DOI:
10.1039/D5SD00170F
(Critical Review)
Sens. Diagn., 2026,
5, 165-183
Adenosine detection technologies: recent advances and applications in the central nervous system
Received
20th September 2025
, Accepted 5th November 2025
First published on 19th November 2025
Abstract
Adenosine, a pivotal endogenous neuromodulator, plays a critical role in maintaining homeostasis related to sleep and emotion regulation. Mounting evidence indicates that dysregulated adenosine homeostasis is intricately involved in the pathological processes of brain disorders. Thus, the quantification of adenosine levels is crucial for evaluating disease states in the brain. Currently, numerous reviews have focused on adenosine detection technologies and their applications in tumor immunology and cardiovascular diseases. However, there have been few systematic reviews of adenosine monitoring in the central nervous system. Here, we first systematically summarize recent advances in adenosine detection technologies. Subsequently, we discuss the implications of adenosine detection in the regulation of central nervous system homeostasis. Finally, we highlight current challenges and future prospects, aiming to provide insights for the diagnosis, treatment, and prognosis of neurological disorders.
 Yuqin Liao | Yuqin Liao is currently a master's student at the College of Life Sciences, Fujian Agriculture and Forestry University, under the supervision of Prof. Xiangdong Tian. She obtained her B.E. from Fujian Normal University in 2023. Her present research focuses on designing nanomaterials for bioimaging and bioassay applications. |
 Jiayuan Jing | Jiayuan Jing is currently a master's student at the Fujian Institute of Research on the Structure of Matter, University of Chinese Academy of Sciences, under the supervision of Prof. Yun Zhang. She obtained her Bachelor's degree from Shihezi University in 2023. Her current research focuses on the biomedical imaging and therapy of nanomaterials. |
 Wenkai Jin | Wenkai Jin is currently a master's student at the College of Chemistry and Chemical Engineering, Hunan University, under the supervision of Prof. Quan Yuan. He received his Bachelor's degree in Chemistry from Jishou University in 2024. His current research focuses on the design of dynamic DNA probes for bioimaging and bioanalysis. |
 Xiangdong Tian | Xiangdong Tian is an associate professor at the Fujian Institute of Research on the Structure of Matter, Chinese Academy of Sciences. He received his B.S. in Life Science and M.S. in Biomedical Engineering from Xiamen University, and earned his Ph.D. in Physical Chemistry at Xiamen University under the supervision of Prof. Zhong-Qun Tian. His research focuses on self-assembled plasmonic nanostructures and SERS for biomedical applications. |
 Tianhuan Peng | Tianhuan Peng is an associate professor in the College of Biology at Hunan University. He received his B.S. degree in 2012 from Beijing University of Chemical Technology and his Ph.D. degree in 2017 from the Shanghai Institute of Applied Physics, Chinese Academy of Sciences. He pursued postdoctoral research at Hunan University from 2017 to 2020. His research interests concentrate on the development and application of functional nucleic acid molecules. |
 Lei Zhang | Lei Zhang obtained her B.S. degree from Henan Normal University and her Ph.D. degree from Hunan University. Currently, she is working as a postdoctoral researcher at Wuhan University. Her research focuses on the application of nanoprobes in biomolecular detection. |
 Quan Yuan | Quan Yuan is a professor at Hunan University. She received her B.S. degree from Wuhan University in 2004 and her Ph.D. degree from Peking University in 2009. Afterwards, she worked as a postdoctoral researcher at the University of Florida from 2009 to 2011. Her research focuses on the bioassays of complex samples. |
1. Introduction
Normal physiological functions of the brain rely on the precise regulation of neuronal networks.1–5 Adenosine, as a brain homeostatic modulator, exerts its effects by activating specific receptor-mediated signaling pathways.6–12 Specifically, adenosine protects neurons by hindering neuroinflammation, oxidative stress, and apoptosis; however, an imbalance in adenosine levels abolishes this benefit and may even promote neuroinflammation, hastening the progression of brain diseases.13–17 Accurate measurement of adenosine levels in the brain is essential for understanding its neuromodulatory functions. Nevertheless, the rapid metabolism of adenosine presents a considerable challenge for achieving reliable measurements in the brain.18 The development of monitoring technologies with high spatiotemporal resolution is therefore imperative.
