A consensus protocol for the in silico optimisation of antibody fragments†
Abstract
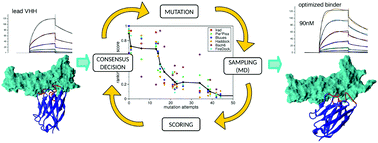
We present an in silico mutagenetic protocol for improving the binding affinity of single domain antibodies (or nanobodies, VHHs). The method iteratively attempts random mutations in the interacting region of the protein and evaluates the resulting binding affinity towards the target by scoring, with a collection of scoring functions, short explicit solvent molecular dynamics trajectories of the binder-target complexes. The acceptance/rejection of each attempted mutation is carried out by a consensus decision-making algorithm, which considers all individual assessments derived from each scoring function. The method was benchmarked by evolving a single complementary determining region (CDR) of an anti-HER2 VHH hit obtained by direct panning of a phage display library. The optimised VHH mutant showed significantly enhanced experimental affinity with respect to the original VHH it matured from. The protocol can be employed as it is for the optimization of peptides, antibody fragments, and (given enough computational power) larger antibodies.


 Please wait while we load your content...
Please wait while we load your content...