Accelerating high-throughput virtual screening through molecular pool-based active learning†
Abstract
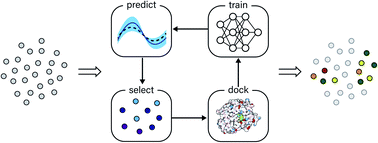
Structure-based virtual screening is an important tool in early stage drug discovery that scores the interactions between a target protein and candidate ligands. As virtual libraries continue to grow (in excess of 108 molecules), so too do the resources necessary to conduct exhaustive virtual screening campaigns on these libraries. However, Bayesian optimization techniques, previously employed in other scientific discovery problems, can aid in their exploration: a surrogate structure–property relationship model trained on the predicted affinities of a subset of the library can be applied to the remaining library members, allowing the least promising compounds to be excluded from evaluation. In this study, we explore the application of these techniques to computational docking datasets and assess the impact of surrogate model architecture, acquisition function, and acquisition batch size on optimization performance. We observe significant reductions in computational costs; for example, using a directed-message passing neural network we can identify 94.8% or 89.3% of the top-50 000 ligands in a 100M member library after testing only 2.4% of candidate ligands using an upper confidence bound or greedy acquisition strategy, respectively. Such model-guided searches mitigate the increasing computational costs of screening increasingly large virtual libraries and can accelerate high-throughput virtual screening campaigns with applications beyond docking.


 Please wait while we load your content...
Please wait while we load your content...