Themed collection Intrinsically Disordered Proteins

Front cover

Inside front cover

Back cover

Contents list
Back matter
Intrinsically disordered proteins
M. Madan Babu introduces this Molecular BioSystems themed issue on intrinsically disordered proteins.

Mol. BioSyst., 2012,8, 21-21
https://doi.org/10.1039/C1MB90045E
Disease mutations in disordered regions—exception to the rule?
Here, we challenge the conventional structure-centric view of missense mutations. We suggest that disease-associated mutations located in the intrinsically disordered protein regions are more prevalent and have a larger functional impact than previously thought.
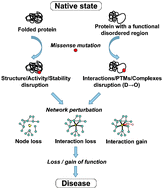
Mol. BioSyst., 2012,8, 27-32
https://doi.org/10.1039/C1MB05251A
How do dynamic cellular signals travel long distances?
This opinion proposes that nature exploited the dynamics of macromolecules to promote and optimize long-distance signaling; signaling is helped by pre-encoded loops and linkers and intrinsic disorder.
Mol. BioSyst., 2012,8, 22-26
https://doi.org/10.1039/C1MB05205E
An omics perspective of protein disorder
We review the literature regarding genome-wide studies of disorder and examine how these studies give rise to new characterizations and categories of this elusive phenomenon.

Mol. BioSyst., 2012,8, 185-193
https://doi.org/10.1039/C1MB05235G
The role of protein disorder in the 14-3-3 interaction network
14-3-3 proteins are ordered hubs that interact with multiple and diverse intrinsically disordered phosphorylated targets opening the possibility to be a master regulator of several signaling networks.
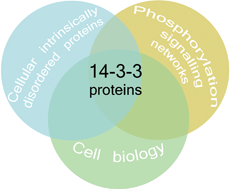
Mol. BioSyst., 2012,8, 178-184
https://doi.org/10.1039/C1MB05216K
Structural analysis of intrinsically disordered proteins by small-angle X-ray scattering
SAXS is a well-established method to characterize disordered biomolecules. Novel ensemble approaches provide quantitative information about the distribution of low-resolution structural parameters, these going beyond traditional single-value descriptors.

Mol. BioSyst., 2012,8, 151-167
https://doi.org/10.1039/C1MB05275F
Order and disorder in large multi-site docking proteins of the Gab family—implications for signalling complex formation and inhibitor design strategies
The first review discussing Gab proteins from the perspective of structural disorder with implications for their autoregulation and therapeutic strategies.
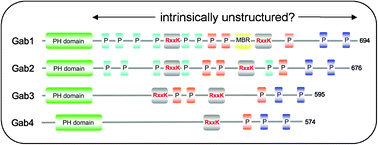
Mol. BioSyst., 2012,8, 33-46
https://doi.org/10.1039/C1MB05272A
Fuzziness: linking regulation to protein dynamics
Fuzziness puts forward a novel model for protein allostery.

