Real-time tracking, retrieval and gene expression analysis of migrating human T cells†
Abstract
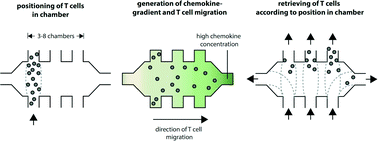
Dynamical analysis of single-cells allows assessment of the extent and role of cell-to-cell variability, however traditional dish-and-pipette techniques have hindered single-cell analysis in quantitative biology. We developed an automated microfluidic cell culture system that generates stable diffusion-based chemokine gradients, where cells can be placed in predetermined positions, monitored via single-cell time-lapse microscopy, and subsequently be retrieved based on their migration speed and directionality for further off-chip gene expression analysis, constituting a powerful platform for multiparameter quantitative studies of single-cell chemotaxis. Using this system we studied CXCL12-directed migration of individual human primary T cells. Spatiotemporally deterministic retrieval of T cell subsets in relation to their migration speed, and subsequent analysis with microfluidic droplet digital-PCR showed that the expression level of CXCR4 – the receptor of CXCL12 – underlies enhanced human T cell chemotaxis.

 Please wait while we load your content...
Please wait while we load your content...