2.7150 ± 0.5477 |
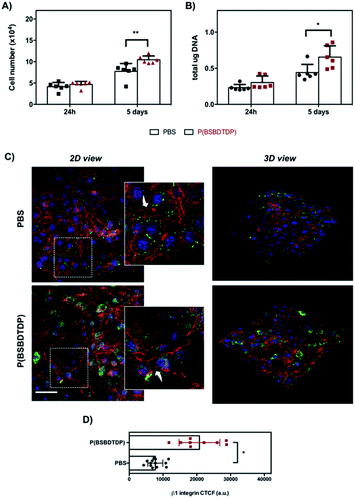
To assess the relevance of the parameters obtained by Haralick's features for the prediction of cell behavior, NIH-3T3 fibroblast proliferation was evaluated on both material groups and in terms of the cell number, DNA content, cytoskeleton organization and focal adhesion protein distribution. Interestingly, the preceding texture analysis appears to agree with the cell proliferation as measured via metabolic viability-based assay, DNA content and SEM observation for both tested groups (Fig. 2, 3 and S1†). In particular, for the materials of group #1, a higher level of both texture contrast and texture variance seems to be a good predictor of higher proliferation (Fig. 2A and B). Indeed, when compared to PBS, P(BSBDTDP) better supports cell proliferation over the culture time. Despite a similar number of cells at day 1, a higher cell number was observed on P(BSBDTDP) than on PBS (**p < 0.01) at day 5 of culture (Fig. 2A and ref. 19), and also a greater DNA content was found in the former nanomaterial at the same time point (Fig. 2B; *p < 0.05), thus suggesting the occurrence of higher cell proliferation compared to PBS. SEM images at day 5 supported the above data, showing a noticeable cellular colonization and a robust spreading of flattened-shape cells mainly covering P(BSBDTDP) surfaces (Fig. S1A† and ref. 19). Similar to what has been observed for proliferation, greater levels of both texture contrast and texture variance are suitable predictors of a different cell adhesion degree; although cells exhibited a mixed morphology of mainly round or elongated polygonal cells on both PBS and P(BSBDTDP), with F-actin (in red) polymerized in a dense meshwork of well-defined stress fibres (Fig. 2C), a strong signal of β1 integrin (the main transmembrane linker protein of focal adhesion,20 in green) was observed mostly on P(BSBDTDP), whereas the integrin β1 fluorescence intensity was significantly reduced in PBS (Fig. 2C and D; *p < 0.05), suggesting a different degree of cell adhesion process on this surface than P(BSBDTDP).
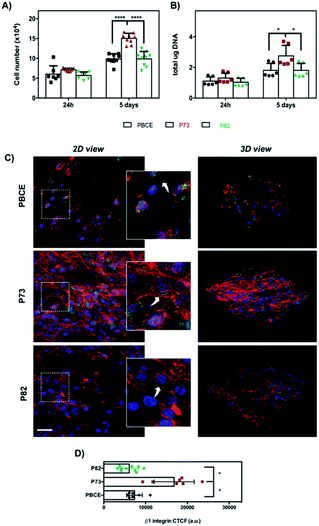
For the materials of group #2, although the statistical significance with p < 0.05 was not obtained for the variance feature, we were able to see that the trend of the Haralick's parameters was well reproduced in the trend of the cell proliferation (Fig. 3A and B). As can be seen in Fig. 3A, a significant difference of cell growth was observed at day 5 in P73 in comparison with PBCE and P82 (****p < 0.0001). Remarkably, both DNA measurement (Fig. 3B) and SEM analysis (Fig. S1B†) corroborated the above data. As was the case with group #1, Haralick's features appear in accordance with a higher cell adhesion and spreading on P73 associated with a highly organized F-actin filament meshwork and a significant enhancement of β1 integrin signal fluorescence than on P82 and PBCE surfaces (Fig. 3C and D). Indeed, in the latter materials, cells exhibited a less spread morphology that was coupled with F-actin stress fibre staining mostly confined to the peripheral areas of the cell (Fig. 3C, white arrows in the insets) and with lower expression of the β1 integrin intensity (Fig. 3C and D). Remarkably, this cellular behavior was consistent with the reduced cell proliferation observed on these surfaces at day 5 of culture compared to growth on the P73 nanomaterial (Fig. 3A and B).
