Open Access Article
Open Access ArticleDecolorization and degradation analysis of Disperse Red 3B by a consortium of the fungus Aspergillus sp. XJ-2 and the microalgae Chlorella sorokiniana XJK
Weihua Tanga,
Xiaolin Xu *a,
Bang-Ce Ye
*a,
Bang-Ce Ye b,
Peng Caoa and
Asghar Alia
b,
Peng Caoa and
Asghar Alia
aKey Laboratory for Green Process of Chemical Engineering of Xinjiang Bingtuan, School of Chemistry and Chemical Engineering, Shihezi University, Shihezi, 832003, People's Republic of China. E-mail: xxl_food@shzu.edu.cn
bSchool of Biological Engineering, East China University of Science and Technology, Shanghai, 200237, People's Republic of China
First published on 9th May 2019
Abstract
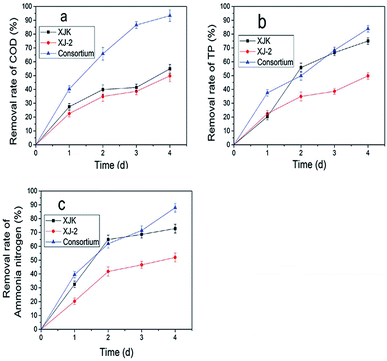
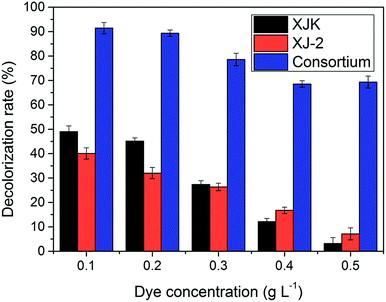
Disperse Red 3B, an anthraquinone dye, was decolorized by a consortium, which was constituted of the fungus (Aspergillus sp. XJ-2) and the microalgae (Chlorella sorokiniana XJK). The consortium performed better than the single system in terms of decolorization and nutrient removal simultaneously in the simulated wastewater of Dispersed Red 3B. The decolorization rate could reach 98.09% by the consortium under the optimized conditions. The removal rate of COD (Chemical Oxygen Demand), TP (Total Phosphorus), and ammonia nitrogen reached 93.9%, 83.9% and 87.6%. Also, the consortium could tolerate higher salt and dye concentration than the single system did. In this co-cultural system, the lignin peroxidase and manganese peroxidase enzyme activities contributed to the degradation of Disperse Red 3B, which reached 86.7 U L−1 and 122.5 U L−1. The result of fermentation liquid analysis with UV-vis, FTIR and GC-MS showed that the colored functional group of the dye was broken and the Dispersed Red 3B was degraded into small molecular compounds with low toxicity. It was suggested that degradation plays a major role during the color removal process. The consortium exhibited greater potential in terms of color removal and water pollutant removal than the separate system did.