Mol. BioSyst., 2012,8, 168-177
https://doi.org/10.1039/C1MB05234A
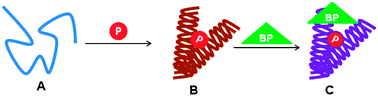
Intrinsically disordered regions as affinity tuners in protein –DNA interactions
The disordered tail ,which may be viewed as DNA recognizing subdomain, may support sliding dynamics and facilitate jumping via a "monkey bar" mechanism in which the flexible tail enhances the brachiation dynamics between two distant DNA fragments.
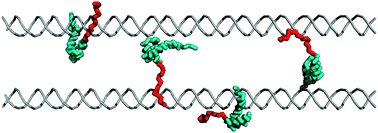
Mol. BioSyst., 2012,8, 47-57
https://doi.org/10.1039/C1MB05273J
Conformational propensities and residual structures in unfolded peptides and proteins
Conformational distributions of amino acid residues in unfolded peptides differ in their sampling of PPII, β-strand, helix and turn conformations.
Mol. BioSyst., 2012,8, 122-133
https://doi.org/10.1039/C1MB05225J
Roles of intrinsic disorder in protein –nucleic acid interactions
This review article focuses on disorder in proteins that bind DNA and RNA and on how this disorder promotes function.
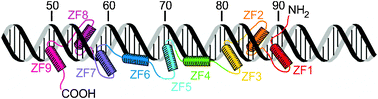
Mol. BioSyst., 2012,8, 97-104
https://doi.org/10.1039/C1MB05258F
Towards a robust description of intrinsic protein disorder using nuclear magnetic resonance spectroscopy
We present recent progress to develop a molecular representation of the disordered state, combining complementary data sets to extract a meaningful description of the conformational behaviour of intrinsically disordered proteins.
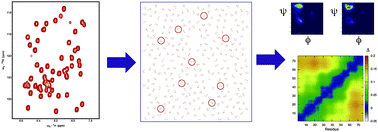
Mol. BioSyst., 2012,8, 58-68
https://doi.org/10.1039/C1MB05291H
A comprehensive overview of computational protein disorder prediction methods
In this review we present an overview of protein disorder prediction methods including an analysis of their advantages and shortcomings and an evaluation of their performance on the CASP9 dataset.
Mol. BioSyst., 2012,8, 114-121
https://doi.org/10.1039/C1MB05207A
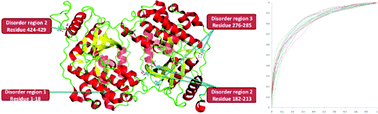
Role of an intrinsically disordered conformation in AMPK-mediated phosphorylation of ULK1 and regulation of autophagy
Phosphorylation-induced conformational changes in an intrinsically disordered region of ULK1 may provide a mechanism for its action in the regulation of autophagy.
Mol. BioSyst., 2012,8, 91-96
https://doi.org/10.1039/C1MB05265A
Mutual effects of disorder and order in fusion proteins between intrinsically disordered domains and fluorescent proteins
In this review we focus on the conformational and functional impact of fusions between fluorescent proteins and intrinsically disordered domains of various lengths.
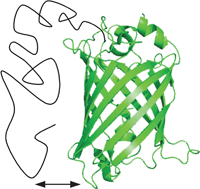
Mol. BioSyst., 2012,8, 105-113
https://doi.org/10.1039/C1MB05244F
Intrinsic disorder in the androgen receptor: identification, characterisation and drugability
In this review the role of intrinsic disorder in AR function is discussed along with the potential to develop new drugs that will target the structurally plastic NTD.
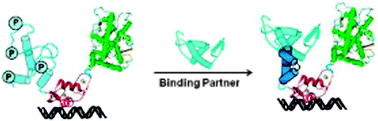
Mol. BioSyst., 2012,8, 82-90
https://doi.org/10.1039/C1MB05249G
Structural disorder within paramyxovirus nucleoproteins and phosphoproteins
In this review we focus on the experimental data showing the abundance of structural disorder within the nucleoprotein and phosphoprotein from three paramyxoviruses, namely Nipah, Hendra and measles viruses.

Mol. BioSyst., 2012,8, 69-81
https://doi.org/10.1039/C1MB05204G
Protein intrinsic disorder and induced pluripotent stem cells
The reprogramming transcription factors essential for the induced pluripotent stem cells formation are shown to be highly enriched in intrinsic disorder.
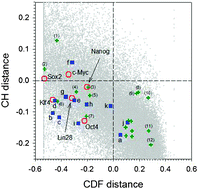
Mol. BioSyst., 2012,8, 134-150
https://doi.org/10.1039/C1MB05163F
Dynamic optimization of signal transductionvia intrinsic disorder
We report the mechanism by which an intrinsic disordered protein, p27kip1, dynamically optimizes signal transduction through a synergy between intrinsic disorder and functional collective motions.
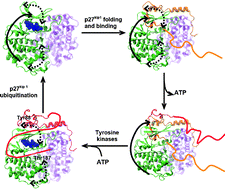
Mol. BioSyst., 2012,8, 194-197
https://doi.org/10.1039/C1MB05412K
Correlation of disorder between S. cerevisiae interacting proteins
We examine the percentage of predicted disorder residues within binary and complex interacting proteins (physical and functional interactions respectively) to investigate how the disorder of a protein relates to that of its interacting partners.
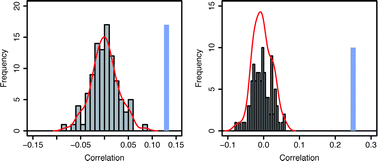
Mol. BioSyst., 2012,8, 417-425
https://doi.org/10.1039/C1MB05214D
Meta-structure correlation in protein space unveils different selection rules for folded and intrinsically disordered proteins
Correlation of meta-structure parameters for globular proteins, IDPs and sequence randomized IDPs is different, highlighting a trend in IDP sequences.

Mol. BioSyst., 2012,8, 411-416
https://doi.org/10.1039/C1MB05367A
Compaction and binding properties of the intrinsically disordered C-terminal domain of Henipavirus nucleoprotein as unveiled by deletion studies
These studies revealed the structural state of four putative MoREs within Henipavirus NTAIL, as well as their role in protein compaction and binding to PXD.