Discussion
To properly design and develop functional and responsive biomaterials that successfully interact with tissue components, a deeper understanding of the behavior of cells at the biomaterial interface is very important.21,22 It is widely known that integrin receptors, a family of cell surface adhesion receptors, play a key role in cell–biomaterial interaction and cell mechanotransduction.22–25 The physical (e.g., topography and surface stiffness) and chemical properties of biomaterials can be sensed through focal adhesion sites (FAs, stable complexes composed of numerous proteins, including paxillin and vinculin, directly or indirectly associated with integrin clusters), which represent the main focus of cell–biomaterial interaction.20,26 At these adhesion sites, external cues can be transferred to the cytoskeleton and translated into biochemical and intracellular changes through the activation of integrin-related signaling pathways; these pathways regulate both rapid responses to substrate mechanics and determine the cell behavior and fate (such as proliferation and differentiation) by affecting the transcription of specific genes.24,27 In fact, the rearrangement of the actin cytoskeleton can also act as a direct mechanical link between integrins and the nuclear envelope, enabling and altering numerous downstream nuclear mechanoresponses.28 In addition, integrins can transmit signals from the inside of the cell to the outside (inside-out signaling) to regulate cell adhesion by increasing affinity to extracellular ligands.27 Undoubtedly, the nature of the biomaterials determines the activation of different integrin subunits and a critical level of cytoskeleton tension which is able to force specific cellular responses.29 Surface analysis of a biomaterial is, then, important for understanding and eventually predicting cell behavior on biomaterials. In this context, numerous analytical techniques provide different types of information about the surface of a sample. Surface analysis techniques, such as Auger electron spectroscopy (AES) and X-ray photoelectron spectroscopy (XPS), can be employed to obtain information on the chemical structure of a surface.30,31 Besides, it is of interest to note that different quantification methods, including time-of-flight secondary ion mass spectroscopy (ToF-SIMS), radiolabeling, and enzyme linked immunosorbent assay (ELISA), are currently used for the determination of surface ligand concentrations on biomaterials, such as hydrogel systems,32 providing important data to control or predict biomaterial–cell interaction. Scanning probe techniques, such as scanning electron microscopy and atomic force microscopy (AFM), have also been successfully applied to probe the structure of surfaces, including the measurements of the surface roughness.33,34 Currently, the emergence of nanotechnology has created new demands for surface techniques. In this growing context, a computer vision texture analysis via Haralick's features can contribute to our statistical understanding of the nanoscale world because these features are able to find out the relationships between nanoscale details at the nano-level, thus throwing light on patterns and objects in an image as well as on its nano-topography.6 In particular, the Haralick's features showed, in the biological context, their superiority in comparison to traditional texture analysis methods such as Gabor filters and Fourier transforms.35 This indicates that we can use Haralick's method to assess and eventually predict the behavior of cells when seeded onto a broad range of biomaterial surfaces. Finally, we hypothesize that other scanning techniques, such as FESEM or Cryo-SEM, could be associated with the analysis of Haralick's textures in the study of the surface of hydrogels; in this regard, since hydrogels can also have a complex inner pore structure, it might also be interesting to perform Haralick's analysis from slices obtained by tomographic techniques (e.g., SEM tomography), for example to predict the behaviour of 3D biological structures (such as the capillary growth within a biomaterial).Conclusions
We demonstrated that nanotopography-induced changes in the cell phenotype and proliferation can be predicted by Haralick's texture analysis of the materials. Overall, these results suggest that the Haralick's texture analysis method could help us to evaluate and understand cell behavior and responses to biomaterial surfaces. This conclusion has implications for tissue engineering and regenerative medicine.Experimental section