1. Introduction
From different industries such as textile, paper, photography, comic, leather, the discharge of effluent containing synthetic dyes could cause serious pollution to the environment and ecology, because the complex aromatic molecular structures of these dyes are difficult to degrade.1 Although many physicochemical processes including adsorption, advanced oxidation, photocatalysis etc. are effective to decolorize dye-containing wastewater, high cost and the formation of secondary pollutants restrict the application of these procedures. Comparatively, biological treatments with microorganisms are relatively cost-effective and eco-friendly. However, traditional activated sludge could not degrade these synthetic dyes adequately, for example aromatic amines in the products can inhibit the activity of bacteria. White-rot fungi have been then intensively investigated in recent years which show strong adaptability and efficiency in the removal of certain dyes for the production of ligninolytic enzymes, including laccase, manganese peroxidase and lignin peroxidase.2 For example, 88% of Congo red was decolorized by Trametes pubescens Cui 7571 cultured for 48 hours, also peak laccase activity (15.783 U mL−1), peak LiP (9.832 U mL−1) and MnP activity (8.647 U mL−1) were examined during the decolorization, respectively.3 Similarity, laccase from Polyporus sp. S133 was found to be one powerful tool for the decolorization of anthraquinone dyes.4 It was demonstrated that Crude MnP from Irpex lacteus F17 could decolorize Malachite green, and its metabolites were appreciably less toxic than the parent compound.5 However, these fungal species have been only demonstrated in the degradation of particular dye. In fact, effluents from textile industry are a mixture of various organic and inorganic contaminants with high COD and BOD5 (Biochemical Oxygen Demand).6 The use of a microbial consortium could be useful to the bioremediation applications as a rich enzyme network and can be applied for the biodegradation of co-contaminated matrices.It has been shown that the fungi co-culture system could greatly improve the effectivity of decolorization. Copete-Pertuz et al. reported the 1.2 times increasing of RB5 removal as the combination culture of T. viride and A. terreus, compared to monoculture. Also, the co-culture of fungi Pleurotus florida and Rhizoctonia solani resulted 98.54% of dye decolorization ratio.7,8
For the effective utilization of nitrogen, phosphorus, and other inorganic matters through the photosynthesis, microalgae were important constituent in many co-culture systems for the removal of contaminants from wastewater.9 Mujtaba and Lee found that co-culture system of Chlorella vulgaris and activated sludge considerably eliminated 98–100% nitrogen, 92–100% phosphorus, and 94–96% COD from artificial municipal wastewater.10 Consortium of the fungus Ganoderma lucidum and C. vulgaris removed COD (84.61%), total nitrogen (80.41%), and total phosphorus (92.21%) respectively treating anaerobically digested swine wastewater.11 Furthermore, filamentous fungi had a strong flocculation effect on microalgae due to their large surface area and easy solid–liquid separation, which was convenient for the recycling of microalgae.12 Ndikubwimana et al. reported a yield of 95% for microalgae with the formation of cell pellets,13 and Prajapati et al. found A. lentulus resulting in 98% harvesting at low glucose level (5.0 g L−1) within 52 h.14 In terms of economic costs, effective wastewater treatment performance and the good sources of biofuels, the co-culture system of fungi and microalgae had great potential in wastewater treatment. Generally both fungi and microalgae could decolorize dyes by the adsorption or degradation. The decolorization rate of Reactive Black 5 by T. versicolor strains MUM 94.04 reached 100% by adsorption and degradation.15 The decolorization rate reached 97.1% by Spirulina platensis adsorbed simulated industrial textile effluents.16 However, the functions of each microbe in consortium are not clear during the wastewater treatment.
Aspergillus sp. is one kind of fungus with better colour removal effect on dyes. Kang et al. found Aspergillus sp. TS-A degraded 98.6% of Mordant Yellow 1.17 And Chlorella sorokiniana is one kind of microalgae with better effect on wastewater treatment. Park et al. found total nitrogen, phosphorous and glucose removal rates were 10.5 mg L−1 d−1, 2 mg L−1 d−1, 1000 mg L−1 respectively by Chlorella sorokiniana.18 In this work, Aspergillus sp. XJ-2 (CGMCC12963) and Chlorella sorokiniana XJK were chosen to the construction of consortium, both of them possessing better decolorization ability on anthraquinone dyes.19,20 Pan et al. found Aspergillus sp. XJ-2 could efficiently decolorize various anthraquinone dyes.19 Xie et al. found the decolorization rate of anthraquinone dye Disperse Blue 2BLN was 83% by Chlorella sorokiniana XJK.20 In this work the consortium was used to decolorize Disperse Red 3B (one anthraquinone dye) and remove nutrients from simulated wastewater. The degradation, adsorption capacity, key enzymes responsible for degradation was analyzed, and the performance of each microbe was compared. Degradation products of Disperse Red 3B were examined by UV-vis, FTIR and GC-MS, and degradation pathways were proposed.
