Themed collection Molecular BioSystems 2013 Proteomics

Digital and analogical reality in proteomics investigation
Are protein functions continuous or discretized? Proteomics investigations are starting to address this non-trivial awesome question focusing upon determining the nature of biological molecular relationships.

Mol. BioSyst., 2013,9, 1062-1063
https://doi.org/10.1039/C3MB90013D
Genotype–phenotype relationships in light of a modular protein interaction landscape
Better approaches tackling the modular nature of proteomes will have dramatic effects on the understanding of genotype–phenotype relationships, argue Matthias Gstaiger and Ruedi Aebersold.
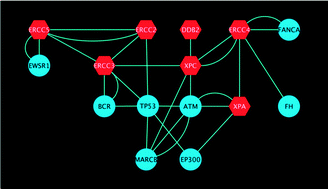
Mol. BioSyst., 2013,9, 1064-1067
https://doi.org/10.1039/C3MB25583B
The mitochondrial Italian Human Proteome Project initiative (mt-HPP)
mt-HPP: a new HUPO initiative on human mitochondrial proteins.

Mol. BioSyst., 2013,9, 1984-1992
https://doi.org/10.1039/C3MB70065H
Application of proteomics technology in adipocyte biology
Obesity and its associated complications have reached epidemic proportions in Western-type societies.
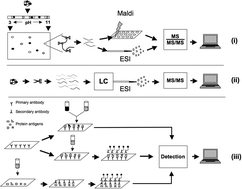
Mol. BioSyst., 2013,9, 1076-1091
https://doi.org/10.1039/C3MB25596D
Some remarks on predicting multi-label attributes in molecular biosystems
Many molecular biosystems and biomedical systems belong to the multi-label systems in which each of their constituent molecules possesses one or more than one function or feature, and hence needs one or more than one label to indicate its attribute(s).
Mol. BioSyst., 2013,9, 1092-1100
https://doi.org/10.1039/C3MB25555G
Essential genes in Bacillus subtilis: a re-evaluation after ten years
An overview of essential genes in Bacillus subtilis with emphasis on changes in the list of essential genes since 2003.
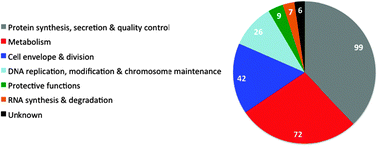
Mol. BioSyst., 2013,9, 1068-1075
https://doi.org/10.1039/C3MB25595F
Mass spectrometry-based identification and characterisation of lysine and arginine methylation in the human proteome
Global identification of 501 in vivo protein methylations, combining hmSILAC with MS-proteomics, indicates predominance amongst complexes related to gene expression.
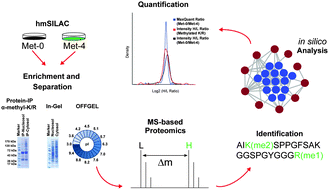
Mol. BioSyst., 2013,9, 2231-2247
https://doi.org/10.1039/C3MB00009E
Proteomic analysis of temperature stress-responsive proteins in Arabidopsis thaliana rosette leaves
Proteomics of Arabidopsis leaves subjected to low or high temperature stress.

Mol. BioSyst., 2013,9, 1257-1267
https://doi.org/10.1039/C3MB70137A
Crosstalk between salicylic acid and jasmonate in Arabidopsis investigated by an integrated proteomic and transcriptomic approach
Crosstalk between SA- and JA-mediated defence pathways is investigated for the first time by both proteomic and transcriptomic approaches.
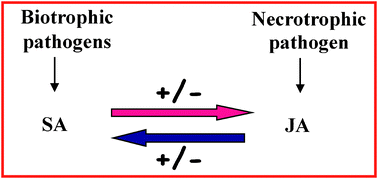
Mol. BioSyst., 2013,9, 1169-1187
https://doi.org/10.1039/C3MB25569G
Proteomic and ionomic profiling reveals significant alterations of protein expression and calcium homeostasis in cystic fibrosis cells
Altered protein expression profiles and Ca2+ dyshomeostasis in cystic fibrosis cells using integrated proteomic and ionomic analysis.
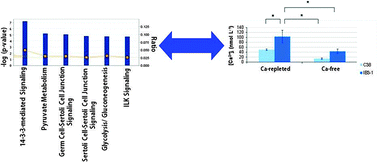
Mol. BioSyst., 2013,9, 1117-1126
https://doi.org/10.1039/C3MB25594H
Integrative quantitation enables a comprehensive proteome comparison of two Mycoplasma pneumoniae genetic perturbations
Combination of six different protein quantitation strategies allowed confidently detection of protein abundance changes among distinct M. pneumoniae strains.

