Computational design of biological circuits: putting parts into context
Abstract
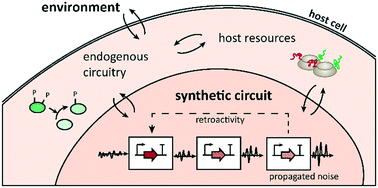
The rational design of synthetic gene circuits has led to many successful applications over the past decade. However, increasingly complex constructs also revealed that analogies to electronics design such as modularity and ‘plug-and-play’ composition are of limited use: biology is less well characterized, more context-dependent, and overall less predictable. Here, we summarize the main conceptual challenges of synthetic circuit design to highlight recent progress towards more tailored, context-aware computational design methods for synthetic biology. Emerging methods to guide the rational design of synthetic circuits that robustly perform desired tasks might reduce the number of experimental trial and error cycles.


 Please wait while we load your content...
Please wait while we load your content...