Abstract
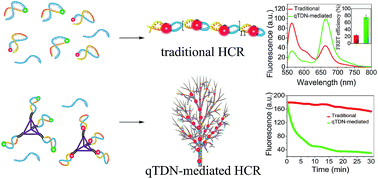
Nonenzymatic nucleic acid amplification techniques (e.g. the hybridization chain reaction, HCR) have shown promising potential for amplified detection of biomarkers. However, the traditional HCR occurs through random diffusion of DNA hairpins, making the kinetics and efficiency quite low. By assembling DNA hairpins at the vertexes of tetrahedral DNA nanostructures (TDNs), the reaction kinetics of the HCR is greatly accelerated due to the synergetic contributions of multiple reaction orientations, increased collision probability and enhanced local concentrations. The proposed quadrivalent TDN (qTDN)-mediated hyperbranched HCR has a ∼70-fold faster reaction rate than the traditional HCR. The approximately 76% fluorescence resonance energy transfer (FRET) efficiency obtained is the highest in the reported DNA-based FRET sensing systems as far as we know. Moreover, qTDNs modified by hairpins can easily load drugs, freely traverse plasma membranes and be rapidly cross-linked via the target-triggered HCR in live cells. The reduced freedom of movement as a result of the large crosslinked structure might constrain the hyperbranched HCR in a confined environment, thus making it a promising candidate for in situ imaging and photodynamic therapy. Hence, we present a paradigm of perfect integration of DNA nanotechnology with nucleic acid amplification, thus paving a promising way to the improved performance of nucleic acid amplification techniques and their wider application.
- This article is part of the themed collection: Celebrating 100 Years of Chemistry at Nankai University


 Please wait while we load your content...
Please wait while we load your content...