2. Materials and methods
2.1 Dye and microorganisms
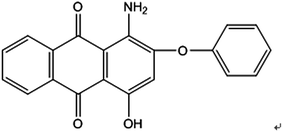
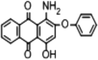
Disperse Red 3B (1-amino-4-hydroxyl-2-phenoxyanthracene-9,10-dione), one of anthraquinone dyes, was purchased from Shanghai Research and Development Biological Technology Limited Company. The structure of Disperse Red 3B is shown in Fig. 1.Chlorella sorokiniana XJK (XJK) was a freshwater oily microalgae isolated from Keketuohai, Xinjiang Provence, China. Aspergillus sp. XJ-2 (XJ-2) was isolated from activated sludge of a textile factory in Xinjiang Provence, China. Aspergillus sp. XJ-2 and Chlorella sorokiniana XJK are conserved in Key Laboratory of Chemical Green Process of Xinjiang Corps, Chemistry and Chemical Engineering College, Shihezi University.
2.2 Culture mediums and conditions
The medium for Chlorella sorokiniana XJK was modified BG11 medium.21 The modified BG11 medium was formulated as follows: 5 g L−1 glucose, 0.83 g L−1 NaNO3, 0.12 g L−1 MgSO4·7H2O, 0.09 g L−1 K2HPO4·3H2O, 0.03 g L−1 CaCl2, 0.02 g L−1 NaCO3, 0.01 g L−1 citric-acid, 0.01 g L−1 Fe(NH4)3(C6H5O7)2, 0.001 g L−1 Na2EDTA·2H2O, 0.2 mL L−1 trace metal mix (2.86 g L−1 H3BO3, 1.86 g L−1 MnCl2·4H2O, 0.39 g L−1 NaMoO4·2H2O, 0.22 g L−1 ZnSO4·7H2O, 0.08 g L−1 CuSO4·5H2O, 0.05 g L−1 Co(NO3)2·6H2O). XJK was cultivated grown up under light intensity 830 mol m−2 s−1 and temperature 30 °C for seven days.22 The medium of Aspergillus sp. XJ-2 was Czapek's medium.19 XJ-2 was cultivated at 30 °C and rotated in a shaker at 170 rpm for 30 h.232.3 Simulated wastewater treatment
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.
1.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1
2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, 2
3, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3
1, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1.5 × 106 cell per mL), inoculation time of Aspergillus sp. XJ-2 (0, 6, 12 and 24 h), incubation temperature (20, 25, 30, 35, 40 °C), pH of simulated wastewater (4, 5, 6, 7, 8) and rotation speed (140, 150, 160, 170, 180 rpm) on the simulated wastewater were investigated. The optimized index was the decolorization rate of simulated wastewater. The determination of decolorization rate was referenced to Kang et al.17 (final absorbance of Disperse Red 3B at 590 nm).
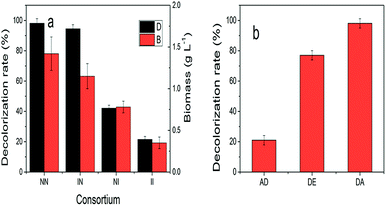
1 1.5 × 106 cell per mL), inoculation time of Aspergillus sp. XJ-2 (0, 6, 12 and 24 h), incubation temperature (20, 25, 30, 35, 40 °C), pH of simulated wastewater (4, 5, 6, 7, 8) and rotation speed (140, 150, 160, 170, 180 rpm) on the simulated wastewater were investigated. The optimized index was the decolorization rate of simulated wastewater. The determination of decolorization rate was referenced to Kang et al.17 (final absorbance of Disperse Red 3B at 590 nm).| DE = DA − AD | (1) |
DE is the degradation capacity of the consortium. DA (the decolorizing capacity of the consortium) was defined as simulated decolorization rate of wastewater when the consortium of XJ-2 and XJK. AD (the adsorption capacity of the consortium) was defined as simulated decolorization rate of wastewater when the consortium of XJ-2 (inactivated) and XJK (inactivated).