Preparation of randomly oriented electrospun nanomaterials
Poly(butylene succinate) (PBS) and poly(butylene succinate/dithiodipropionate) (P(BSBDTDP)) electrospun mats were fabricated (Fig. 1A) as previously described.19 An additional three random electrospun mats, named PBCE, P73 and P82, were obtained starting from the poly(butylene 1,4-cyclohexanedicarboxylate) homopolymer (PBCE), and two random copolymers made of butylenecyclohexanedicarboxylate (BCE) and triethylenecyclohexanedicarboxylate (TECE) co-units, as previously described.36 By varying the electrospinning conditions, randomly oriented electrospun PBCE (fibre dimension: 500 ± 190 nm), P73 (fibre dimension: 400 ± 140 nm) and P82 (fibre dimension: 580 ± 160 nm) were produced (Fig. 1B).Scanning electron microscopy of the scaffolds
The electrospun unseeded scaffolds were observed using a Zeiss EVO-MA10 scanning electron microscope (Carl Zeiss, Oberkochen, Germany) with an accelerating voltage of 20 kV on samples sputter-coated with gold, at 4000× magnification.Scanning electron microscopy of cell-cultured scaffolds
On day 5 of culture the cell-seeded scaffolds were fixed with 2.5% (v/v) glutaraldehyde solution in 0.1 M Na-cacodylate buffer (pH = 7.2) for 1 h at 4 °C, washed with Na-cacodylate buffer, dehydrated at room temperature in an ethanol gradient series up to 100% and then lyophilized for 4 h for complete dehydration. Scaffolds were then sputter coated with gold and observed with a Zeiss EVO-MA10 SEM (Carl Zeiss, Oberkochen, Germany) at 700× magnification. Cells seeded on plastic cell-culture coverslip disks (Thermanox Plastic, Nalge Nunc International, Rochester, NY) were used as the control.Texture analysis of SEM images
Via the calculations of the Haralick's features it is possible to evaluate the texture of a gray-level image and, therefore, to correlate the texture features with the observed biological parameters, namely cell proliferation in our study. For each SEM image of the materials without cells, we have selected at least two regions of interest (ROIs) to measure the gray-level co-occurrence matrix (GLCM). We then, for each GLCM, calculated two Haralick's features: the “contrast” and the “variance”.7 The contrast computes the amount of dissimilarity inside the GLCM and is a measure of the local variation in pixel values, whereas the variance has the same meaning as the statistical variance and is a measure of the pixel heterogeneity.Cell seeding and culture
The murine fibroblast cell line, NIH-3T3 (CRL1658), was obtained from the American Type Culture Collection (ATCC, Manassas, VA). NIH-3T3 cells were cultured in Dulbecco's modified Eagle medium (DMEM) with 4.5 g L−1 glucose (Thermo Scientific, Waltham, MA), supplemented with 10% bovine calf serum (Sigma-Aldrich, St. Louis, MO) and 1% (p/v) L-glutamine (Lonza Ltd, Basel, Switzerland). Cells were incubated at 37 °C with 5% CO2, routinely trypsinized after confluence, then counted, and seeded. Prior to cell seeding, scaffolds were cut into round pieces (2 cm2 area, about 80 μm thick), assembled in 24-well CellCrown supports (Scaffdex, Tampere, Finland) to avoid their floating in the cell culture medium, and inserted into the wells of a 24-well plate. Subsequently, they were sterilised with EtOH 96% for 15 min, then treated with EtOH 70% for 15 min, washed twice in phosphate-buffered saline (PBS) for 10 min, assembled in a 24-well plate, and incubated overnight in PBS containing 1% penicillin/streptomycin under UV light, as previously reported.19Metabolic viability-based assay
On day 0 a drop of cell suspension containing 5 × 104 cells was added onto the top of each scaffold and cell proliferation was assessed by using the quantitative 3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyl tetrazolium bromide-based assay (MTT; Sigma-Aldrich) according to the manufacturer's instructions and as previously described.19 Briefly, at each time, NIH-3T3-seeded scaffolds were transferred into a 24-well culture plate and the medium was replaced by 500 μL of DMEM, followed by the addition of 50 μL MTT (5 mg mL−1). The scaffolds were then incubated for 4 h at 37 °C in a humidified 5% CO2 incubator. Formazan crystals, formed by the interaction of the MTT solution with the live cells, were then dissolved in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) isopropanol – 0.04 M HCl. Aliquots of 100 μL were sampled and the absorbance was measured at 565 nm by using a microplate reader (BioRad Laboratories, Hercules, CA). A standard curve of cell viability was used to obtain the cell number per sample.
