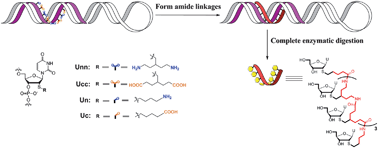
Templated synthesis of nylon nucleic acids and characterization by nuclease digestion†
Abstract
* Corresponding authors
a
Department of Chemistry, New York University, New York, NY 10003, USA
E-mail:
ned.seeman@nyu.edu, james.canary@nyu.edu
Fax: +1 212 995 4367
Tel: +1 212998 8422
b Present address: Department of Chemistry, Northern Michigan University, Marquette, MI 49855, USA

 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?
Y. Liu, R. Wang, L. Ding, R. Sha, N. C. Seeman and J. W. Canary, Chem. Sci., 2012, 3, 1930 DOI: 10.1039/C2SC20129A
To request permission to reproduce material from this article, please go to the Copyright Clearance Center request page.
If you are an author contributing to an RSC publication, you do not need to request permission provided correct acknowledgement is given.
If you are the author of this article, you do not need to request permission to reproduce figures and diagrams provided correct acknowledgement is given. If you want to reproduce the whole article in a third-party publication (excluding your thesis/dissertation for which permission is not required) please go to the Copyright Clearance Center request page.
Read more about how to correctly acknowledge RSC content.
 Fetching data from CrossRef.
Fetching data from CrossRef.
This may take some time to load.
Loading related content