2.4 Analyses
UV-vis analysis (Spectrum lab S22pc, China): decolorization rate was monitored by scanning the spectrum between 400 and 800 nm using a Spectrum lab S22pc UV-vis spectrophotometer. Disperse Red 3B was used as the control.24
FTIR analysis (Magna-IR 750, Thermo Nicolet): a Magna-IR 750 FTIR spectrometer was used for the FTIR analysis of the extracted metabolites in the mid-IR region of 4000–400 cm−1 at a scan speed of 16; Disperse Red 3B was used as the control. The resolution and scan number were set at 8.0 cm−1 and 16 times, respectively.25
GC/MS analysis (Agilent Technologies Inc, USA): GC system (model 7890A, Agilent) and HP-5MS capillary column (30 × 0.32 × 0.25) combined with MS system (model 5975 C, Agilent) constituted the GC-MS system. Carrier gas was helium with carrier flow rate 0.8 mL min−1.17 The initial temperature was 60 °C for 1 min, which was increased to 190 °C with 10 °C min−1. Then the temperature was increased to 235 °C with 3 °C min−1. Finally the temperature was increased to 280 °C with 10 °C min−1 and was maintained for 5 min. Injection port temperature was 250 °C and detector temperature was 280 °C.26
3. Results and discussion
3.1 Simulated wastewater treatment
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
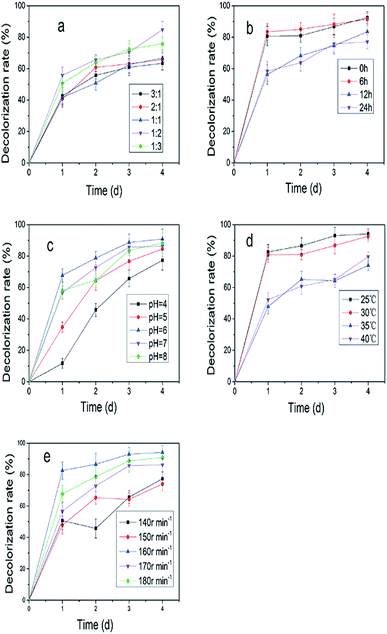
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (Fig. 2a). Fig. 2b showed the decolorization rate was improved to 92.5% on the fourth day when XJ-2 was inoculated at 0 h. Temperature and pH had great influence on the decolorization of simulated wastewater (Fig. 2c and d), which were 25 °C and 6. Higher rotation speed of shaker could also increase the decolorization rate of simulated wastewater (Fig. 2e). The highest decolorization rate reached 98.09% when the rotation speed of shaker was 160 rpm.
2 (Fig. 2a). Fig. 2b showed the decolorization rate was improved to 92.5% on the fourth day when XJ-2 was inoculated at 0 h. Temperature and pH had great influence on the decolorization of simulated wastewater (Fig. 2c and d), which were 25 °C and 6. Higher rotation speed of shaker could also increase the decolorization rate of simulated wastewater (Fig. 2e). The highest decolorization rate reached 98.09% when the rotation speed of shaker was 160 rpm.
According to the above conditions, the consortium of Aspergillus sp. XJ-2 and Chlorella sorokiniana XJK was constructed. The optimal conditions for the co-culture system were shown below: the inoculum ratio of Aspergillus sp. XJ-2 and Chlorella sorokiniana XJK was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, the inoculation time of XJ-2 was 0 h, pH was 6, the temperature was 25 °C and the rotation speed of shaker was 160 rpm.
