Dual-network sparse graph regularized matrix factorization for predicting miRNA–disease associations
Abstract
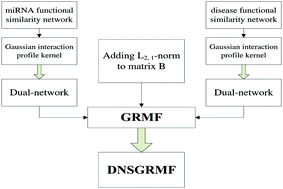
With the development of biological research and scientific experiments, it has been discovered that microRNAs (miRNAs) are closely related to many serious human diseases; however, finding the correct miRNA–disease associations is both time consuming and challenging. Therefore, it is very necessary to develop some new methods. Although the existing methods are very helpful in this regard, they all present some shortcomings; thus, some new methods need to be developed to overcome these shortcomings. In this study, a method based on dual network sparse graph regularized matrix factorization (DNSGRMF) was proposed, which increased the sparsity by adding the L2,1-norm. Moreover, Gaussian interaction profile kernels were introduced. The experiments showed that our method was feasible and had a high AUC value. Additionally, the five-fold cross-validation method was used to evaluate this method. A simulation experiment was used to predict some new associations on the datasets, and the obtained experimental results were satisfactory, which proved that our method was indeed feasible.


 Please wait while we load your content...
Please wait while we load your content...