A wide range of adenosine detection technologies has been developed.19–23 High-performance liquid chromatography (HPLC) is generally recognized as the gold standard due to its exceptional stability and reliability.24 However, the complex workflow hinders adenosine monitoring with high spatiotemporal resolution in vivo.25 Electrochemical methods offer sub-second temporal resolution but are limited by invasiveness and crosstalk interference from adenosine analogues, compromising measurement accuracy.26,27 Compared to non-optical methods above, optical methods offer reduced invasiveness and superior spatiotemporal resolution, enabling high-sensitivity monitoring of adenosine in vivo.28,29
Comprehensive reviews have discussed adenosine detection technologies and elucidated the pathophysiological roles of adenosine in various cancers.30–37 However, recent advances in adenosine monitoring and their applications in neural circuits remain largely unreviewed. In this review, we first present recent progress in adenosine monitoring technologies, followed by a discussion of their roles in sleep–wake regulation, emotional modulation, and neurological disorders. Finally, we discuss the prevailing challenges and future directions of adenosine detection for uncovering brain states (Fig. 1). This review is expected to provide novel insights for future research in this field.
 |
| | Fig. 1 Overview of adenosine detection techniques and their applications in the central nervous system. | |
2. Technological advances of adenosine detection
Given its essential physiological roles, adenosine has been recognized as a valuable biomarker for both disease diagnosis and therapeutic monitoring.38 Therefore, the development of highly sensitive adenosine detection strategies is of great importance (Fig. 2). In this part, we first outline non-optical detection approaches, then discusses optical methods such as fluorescence and chemiluminescence in detail, and finally compares the characteristics of all mentioned techniques.
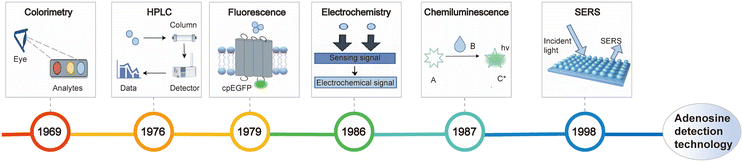 |
| | Fig. 2 Timeline of technologies for adenosine detection. | |
2.1 Non-optical detection methods
2.1.1 High-performance liquid chromatography.
HPLC, a robust technique based on liquid partition chromatography, has been widely utilized for adenosine detection because of its high reliability. The method was initially applied to adenosine detection in 1976.39 Wojcik et al. presented excellent linearity for standard adenosine samples ranging from 100 fmol to 100 pmol using HPLC.40 Integration of HPLC with multidimensional analytical platforms has significantly improved the sensitivity and accuracy of adenosine detection. For instance, Lovatt et al.41 and Marin et al.42 employed HPLC coupled with ultraviolet detection to quantify adenosine at 260 nm. Marin et al. further achieved precise detection using reversed-phase HPLC coupled with ultraviolet detection, reaching a lower limit of detection (LOD) of 6.25 pmol per sample. HPLC is also compatible with microdialysis and mass spectrometry (MS). Using microdialysis-HPLC-tandem MS, Cannazza et al. observed that adenosine levels in mice initially increased, then returned to baseline following amphetamine administration, and remained stable after cocaine administration.43 This method enables the monitoring of dynamic changes in extracellular adenosine within the mouse nucleus accumbens. HPLC-MS/MS technique not only mitigates matrix interference from complex biological samples, allowing detection of adenosine at concentrations as low as picograms per milliliter in blood or tissues, but also offers high-throughput analysis capabilities. It is noteworthy that blood samples require enzyme inhibition and low-temperature processing within seconds of collection to prevent rapid adenosine degradation.44
Despite its widespread use, HPLC still faces challenges such as inadequate analyte retention and limited resolution. To this end, Olesti et al. introduced hydrophilic interaction liquid chromatography as an alternative to reversed-phase high-performance liquid chromatography.45 The hydrophilic interaction liquid chromatography mode, which employs organic solvent mobile phases, markedly enhances adenosine retention and achieves superior separation efficiency. This approach has been applied to the quantitative detection of adenosine in rat plasma and brain homogenates. Subsequently, Virgiliou et al. optimized the method by mixing samples in vacuum blood collection tubes immediately upon collection, effectively inhibiting enzymatic degradation of adenosine.46 This optimized method exhibited exceptional specificity and sensitivity with a LOD of 0.005 μg mL−1, but its time consumption hinders the direct measurement of adenosine from becoming a routine clinical assay. Moreover, this method is limited to measuring adenosine levels in a static state in brain sections, not in vivo.