2, the inoculation time of XJ-2 was 0 h, pH was 6, the temperature was 25 °C and the rotation speed of shaker was 160 rpm.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
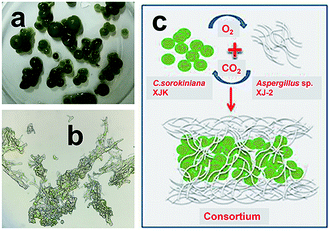
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. Fig. 3a showed fungi and microalgae formed sphere structure. Microstructure of the consortium showed microalgae were closely intertwined with fungi by fungal hyphae (Fig. 3b). Simulation diagram of the consortium formation was described in Fig. 3c. The compact fungal-microalgae ball structure was formed by encapsulating microalgae inside the fungus by fungal hyphae.27 The CO2 produced by the fungi through aerobic respiration can provide nutrients for the growth of microalgae, while the O2 produced by microalgae through photosynthesis can provide nutrients for the growth of fungi.28 This compact ball structure may facilitate the collection of microalgae.14,15
2. Fig. 3a showed fungi and microalgae formed sphere structure. Microstructure of the consortium showed microalgae were closely intertwined with fungi by fungal hyphae (Fig. 3b). Simulation diagram of the consortium formation was described in Fig. 3c. The compact fungal-microalgae ball structure was formed by encapsulating microalgae inside the fungus by fungal hyphae.27 The CO2 produced by the fungi through aerobic respiration can provide nutrients for the growth of microalgae, while the O2 produced by microalgae through photosynthesis can provide nutrients for the growth of fungi.28 This compact ball structure may facilitate the collection of microalgae.14,15
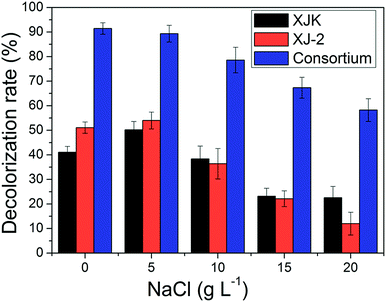 | ||
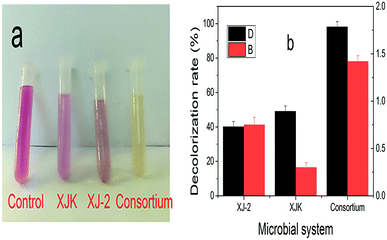
| Fig. 7 Decolorization rate of simulated wastewater under different NaCl concentrations by single Aspergillus sp. XJ-2, single Chlorella sorokiniana XJK and the consortium. | ||
According to these results, the consortium of Aspergillus sp. XJ-2 and Chlorella sorokiniana XJK could have a great advantage over the single system in the treatment of simulated wastewater.
3.2 Degradation pathway analysis
| Enzyme activity (U L−1) | Time (d) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||||||
| E | Er | E | Er | E | Er | E | Er | ||
| a In: intracellular; Ex: extracellular; E: mean; Er: standard deviation. | |||||||||
| Lac | In | 1.4 | 0.1 | 4.3 | 0.3 | 5.7 | 0.4 | 5.9 | 0.3 |
| Ex | 4.3 | 0.3 | 5.7 | 0.4 | 7.2 | 0.7 | 17.2 | 1 | |
| Mnp | In | 9.5 | 0.3 | 50.9 | 1.1 | 69.9 | 0.6 | 71.9 | 1.3 |
| Ex | 41.3 | 2.1 | 80.9 | 2.4 | 107.2 | 3.2 | 122.5 | 4.2 | |
| Lip | In | 6.7 | 0.2 | 8.9 | 0.3 | 20 | 0.4 | 40 | 0.4 |
| Ex | 6.7 | 0.1 | 28.9 | 0.4 | 60.7 | 0.9 | 86.7 | 1.1 | |
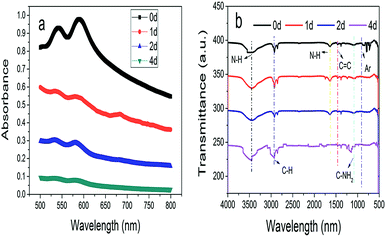
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C
O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C. Moreover, it could be understood that many new peaks were formed on the fourth day. Therefore, it could be inferred that the Dispersed Red 3B was degraded by the optimized consortium to form some small molecule compound.
