Systems biology of neurodegenerative diseases
Abstract
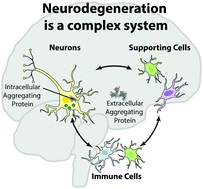
Neurodegenerative diseases (NDs) collectively afflict more than 40 million people worldwide. The majority of these diseases lack therapies to slow or stop progression due in large part to the challenge of disentangling the simultaneous presentation of broad, multifaceted pathophysiologic changes. Present technologies and computational capabilities suggest an optimistic future for deconvolving these changes to identify novel mechanisms driving ND onset and progression. In particular, integration of highly multi-dimensional omic analytical techniques (e.g., microarray, mass spectrometry) with computational systems biology approaches provides a systematic methodology to elucidate new mechanisms driving NDs. In this review, we begin by summarizing the complex pathophysiology of NDs associated with protein aggregation, emphasizing the shared complex dysregulation found in all of these diseases, and discuss available experimental ND models. Next, we provide an overview of technological and computational techniques used in systems biology that are applicable to studying NDs. We conclude by reviewing prior studies that have applied these approaches to NDs and comment on the necessity of combining analysis from both human tissues and model systems to identify driving mechanisms. We envision that the integration of computational approaches with multiple omic analyses of human tissues, and mouse and in vitro models, will enable the discovery of new therapeutic strategies for these devastating diseases.
- This article is part of the themed collection: In Vivo Systems Biology

 Please wait while we load your content...
Please wait while we load your content...