1 (v/v) isopropanol – 0.04 M HCl. Aliquots of 100 μL were sampled and the absorbance was measured at 565 nm by using a microplate reader (BioRad Laboratories, Hercules, CA). A standard curve of cell viability was used to obtain the cell number per sample.
DNA content
To obtain the total DNA content after 1 and 5 days of culture, samples were processed through three freeze/thaw cycles in sterile, deionized, distilled water. Between each freeze/thaw cycle, scaffolds were roughly vortexed. The released DNA content was measured with a fluorometric DNA quantification kit (PicoGreen; Molecular Probes, Eugene, OR), following the manufacturer's protocol and as previously described.37 The amount of DNA was expressed as the weight of DNA per scaffold (μg per scaffold).Confocal laser scanning microscopy
After 24 h of culture, cell-seeded scaffolds were washed with PBS, fixed with 4% (w/v) paraformaldehyde solution (PFA) for 30 min at 4 °C, and then permeabilized with 0.1% Triton X-100 for 5 min. To visualize the F-actin cytoskeleton organization, cells were stained with tetramethylrhodamine B isothiocyanate (TRITC) phalloidin conjugate solution (10 μg mL−1, EX/EM maxima ∼540/575, Sigma-Aldrich) in PBS for 40 min at RT. For focal adhesion detection, cells were incubated with primary mouse anti-β1 integrin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in 1% BSA, NSJ Bioreagents, San Diego, CA). Afterwards, samples were incubated with specific secondary antibodies for immunofluorescence, all used at a concentration of 1
100 in 1% BSA, NSJ Bioreagents, San Diego, CA). Afterwards, samples were incubated with specific secondary antibodies for immunofluorescence, all used at a concentration of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 in 1% BSA. Hoechst 33342 (2 μg mL−1) was used for nuclei staining. The images were taken using a TCS SP8 confocal microscope (Leica Microsystems, Bensheim, Germany) equipped with a digital image capture system at 40× magnification. 3D views of images were also taken. Additionally, the intensity of the β1-integrin fluorescence was calculated and quantified as corrected total cell fluorescence (CTCF) with the formula CTCF = integrated density − (area of selected cell × mean fluorescence of background readings) using image analysis software (Fiji Is Just ImageJ, version: 2.0.0-rc-69/1.52p), according to previous studies.38
500 in 1% BSA. Hoechst 33342 (2 μg mL−1) was used for nuclei staining. The images were taken using a TCS SP8 confocal microscope (Leica Microsystems, Bensheim, Germany) equipped with a digital image capture system at 40× magnification. 3D views of images were also taken. Additionally, the intensity of the β1-integrin fluorescence was calculated and quantified as corrected total cell fluorescence (CTCF) with the formula CTCF = integrated density − (area of selected cell × mean fluorescence of background readings) using image analysis software (Fiji Is Just ImageJ, version: 2.0.0-rc-69/1.52p), according to previous studies.38
Statistical analysis
All calculations were performed using GraphPad Prism 6.0 (GraphPad Inc., San Diego, CA). The results were expressed as mean ± standard deviation. The different experimental groups were compared using Student's unpaired t-test or one-way variance analysis (ANOVA), followed by a post hoc Bonferroni test for multiple comparisons (significance level of 0.05).Author contributions
LV, LF, and NB conceived the study, were responsible for the correctness of analyses, and contributed to the writing and editing of the manuscript. NL and MLF were responsible for the electrospun material production and participated in the writing of this manuscript. All authors approved the final manuscript.Conflicts of interest
The authors declare that there are not conflicting interests.Acknowledgements
The authors are grateful to G. Quattrone for the texture analysis of SEM images. N. Bloise and L. Visai acknowledge the grant of the Italian Ministry of Education, University and Research (MIUR) to the Department of Molecular Medicine (DMM) of the University of Pavia under the initiative “Dipartimenti di Eccellenza (2018–2022)”. The authors wish to thank P. Vaghi (Centro Grandi Strumenti, University of Pavia, Italy, https://cgs.unipv.it/) for technical assistance in the CLSM studies and G. Bruni, Department of Chemistry, University of Pavia, Italy, for SEM investigation support. A special thanks to S. Burgess (University of Pavia, Italy) for correcting the English text of the manuscript.References
- N. M. Alves, I. Pashkuleva, R. L. Reis and J. F. Mano, Small, 2010, 6, 2208–2220 CrossRef CAS PubMed.
