Mass spectrometric analysis of core fucosylation and sequence variation in a human–camelid monoclonal antibody
Abstract
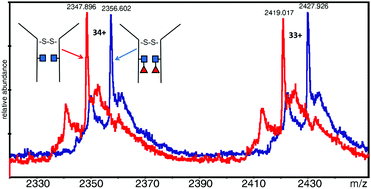
Electrospray mass spectrometry (ESI-MS) was used to measure the masses of an intact dimeric monoclonal antibody (Mab) and assess the fucosylation level. The Mab under study was EG2-hFc, a chimeric human–camelid antibody of about 80 kDa (A. Bell et al., Cancer Lett., 2010, 289(1), 81–90). It was obtained from cell culture with and without a fucosylation inhibitor, and treated with EndoS which cleaves between the two core N-acetyl glucosamine (GlcNAc) residues. It is the first time that this combined approach with a unique mass spectrometer was used to measure 146 Da differences as part of a large intact dimeric antibody. Results showed that in the dimer, both heavy chains were fucosylated on the core GlcNAc of the Fc Asn site equivalent to Asn297. In the presence of the fucosylation inhibitor, fucosylation was lost on both subunits. Following reduction, monomers were analyzed and the masses obtained corroborated the dimer results. Dimeric EG2-hFc Mab treated with PNGase F, to deglycosylate the protein, was also measured by MS for mass comparison. In spite of the success of fucosylation level measurements, the experimental masses of deglycosylated dimers and GlcNAc–Fuc bearing dimers did not correspond to masses of our sequence of reference (A. Bell et al., Cancer Lett., 2010, 289(1), 81–90; http://www.uniprot.org; http://www.expasy.org), which prompted experiments to determine the protein backbone sequence. Digest mixtures from trypsin, GluC, as well as trypsin + GluC proteolysis were analyzed by matrix-assisted laser desorption/ionization (MALDI) MS and MS/MS. A few variations were found relative to the reference sequence, which are discussed in detail herein. These measurements allowed us to build a new “experimental” sequence for the EG2-hFc samples investigated in this work, although there are still ambiguities to be resolved in this new sequence. MALDI-MS/MS also confirmed the fucosylation pattern in the Fc tryptic peptide EEQYNSTYR.
- This article is part of the themed collection: Glycomics & Glycoproteomics: From Analytics to Function


 Please wait while we load your content...
Please wait while we load your content...