Mol. BioSyst., 2013,9, 1249-1256
https://doi.org/10.1039/C3MB25581F
Shotgun proteomics reveals specific modulated protein patterns in tears of patients with primary open angle glaucoma naïve to therapy
LC-MSE proteomics revealed 27 differential tear proteins in naïve POAG vs. CTRL,13 of them were modulated in medically controlled POAG indicating a probable influence of therapy on the same molecular pattern.

Mol. BioSyst., 2013,9, 1108-1116
https://doi.org/10.1039/C3MB25463A
Complementary biochemical approaches applied to the identification of plastidial calmodulin-binding proteins
Using a proteomic approach, we identified new plastidial CaM-binding candidates and validated a subset of them by overlay assays and fluorescence anisotropy.

Mol. BioSyst., 2013,9, 1234-1248
https://doi.org/10.1039/C3MB00004D
Differential protein profiling of renal cell carcinoma urinary exosomes
The proteome of urinary exosomes from renal cell carcinoma patients is reproducibly different from healthy controls, as shown by a radar plot comparing the differential levels of selected proteins.

Mol. BioSyst., 2013,9, 1220-1233
https://doi.org/10.1039/C3MB25582D
Postnatal cardiomyocyte growth and mitochondrial reorganization cause multiple changes in the proteome of human cardiomyocytes
Comparison of fetal and adult human heart proteomes reflects the structural and biochemical differences between the two.

Mol. BioSyst., 2013,9, 1210-1219
https://doi.org/10.1039/C3MB25556E
Red blood cell metabolism under prolonged anaerobic storage
MS-based untargeted metabolomics confirmed oxygen dependent modulation of red blood cell metabolism.

Mol. BioSyst., 2013,9, 1196-1209
https://doi.org/10.1039/C3MB25575A
Unravelling the bull fertility proteome
A representative 2-DE map of a cryopreserved bull semen sample. Differentially expressed proteins are indicated in the map.

Mol. BioSyst., 2013,9, 1188-1195
https://doi.org/10.1039/C3MB25494A
Alteration of proteomic profiles in PBMC isolated from patients with Fabry disease: preliminary findings
In Fabry patients, we found, by a proteomic approach, some down-regulated proteins (CALX, Rho GDI2, Rho GDI1, NCC27) and others upregulated (NSE, 1433T, 1433Z, Gal-1). These data could be related to pathophysiological aspects of disease.

Mol. BioSyst., 2013,9, 1162-1168
https://doi.org/10.1039/C3MB25402J
Effect of IR laser on myoblasts: a proteomic study
IR laser treatment induces a general cytoskeleton reorganization and promotes the myoblast differentiation in vitro.

Mol. BioSyst., 2013,9, 1147-1161
https://doi.org/10.1039/C2MB25398D
Urinary exosomes and diabetic nephropathy: a proteomic approach
Label-free LC-MS/MS assessment of differential proteome in urinary exosomes from ZDF diabetic rats, and immunoblotting validation, as exemplified by proteins in the inserts (MUP1, CD10, and PEPD).
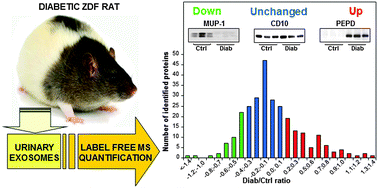
Mol. BioSyst., 2013,9, 1139-1146
https://doi.org/10.1039/C2MB25396H
Comparative proteome profiling of breast tumor cell lines by gel electrophoresis and mass spectrometry reveals an epithelial mesenchymal transition associated protein signature
In this work, we present a comparative proteome analysis of two breast cancer cell lines providing information about a set of proteins involved in the process of epithelial mesenchymal transition.
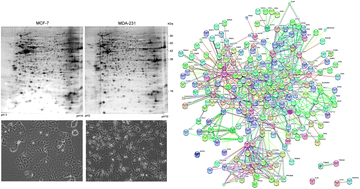
Mol. BioSyst., 2013,9, 1127-1138
https://doi.org/10.1039/C2MB25401H
Detection of high molecular weight proteins by MALDI imaging mass spectrometry
Detection of high molecular weight proteins on thin tissue sections by imaging mass spectrometry is improved by the use of ferulic acid.
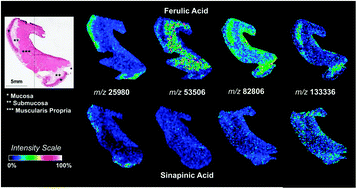
Mol. BioSyst., 2013,9, 1101-1107
https://doi.org/10.1039/C2MB25296A
About this collection
Molecular BioSystems is delighted to present this collection of papers from the themed issue Proteomics 2013, guest edited by Andrea Urbani, President of the Italian Proteomics Association.