Mass spectrometry and computational study of collision-induced dissociation of 9-methylguanine–1-methylcytosine base-pair radical cation: intra-base-pair proton transfer and hydrogen transfer, non-statistical dissociation, and reaction with a water ligand†
Abstract
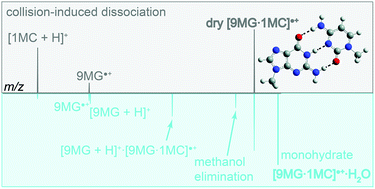
A combined experimental and theoretical study is presented on the collision-induced dissociation (CID) of 9-methylguanine–1-methylcytosine base-pair radical cation (abbreviated as [9MG·1MC]˙+) and its monohydrate ([9MG·1MC]˙+·H2O) with Xe and Ar gases. Product ion mass spectra were measured as a function of collision energy using guided-ion beam tandem mass spectrometry, from which cross sections and threshold energies for various dissociation pathways were determined. Electronic structure calculations were performed at the DFT, RI-MP2 and DLPNO-CCSD(T) levels of theory to identify product structures and map out reaction potential energy surfaces. [9MG·1MC]˙+ has two structures: a conventional structure 9MG˙+·1MC (population 87%) consisting of hydrogen-bonded 9-methylguanine radical cation and neutral 1-methylcytosine, and a proton-transferred structure [9MG − H]˙·[1MC + H]+ (less stable, population 13%) formed by intra-base-pair proton transfer from the N1 of 9MG˙+ to the N3 of 1MC within 9MG˙+·1MC. The two structures have similar dissociation energies but can be distinguished in that 9MG˙+·1MC dissociates into 9MG˙+ and 1MC whereas [9MG – H]˙·[1MC + H]+ dissociates into neutral [9MG – H]˙ radical and protonated [1MC + H]+. An intriguing finding is that, in both Xe- and Ar-induced CID of [9MG·1MC]˙+, product ions were overwhelmingly dominated by [1MC + H]+, which is contrary to product distributions predicted using a statistical reaction model. Monohydration of [9MG·1MC]˙+ reversed the populations of the conventional structure (43%) vs. the proton-transferred structure (57%) and induced new reactions upon collisional activation, of which intra-base-pair hydrogen transfer produced [9MG + H]+ and the reaction of the water ligand with a methyl group in [9MG·1MC]˙+ led to methanol elimination from [9MG·1MC]˙+·H2O.


 Please wait while we load your content...
Please wait while we load your content...