A dual identification strategy based on padlock ligation and CRISPR/Cas14a for highly specific detection of BRAF V600E mutation in clinical samples†
Abstract
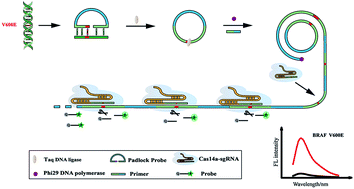
BRAF V600E mutation is a single-nucleotide variation (SNV) that is widely found in various cancers and has been demonstrated to have a strong association with the prognosis and development of some diseases. Thus, we developed a strategy based on rolling circle amplification (RCA) and CRISPR/Cas14a to meet the great need for detecting highly specific BRAF V600E mutation in fine-needle biopsy samples. In this study, a padlock probe was designed to recognize and trigger subsequent ligase chain reactions (LCR). And due to the Taq DNA ligase, a great number of ligated annular padlock probes were generated in the presence of BRAF V600E mutation, subsequently generating long repeated single-strand DNA by RCA. The obtained amplicons were activators triggering the trans-cleavage of CRISPR/Cas14a. CRISPR/Cas14a shows outstanding performance in identifying ssDNA with single base mutation, which significantly increases the specificity of mutation discrimination. Under the optimal conditions, our strategy can identify BRAF V600E mutation down to 0.307 fM with a wide linear range from 1 fM to 10 pM. On the other hand, the dual identification strategy endows the method with terrific specificity for the detection of SNV. Furthermore, our method has been successfully employed to identify BRAF V600E mutation in clinical fine-needle aspiration samples, proving great potential for ultra-specific identification of low abundance BRAF V600E mutation and providing a novel method for diagnosis and treatment of cancer.


 Please wait while we load your content...
Please wait while we load your content...