- B. Majhy, P. Priyadarshini and A. K. Sen, RSC Adv., 2021, 11, 15467–15476 RSC.
- P. Decuzzi and M. Ferrari, Biomaterials, 2010, 31, 173–179 CrossRef CAS PubMed.
- F. Gentile, L. Tirinato, E. Battista, F. Causa, C. Liberale, E. M. di Fabrizio and P. Decuzzi, Biomaterials, 2010, 31, 7205–7212 CrossRef CAS PubMed.
- F. Gentile, R. La Rocca, G. Marinaro, A. Nicastri, A. Toma, F. Paonessa, G. Cojoc, C. Liberale, F. Benfenati, E. di Fabrizio and P. Decuzzi, ACS Appl. Mater. Interfaces, 2012, 4, 2903–2911 CrossRef CAS PubMed.
- T. Löfstedt, P. Brynolfsson, T. Asklund, T. Nyholm and A. Garpebring, PLoS One, 2019, 14, e0212110 CrossRef PubMed.
- R. M. Haralick, K. Shanmugam and I. Dinstein, IEEE Transactions on Systems, Man, and Cybernetics, 1973, SMC-3, 610–621 Search PubMed.
- F. T. Ulaby, F. Kouyate, B. Brisco and T. H. L. Williams, IEEE Trans. Geosci. Electron., 1986, GE-24, 235–245 Search PubMed.
- J. V. Marcos, R. Nava, G. Cristóbal, R. Redondo, B. Escalante-Ramírez, G. Bueno, Ó. Déniz, A. González-Porto, C. Pardo, F. Chung and T. Rodríguez, Micron, 2015, 68, 36–46 CrossRef PubMed.
- J. Raheja, S. Kumar and A. Chaudhary, Optik, 2013, 124, 6469–6474 CrossRef.
- B. VijayaLakshmi and V. Mohan, Comput. Electron. Agric., 2016, 125, 99–112 CrossRef.
- X. Ou, W. Pan and P. Xiao, Int. J. Pharm., 2014, 460, 28–32 CrossRef CAS PubMed.
- M. E. Mayerhoefer, W. Schima, S. Trattnig, K. Pinker, V. Berger-Kulemann and A. Ba-Ssalamah, J. Magn. Reson. Imaging, 2010, 32, 352–359 CrossRef PubMed.
- H. Li, M. L. Giger, L. Lan, J. Bancroft Brown, A. MacMahon, M. Mussman, O. I. Olopade and C. Sennett, J. Digit. Imaging, 2012, 25, 591–598 CrossRef PubMed.
- K. Nie, J.-H. Chen, H. J. Yu, Y. Chu, O. Nalcioglu and M.-Y. Su, Acad. Radiol., 2008, 15, 1513–1525 CrossRef PubMed.
- P. Brynolfsson, D. Nilsson, R. Henriksson, J. Hauksson, M. Karlsson, A. Garpebring, R. Birgander, J. Trygg, T. Nyholm and T. Asklund, Med. Phys., 2014, 41, 101903 CrossRef PubMed.
- H. J. W. L. Aerts, E. R. Velazquez, R. T. H. Leijenaar, C. Parmar, P. Grossmann, S. Carvalho, S. Cavalho, J. Bussink, R. Monshouwer, B. Haibe-Kains, D. Rietveld, F. Hoebers, M. M. Rietbergen, C. R. Leemans, A. Dekker, J. Quackenbush, R. J. Gillies and P. Lambin, Nat. Commun., 2014, 5, 4006 CrossRef CAS PubMed.
- H. Cho and H. Park, 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), DOI: DOI:10.1109/EMBC.2017.8037508.
- G. Guidotti, M. Soccio, M. Gazzano, N. Bloise, G. Bruni, A. Aluigi, L. Visai, A. Munari and N. Lotti, Polym. Degrad. Stab., 2020, 182, 109403 CrossRef CAS.
