ReacNetGenerator: an automatic reaction network generator for reactive molecular dynamics simulations†
Abstract
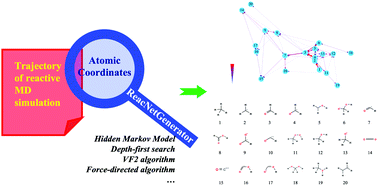
Reactive molecular dynamics (MD) simulation makes it possible to study the reaction mechanism of complex reaction systems at the atomic level. However, the analysis of MD trajectories which contain thousands of species and reaction pathways has become a major obstacle to the application of reactive MD simulation in large-scale systems. Here, we report the development and application of the Reaction Network Generator (ReacNetGenerator) method. It can automatically extract the reaction network from the reaction trajectory without any predefined reaction coordinates and elementary reaction steps. Molecular species can be automatically identified from the cartesian coordinates of atoms and the hidden Markov model is used to filter the trajectory noises which makes the analysis process easier and more accurate. The ReacNetGenerator has been successfully used to analyze the reactive MD trajectories of the combustion of methane and 4-component surrogate fuel for rocket propellant 3 (RP-3), and it has great advantages in terms of efficiency and accuracy compared to traditional manual analysis.


 Please wait while we load your content...
Please wait while we load your content...