Network-based approach to identify principal isoforms among four cancer types†
Abstract
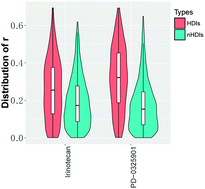
Protein isoforms are structurally similar proteins produced by alternative splicing of a single gene or genes from the same family. Isoforms of a protein can perform the same, similar, or even opposite biological functions. A previous study identified principal isoforms of proteins based on the extent of interactions per isoform in a functional relationship network, focusing on data from normal tissues. Additionally, the expression levels of specific isoforms of various genes associated with tumorigenesis and prognosis are frequently altered in tumors compared with those in normal tissues. In this study, we aimed to identify higher degree isoforms (HDIs) of multi-isoform genes (MIGs) in cancer by applying a meta-analytical framework to calculate co-expression between each pair of isoforms in two large datasets of RNA-seq profiles from breast cancer, lung cancer, leukemia, and colon cancer cell lines. Then, we compared HDIs with isoforms identified by proteomic data and prognostic and predictive evidence in various cancers. In addition, we separately analyzed the associations between HDIs and non-HDIs (nHDIs) of the same genes according to transcript expression and drug responses in various cancer type cell lines. Collectively, these results indicated the complex properties of HDIs per gene identified by cancer type-based isoform–isoform co-expression networks and showed the potential of HDIs as novel therapeutic targets for cancer treatment.


 Please wait while we load your content...
Please wait while we load your content...