- P. Tomakidi, S. Schulz, S. Proksch, W. Weber and T. Steinberg, Cell Tissue Res., 2014, 357, 515–526 CrossRef CAS PubMed.
- W. Y. Tong, Y. M. Liang, V. Tam, H. K. Yip, Y. T. Kao, K. M. C. Cheung, K. W. K. Yeung and Y. W. Lam, Mol. Cell. Proteomics, 2010, 9, 2089–2098 CrossRef CAS PubMed.
- E. Barcelona-Estaje, M. J. Dalby, M. Cantini and M. Salmeron-Sanchez, Adv. Healthcare Mater., 2021, 10, e2002048 CrossRef PubMed.
- H. Amani, H. Arzaghi, M. Bayandori, A. S. Dezfuli, H. Pazoki-Toroudi, A. Shafiee and L. Moradi, Adv. Mater. Interfaces, 2019, 6, 1900572 CrossRef.
- B. Özkale, M. S. Sakar and D. J. Mooney, Biomaterials, 2021, 267, 120497 CrossRef PubMed.
- R. J. McMurray, M. J. Dalby and P. M. Tsimbouri, J. Tissue Eng. Regener. Med., 2015, 9, 528–539 CrossRef PubMed.
- B. K. K. Teo, S. T. Wong, C. K. Lim, T. Y. S. Kung, C. H. Yap, Y. Ramagopal, L. H. Romer and E. K. F. Yim, ACS Nano, 2013, 7, 4785–4798 CrossRef CAS PubMed.
- D. Mohammed, M. Versaevel, C. Bruyère, L. Alaimo, M. Luciano, E. Vercruysse, A. Procès and S. Gabriele, Front. Bioeng. Biotechnol., 2019, 162 DOI:10.3389/fbioe.2019.00162.
- A. Isomursu, M. Lerche, M. E. Taskinen, J. Ivaska and E. Peuhu, Exp. Cell Res., 2019, 378, 217–225 CrossRef CAS PubMed.
- R. Harjumäki, X. Zhang, R. W. N. Nugroho, M. Farooq, Y.-R. Lou, M. Yliperttula, J. J. Valle-Delgado and M. Österberg, ACS Appl. Bio Mater., 2020, 3, 1406–1417 CrossRef PubMed.
- Y. Alqaheem and A. A. Alomair, Membranes, 2020, 10, E33 CrossRef PubMed.
- T. Yoneyama and T. Hanawa, J. Oral Sci., 2020, 63, 50–53 CrossRef PubMed.
- M. V. Beer, K. Hahn, S. Diederichs, M. Fabry, S. Singh, S. J. Spencer, J. Salber, M. Möller, A. G. Shard and J. Groll, Biointerphases, 2015, 10, 021007 CrossRef PubMed.
- F. Variola, Phys. Chem. Chem. Phys., 2015, 17, 2950–2959 RSC.
- J. Rydz, A. Šišková and A. Andicsová Eckstein, Adv. Mater. Sci. Eng., 2019, 2019, e6871785 Search PubMed.
- M. D. Sutcliffe, P. M. Tan, A. Fernandez-Perez, Y.-J. Nam, N. V. Munshi and J. J. Saucerman, Sci. Rep., 2018, 8, 1258 CrossRef PubMed.
- N. Bloise, E. Berardi, C. Gualandi, E. Zaghi, M. Gigli, R. Duelen, G. Ceccarelli, E. E. Cortesi, D. Costamagna, G. Bruni, N. Lotti, M. L. Focarete, L. Visai and M. Sampaolesi, Int. J. Mol. Sci., 2018, 19, E3212 CrossRef PubMed.
- N. Bloise, A. Patrucco, G. Bruni, G. Montagna, R. Caringella, L. Fassina, C. Tonin and L. Visai, Materials, 2020, 13, E3052 CrossRef PubMed.
- T. Topal, X. Hong, X. Xue, Z. Fan, N. Kanetkar, J. T. Nguyen, J. Fu, C. X. Deng and P. H. Krebsbach, Sci. Rep., 2018, 8, 12977 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: SEM analysis of cell morphology. See DOI: 10.1039/d1na00890k |
| This journal is © The Royal Society of Chemistry 2022 |