Tri- and tetra-nuclear polypyridyl ruthenium(ii) complexes as antimicrobial agents
Abstract
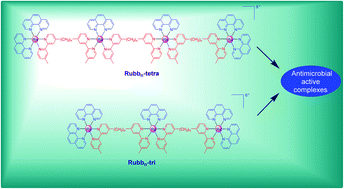
A series of inert tri- and tetra-nuclear polypyridylruthenium(II) complexes that are linked by the bis[4(4′-methyl-2,2′-bipyridyl)]-1,n-alkane ligand (“bbn” for n = 10, 12 and 16) have been synthesised and their potential as antimicrobial agents examined. Due to the modular nature of the synthesis of the oligonuclear complexes, it was possible to make both linear and non-linear tetranuclear ruthenium species. The minimum inhibitory concentrations (MIC) of the ruthenium(II) complexes were determined against four strains of bacteria − Gram positive Staphylococcus aureus (S. aureus) and methicillin-resistant S. aureus (MRSA), and Gram negative Escherichia coli (E. coli) and Pseudomonas aeruginosa (P. aeruginosa). In order to gain an understanding of the relative antimicrobial activities, the cellular uptake and water–octanol partition coefficients (log P) were determined for a selection of the ruthenium complexes. Although the trinuclear complexes were the most lipophilic based upon log P values and showed the greatest cellular uptake, the linear tetranuclear complexes were generally more active, with MIC values <1 μM against the Gram positive bacteria. Similarly, although the non-linear tetranuclear complexes were slightly more lipophilic and were taken up to a greater extent by the bacteria, they were consistently less active than their linear counterparts. Of particular note, the cellular accumulation of the oligonuclear ruthenium complexes was greater in the Gram negative strains compared to that in the Gram positive S. aureus and MRSA. The results demonstrate that the lower antimicrobial activity of polypyridylruthenium(II) complexes towards Gram negative bacteria, particularly P. aeruginosa, is not strongly correlated to the cellular accumulation but rather to a lower intrinsic ability to kill the Gram negative cells.

 Please wait while we load your content...
Please wait while we load your content...