Crystallizing protein assemblies via free and grafted linkers
Abstract
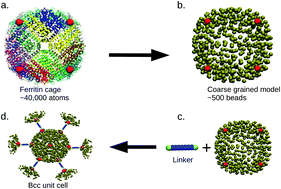
Porous protein superlattices have plausible catalytic applications in biotechnology and nanotechnology. They are solid yet open structures with the potential for preserving the activity of enzymes. However, there is still a lack of understanding of the design parameters that are required to arrange proteins in a periodic porous fashion. Here, we introduce a coarse-grained molecular dynamics (MD) simulation approach to study the effects of the lengths and geometries of linkers on the stability of 3D crystalline assemblies of metal ion anchored ferritin proteins. By simulating a system of proteins (eight metal ion anchored sites per protein) and linkers (two free ends per linker), we find that there is a range of optimal linker lengths for crystalline order. The optimal linker length is found to depend on the linker to protein concentration ratio and binding energy. We also examine the case of grafted flexible linkers on the protein surface as an alternative route for constructing highly porous crystalline structures. Our study demonstrates that the length of grafted linkers is a better tunable parameter than the length of free linkers to achieve high porosity protein superlattices. The computational study developed here provides guidelines to assemble biomolecules into crystals with high porosity.


 Please wait while we load your content...
Please wait while we load your content...