Mol. BioSyst., 2012,8, 392-410
https://doi.org/10.1039/C1MB05401E
Increased structural disorder of proteins encoded on human sex chromosomes
We show that structural disorder of human proteins coded on the sex chromosomes is significantly higher than the autosome-coded one.

Mol. BioSyst., 2012,8, 229-236
https://doi.org/10.1039/C1MB05285C
Uncertainty analysis in protein disorder prediction
The uncertainty in the reference models and in data is reduced by using appropriate meta predictors of protein disorder. The conservation of disorder is less stable than conservation of structure.
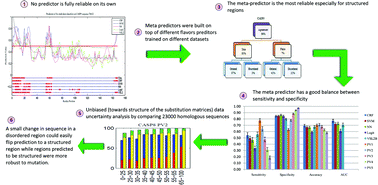
Mol. BioSyst., 2012,8, 381-391
https://doi.org/10.1039/C1MB05373F
Protein disorder in the centrosome correlates with complexity in cell types number
Centrosomal proteins are more disordered and coiled-coil than control proteins of the same organism, and more so in more complex organisms. Disordered regions increase in evolution through large indels, at a rate that is faster in centrosomal than in control proteins, and they evolved faster in phylogenetic branches that experimented a significant growth of cells number.
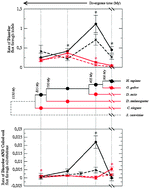
Mol. BioSyst., 2012,8, 353-367
https://doi.org/10.1039/C1MB05199G
Thermo-resistant intrinsically disordered proteins are efficient 20S proteasome substrates
The 20S proteasome susceptible intrinsically disordered proteins (IDPs) are highly soluble following heat treatment.

Mol. BioSyst., 2012,8, 368-373
https://doi.org/10.1039/C1MB05283G
Location of disorder in coiled coil proteins is influenced by its biological role and subcellular localization : a GO-based study on human proteome
Coiled coils have been often linked with intrinsic disorder. Current work focusses on preferences of ordered and disordered coiled coils in various cellular components, molecular functions and biological process.
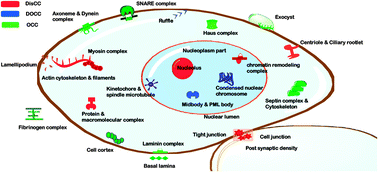
Mol. BioSyst., 2012,8, 346-352
https://doi.org/10.1039/C1MB05210A
Intrinsic protein disorder in human pathways
We identify the role of disordered proteins within the framework of pathway relationships. Three categories of relations are significantly enriched in disordered proteins: gene expression, protein binding and protein phosphorylation.

Mol. BioSyst., 2012,8, 320-326
https://doi.org/10.1039/C1MB05274H
Occurrence of disordered patterns and homorepeats in eukaryotic and bacterial proteomes
Occurrence of 20 homorepeats of 6 residues long in 123 proteomes.
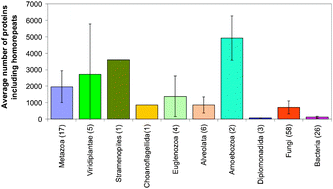
Mol. BioSyst., 2012,8, 327-337
https://doi.org/10.1039/C1MB05318C
Kinetic measurements give new insights into lipid membrane permeabilization by α-synuclein oligomers
Kinetic measurements of the influx of dithionite anions into large unilamellar vesicles show that an impaired membrane integrity caused by α-synuclein oligomers cannot be explained by a simple pore formation process.
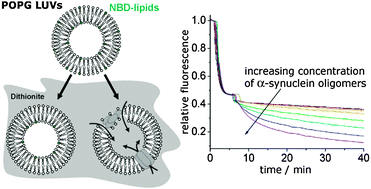
Mol. BioSyst., 2012,8, 338-345
https://doi.org/10.1039/C1MB05293D
Understanding the structural ensembles of a highly extended disordered protein
Destabilizing osmolytes cause depletion of the most compact structures for intrinsically disordered proteins.
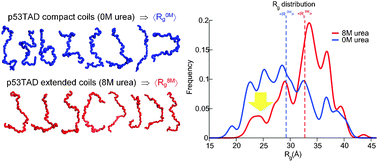
Mol. BioSyst., 2012,8, 308-319
https://doi.org/10.1039/C1MB05243H
Sedimentation velocity of intrinsically disordered proteins : what information can we actually obtain?
We show that although velocity measurements do yield information on gross conformation, the information is restricted to only the weight averaged sedimentation and diffusion coefficients of the conformational ensemble.
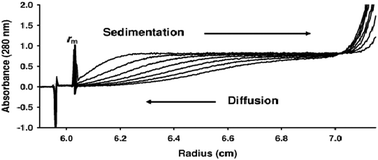
Mol. BioSyst., 2012,8, 378-380
https://doi.org/10.1039/C1MB05262D
Insights in the (un)structural organization of Bacillus pasteurii UreG, an intrinsically disordered GTPase enzyme
BpUreG, a naturally occurring intrinsically disordered enzyme, undergoes transitions between three major types of conformational ensembles with different degrees of residual structure, depending on temperature and denaturant concentration.

