Optimizing peptide nucleic acid probes for hybridization-based detection and identification of bacterial pathogens†
Abstract
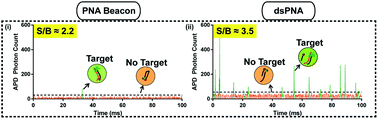
Point-of-care (POC) diagnostics for infectious diseases have the potential to improve patient care and antibiotic stewardship. Nucleic acid hybridization is at the core of many amplification-free molecular diagnostics and detection probe configuration is key to diagnostic performance. Modified nucleic acids such as peptide nucleic acid (PNA) offer advantages compared to conventional DNA probes allowing for faster hybridization, better stability and minimal sample preparation for direct detection of pathogens. Probes with tethered fluorophore and quencher allow for solution-based assays and eliminate the need for washing steps thereby facilitating integration into microfluidic devices. Here, we compared the sensitivity and specificity of double stranded PNA probes (dsPNA) and PNA molecular beacons targeting E. coli and P. aeruginosa for direct detection of bacterial pathogens. In bulk fluid assays, the dsPNAs had an overall higher fluorescent signal and better sensitivity and specificity than the PNA beacons for pathogen detection. We further designed and tested an expanded panel of dsPNA probes for detection of a wide variety of pathogenic bacteria including probes for universal detection of eubacteria, Enterobacteriaceae family, and P. mirablis. To confirm that the advantage translated to other assay types we compared the PNA beacon and dsPNA in a prototype droplet microfluidic device. Beyond the bulk fluid assay and droplet devices, use of dsPNA probes may be advantageous in a wide variety of assays that employ homogenous nucleic acid hybridization.


 Please wait while we load your content...
Please wait while we load your content...