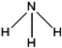
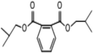
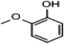
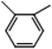
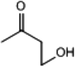
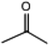
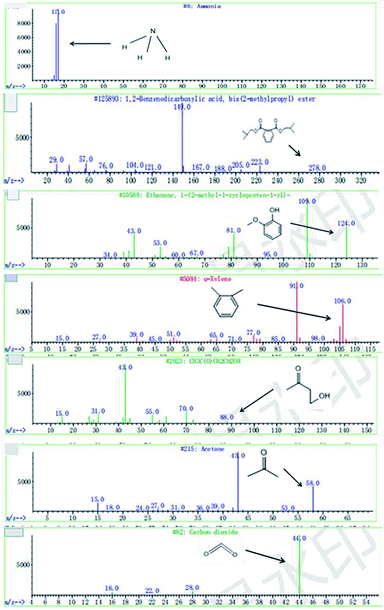
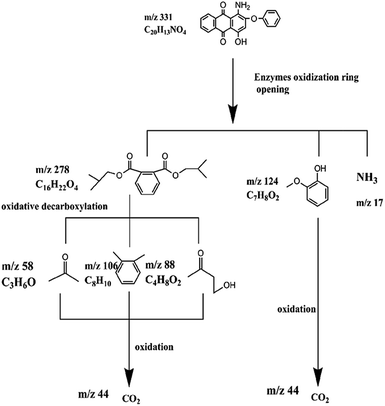
C. Moreover, it could be understood that many new peaks were formed on the fourth day. Therefore, it could be inferred that the Dispersed Red 3B was degraded by the optimized consortium to form some small molecule compound.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C double bond.5 While Disperse Blue 2BLN was degradation, Xie and Pan et al. suggested that the weaker the bonding sequence was, the more easily the bond was destroyed.19,20 The presumed degradation pathway was shown in Fig. 11. First, Disperse Red 3B was degraded into NH3, diisobutyl phthalate and guaiacol due to the two hetero cyclic were broken in the middle. Then, diisobutyl phthalate was degraded into o-xylene, acetone and 4-hydroxy-2-butanone by oxidative decarboxylation. Finally, all intermediate reaction products were degraded into CO2 and other final products.
C double bond.5 While Disperse Blue 2BLN was degradation, Xie and Pan et al. suggested that the weaker the bonding sequence was, the more easily the bond was destroyed.19,20 The presumed degradation pathway was shown in Fig. 11. First, Disperse Red 3B was degraded into NH3, diisobutyl phthalate and guaiacol due to the two hetero cyclic were broken in the middle. Then, diisobutyl phthalate was degraded into o-xylene, acetone and 4-hydroxy-2-butanone by oxidative decarboxylation. Finally, all intermediate reaction products were degraded into CO2 and other final products.
4. Conclusions
In this work, the consortium of the fungus Aspergillus sp. XJ-2 and the microalgae Chlorella sorokiniana XJK was constructed. The consortium exhibited stronger efficiency in terms of decolorization, COD removal and nutrients removal than the single system did. The consortium could adapt to wastewater with higher dye and higher salt concentration. The result of fermentation liquid analysis showed that Dispersed Red 3B eventually was mineralized. The consortium existed great potential to adapt to wastewater of different types and sources.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded by the National Natural Science Foundation of China (grant number 21466032). And this work was also partially supported by Scientific Research Foundation for Changjiang Scholars of Shihezi University (grant number CJXZ201501).References
- M. C. Tomei, D. M. Angelucci and A. J. Daugulis, Sci. Total Environ., 2016, 573, 585–593 CrossRef CAS PubMed.
- S. Popli and U. D. Patel, Int. J. Environ. Sci. Technol., 2015, 12, 405–420 CrossRef CAS.
- J. Si, B.-K. Cui and Y.-C. Dai, Ann. Microbiol., 2013, 63, 1099–1108 CrossRef CAS.
- T. Hadibarata, A. R. M. Yusoff and R. A. Kristanti, Water, Air, Soil Pollut., 2012, 223, 933–941 CrossRef CAS.
- P. Yang, W. Shi, H. Wang and H. Liu, Braz. J. Microbiol., 2016, 47, 828–834 CrossRef CAS PubMed.
- R. G. Saratale, S. S. Gandhi, M. V. Purankar, M. B. Kurade, S. P. Govindwar, S. E. Oh and G. D. Saratale, J. Biosci. Bioeng., 2013, 115, 658–667 CrossRef CAS PubMed.
- L. S. Copete-Pertuz, F. Alandete-Novoa, J. Plácido, G. A. Correa-Londoño and A. L. Mora-Martínez, Sci. Total Environ., 2019, 646, 1536–1545 CrossRef CAS PubMed.