Mol. BioSyst., 2012,8, 220-228
https://doi.org/10.1039/C1MB05227F
Is there a biological cost of protein disorder? Analysis of cancer-associated mutations
In this work, we tested if there is any biological risk associated with protein disorder at the level of single nucleotide mutations.
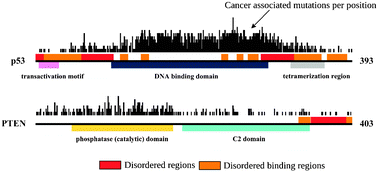
Mol. BioSyst., 2012,8, 296-307
https://doi.org/10.1039/C1MB05246B
Attributes of short linear motifs
The curated instances of the Eukaryotic Linear Motif (ELM) database are analysed to provide a comprehensive overview of the defining attributes of short, linear motifs (SLiMs).
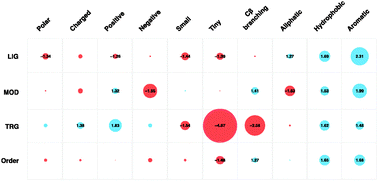
Mol. BioSyst., 2012,8, 268-281
https://doi.org/10.1039/C1MB05231D
Intrinsically disordered proteins as molecular shields
The broad family of LEA proteins are intrinsically disordered proteins (IDPs) with several potential roles in desiccation tolerance, or anhydrobiosis, one of which is to limit desiccation-induced aggregation of cellular proteins.
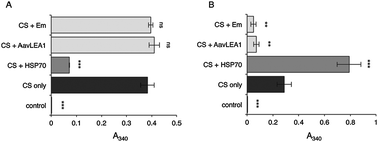
Mol. BioSyst., 2012,8, 210-219
https://doi.org/10.1039/C1MB05263B
Spontaneous symmetry breaking in a non-rigid molecule approach to intrinsically disordered proteins
An analog to Longuet-Higgins' non-rigid molecular group theory arguments can be applied to the structure and reaction dynamics of intrinsically disordered proteins via a somewhat counterintuitive Morse Function treatment inspired by statistical mechanics, providing possible symmetry classifications of the molecular ‘fuzzy lock-and-key’.

Mol. BioSyst., 2012,8, 374-377
https://doi.org/10.1039/C1MB05256J
Modulation of an IDP binding mechanism and rates by helix propensity and non-native interactions: association of HIF1α with CBP
Coarse-grained models predict the Hif1α–CBP binding rate, and suggest an induced fit binding mechanism, influenced by non-native interactions.
Mol. BioSyst., 2012,8, 256-267
https://doi.org/10.1039/C1MB05252G
Interactome-wide prediction of short, disordered protein interaction motifs in humans
Comprehensive analysis of the human interactome predicts thousands of conserved, disordered protein motifs of potential functional importance.
Mol. BioSyst., 2012,8, 282-295
https://doi.org/10.1039/C1MB05212H
Intrinsically disordered regions have specific functions in mitochondrial and nuclear proteins
Generally mitochondrial proteins have an N-terminal gradient of positive charges and the disordered regions of DNA-binding proteins are negatively charged.
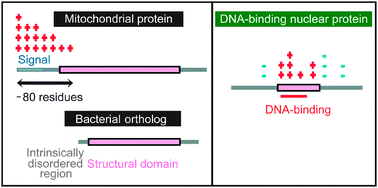
Mol. BioSyst., 2012,8, 247-255
https://doi.org/10.1039/C1MB05208J
Aromatic residues link binding and function of intrinsically disordered proteins
We report the role of π–π interactions in linking binding and function of intrinsically disordered proteins.

Mol. BioSyst., 2012,8, 237-246
https://doi.org/10.1039/C1MB05239J
Synergistic folding of two intrinsically disordered proteins : searching for conformational selection
Topology-based modeling reveals that NCBD interacts with ACTR via "extended conformational selection" and involves multiple stages of selection and induced folding, which appears highly consistent with H/D-MS and atomistic simulations.
Mol. BioSyst., 2012,8, 198-209
https://doi.org/10.1039/C1MB05156C