Identification of an auxiliary druggable pocket in the DNA gyrase ATPase domain using fragment probes†
Abstract
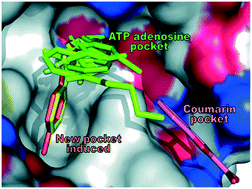
Discovery of new drug binding sites on well-established targets is of great interest as it facilitates the design of new mechanistic inhibitors to overcome the acquired drug resistance. Small chemical fragments can easily enter and bind to the cavities on the protein surface. Thus, they can be used to probe new druggable pockets in proteins. DNA gyrase plays indispensable roles in DNA replication, and both its GyrA and GyrB subunits are clinically validated antibacterial targets. New mechanistic GyrB inhibitors are urgently desired since the withdrawal of novobiocin from the market by the FDA due to its reduced efficiency and other reasons. Here, a fragment library was screened against the E. coli GyrB ATPase domain by combining affinity- and bioactivity-based approaches. The following X-ray crystallographic efforts were made to determine the cocrystal structures of GyrB with ten fragment hits, and three different binding modes were disclosed. Fortunately, a hydrophobic pocket which is previously unknown was identified by two fragments. Fragments that bind to this pocket were shown to inhibit the ATPase activity as well as the DNA topological transition activity of DNA gyrase in vitro. A set of fragment analogs were screened to explore the binding capacity of this pocket and identify the better starting fragments for lead development. Phylogenetic analysis revealed that this pocket is conserved in most Gram-negative and also many Gram-positive human pathogenic bacteria, implying a broad-spectrum antibacterial potential and a lower risk of mutation. Thus, the novel druggable pocket and the starting fragments provide a novel basis for designing new GyrB-targeting therapeutics.


 Please wait while we load your content...
Please wait while we load your content...