TBA loop mapping with 3′-inverted-deoxythymidine for fine-tuning of the binding affinity for α-thrombin†
Abstract
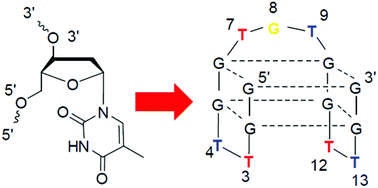
TBA is a 15-mer DNA aptamer for human α-thrombin, and its three T-rich loops are involved in the binding interactions with thrombin differently. In order to clarify their specific spatial locations in the binding interactions and search for more favourable positions, here a systematic investigation of all the loop residues was conducted with 3′-inverted thymidine (iT), by which both unnatural 3′–3′- and 5′–5′-linkages for each incorporation were introduced in the tertiary structure. The changes in Tm values and CD spectra revealed that motifs T3T12 and T4T13 are structurally distinct. Longer anti-clotting time was obtained for the T3 and T12 modifications, respectively, while T4 and T13 were completely intolerant with such changes, in terms of stability and binding to thrombin. In particular, the increased affinity bindings and longer anti-clotting time were obtained with the replacement on the central loop T7G8T9, which were closely related to the existence of a monovalent ion, K+ or Na+, consistently with the supposed binding site of these ions in TBA. It is worthwhile to note that both the subtle variations of the loop residues induced by iT and the monovalent ions determined the interacting residues of TBA and the binding strength rather than the thermal stability of the TBA structure.
- This article is part of the themed collection: Chemical Biology in OBC


 Please wait while we load your content...
Please wait while we load your content...