Tryptophan scanning mutagenesis as a way to mimic the compound-bound state and probe the selectivity of allosteric inhibitors in cells†
Abstract
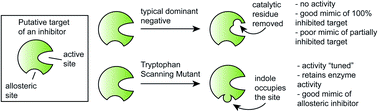
Understanding the selectivity of a small molecule for its target(s) in cells is an important goal in chemical biology and drug discovery. One powerful way to address this question is with dominant negative (DN) mutants, in which an active site residue in the putative target is mutated. While powerful, this approach is less straightforward for allosteric sites. Here, we introduce tryptophan scanning mutagenesis as an expansion of this idea. As a test case, we focused on the challenging drug target, heat shock cognate protein 70 (Hsc70), and its allosteric inhibitor JG-98. Structure-based modelling predicted that mutating Y149W in human Hsc70 or Y145W in the bacterial ortholog DnaK would place an indole side chain into the allosteric pocket normally occupied by the compound. Indeed, we found that the tryptophan mutants acted as if they were engaged with JG-98. We then used DnaK Y145W to suggest that this protein may be an anti-bacterial target. Indeed, we found that DnaK inhibitors have minimum inhibitory concentration (MIC) values <0.125 μg mL−1 against several pathogens, including multidrug-resistant Staphylococcus aureus (MRSA) strains. We propose that tryptophan scanning mutagenesis may provide a distinct way to address the important problem of target engagement.


 Please wait while we load your content...
Please wait while we load your content...