- S. Kumari and R. Naraian, J. Environ. Manage., 2016, 180, 172–179 CrossRef CAS PubMed.
- C. Tsioptsias, G. Lionta and P. Samaras, Environ. Technol., 2017, 38, 1120–1126 CrossRef CAS PubMed.
- G. Mujtaba and K. Lee, Water Res., 2017, 120, 174–184 CrossRef CAS PubMed.
- G. Guo, W. Cao, S. Sun, Y. Zhao and C. Hu, J. Appl. Phycol., 2017, 29, 2857–2866 CrossRef CAS.
- A. Bhattacharya, M. Mathur, P. Kumar, S. K. Prajapati and A. Malik, Algal Res., 2017, 21, 42–51 CrossRef.
- T. Ndikubwimana, X. Zeng, Y. Liu, J.-S. Chang and Y. Lu, Algal Res., 2014, 6, 186–193 CrossRef.
- S. K. Prajapati, P. Kumar, A. Malik and P. Choudhary, BioEnergy Res., 2014, 7, 1430–1440 CrossRef CAS.
- C. A. Ottoni, C. Santos, Z. Kozakiewicz and N. Lima, Folia Microbiol., 2013, 58, 187–193 CrossRef CAS PubMed.
- N. F. Cardoso, E. C. Lima, B. Royer, M. V. Bach, G. L. Dotto, L. A. Pinto and T. Calvete, J. Hazard. Mater., 2012, 241, 146–153 CrossRef PubMed.
- Y. Kang, X. Xu, H. Pan, J. Tian, W. Tang and S. Liu, Bioengineered, 2018, 9, 222–232 CrossRef CAS PubMed.
- J.-E. Park, Y.-B. Cho, S. Zhang and S.-J. Hwang, Journal of Korean Society of Water and Wastewater, 2013, 27, 703–709 CrossRef.
- H. Pan, X. Xu, Z. Wen, Y. Kang, X. Wang, Y. Ren and D. Huang, Bioengineered, 2017, 8, 630–641 CrossRef CAS PubMed.
- L. Xie, L. Zhou, T. Liu and X. Xu, RSC Adv., 2016, 6, 106935–106944 RSC.
- L. Jiang, L. Zhang, C. Nie and H. Pei, Biotechnol. Biofuels, 2018, 11, 68 CrossRef PubMed.
- S. K. Prajapati, P. Kaushik, A. Malik and V. K. Vijay, Bioresour. Technol., 2013, 135, 232–238 CrossRef CAS PubMed.
- M. N. Aydogan and N. P. Arslan, Desalin. Water Treat., 2015, 56, 2258–2266 CrossRef CAS.
- J. P. Rodriguez, D. E. Williams, I. D. Sabater, R. C. Bonugli-Santos, L. D. Sette, R. J. Andersen and R. G. Berlinck, RSC Adv., 2015, 5, 66360–66366 RSC.
- S. Ilk, D. Demircan, S. SAĞLAM, N. SAĞLAM and Z. M. Rzayev, Turk. J. Chem., 2016, 40, 262–276 CrossRef CAS.
- D. Rajkumar, B. J. Song and J. G. Kim, Dyes Pigm., 2007, 72, 1–7 CrossRef.
- Y. Li, Y. Xu, L. Liu, P. Li, Y. Yan, T. Chen, T. Zheng and H. Wang, Algal Res., 2017, 25, 402–412 CrossRef.
- W. Zhou, Y. Cheng, Y. Li, Y. Wan, Y. Liu, X. Lin and R. Ruan, Appl. Biochem. Biotechnol., 2012, 167, 214–228 CrossRef CAS PubMed.
- N. Muradov, M. Taha, A. F. Miranda, D. Wrede, K. Kadali, A. Gujar, T. Stevenson, A. S. Ball and A. Mouradov, Biotechnol. Biofuels, 2015, 8, 24 CrossRef PubMed.
- P. R. Chenaux, N. Lalji and D. D. Lefebvre, AMB Express, 2014, 4, 74 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2019 |