2.1.2 Electrochemical methods.
Owing to their advantages of rapid response and excellent temporal resolution, electrochemical methods have become standard techniques for adenosine monitoring. The year 1986 marked the first use of this method in demonstrating adenosine's regulation of striatal neurotransmitter release.47 To investigate the relationship between adenosine release and neuronal activity in distinct brain regions, Cechova et al. employed fast-scan cyclic voltammetry (FSCV) to demonstrate that transient neuronal activity induced a large increase in extracellular adenosine, reaching a concentration of 0.94 ± 0.09 μM in the rat caudate-putamen following electrical stimulation.48 Similarly, Venton's group explored the relationship between adenosine release and cerebral blood flow.49 Using FSCV, the authors investigated transient adenosine release in the caudate-putamen and showed that adenosine modulates cerebral blood flow on a sub-second timescale. Furthermore, the mechanisms of rapid adenosine release vary across brain regions. Pajski et al. utilized FSCV to confirm these differences.50 Through electrical stimulation experiments in distinct brain regions, they found that adenosine efflux within the caudate-putamen is dependent on the activation of ionotropic glutamate receptors, whereas release in the nucleus accumbens, cortex, and hippocampus is primarily mediated by the breakdown of extracellular ATP. Chang's group further advanced this approach by developing dual-channel FSCV.51 They characterized the spatial extent of rapid adenosine signaling by varying the distance between the working electrodes within brain slices. This dual-electrode configuration enabled simultaneous recordings from two electrodes, allowing synchronous monitoring of adenosine release within the hippocampal CA1 region on a microscopic timescale. Notably, comparisons of signal amplitude and frequency enhanced the resolution of adenosine release. Despite the success of FSCV in resolving the rapid dynamics of adenosine in the brain, its instability and limited selectivity in complex environments impede its application for long-term monitoring. To achieve highly selective and stable monitoring, Chang et al. employed a multi-enzyme cascade modification strategy integrating nucleoside phosphorylase (NP), adenosine deaminase (ADA), and xanthine oxidase (XO).52 This tri-enzyme cascade catalytic reaction ultimately converted adenosine into hydrogen peroxide (H2O2), with adenosine (ADO) levels indirectly being measured by changes in H2O2 current (Fig. 3A). Then, continuous electrochemical monitoring of adenosine in anesthetized rats showed that implanted microsensors in the gastrocnemius muscle generated a significant current response upon injection of 4 mM adenosine (Fig. 3B). In addition, injections of 4 mM and 10 mM adenosine induced a dose-dependent increase in current response within the dorsal striatum (Fig. 3C). These data confirmed that the sensor is capable of monitoring exogenous adenosine levels in vivo. On this basis, Shim's team designed a dual-electrode sensor for adenosine detection.53 They immobilized ADA and NP onto a gold nanoparticle-modified electrode, whereas XO and hydrazine were immobilized onto a porous gold-modified electrode. This design enhanced enzyme loading and activity by immobilizing distinct enzymes on two independent electrodes, ultimately yielding an 8.2-fold sensitivity improvement compared to a single-electrode sensor. In 2020, Venton et al. recorded cyclic voltammograms to reveal transient adenosine release.54 This method visualized adenosine levels by mapping current values at different time points and potentials to electrochemical signatures, represented by color-coded representations. To further expand clinical application potential, Lan et al. constructed an adenosine electrochemical sensor incorporating PtCu nanoparticles anchored on multi-walled carbon nanotubes, exploiting the high surface area and excellent conductivity of multi-walled carbon nanotubes to increase the active site density of PtCu nanoparticles.55 This configuration increased the electrocatalytic efficiency towards H2O2 production generated from adenosine oxidation, thereby indirectly amplifying the adenosine signal. With a LOD of 1.0 nM under optimal conditions and successful application in real serum samples, this sensor showed potential for clinical diagnostics. Additionally, Tian et al. constructed a label-free electrochemical aptasensor for adenosine detection using hollow bimetallic ZnNi MOF microspheres. By optimizing the molar ratio of Zn2+ to Ni2+, the material was endowed with enhanced electrochemical activity and strong aptamer binding affinity of adenosine. The developed sensor achieved a LOD of 20.32 fg mL−1 with excellent reproducibility, demonstrating its reliability for precise clinical adenosine detection and showing significant potential in early tumor diagnosis and auxiliary diagnosis of brain disorders.56 It should be noted that the implantation of microelectrodes induces inevitable physical injury to local brain tissue and microvasculature, while also triggering a series of neuroinflammatory responses. This pathological cascade, characterized by glial activation and immune cell infiltration, fundamentally alters the pericellular microenvironment. The resulting inflammation disrupts purinergic signaling pathways and distorts the adenosine levels that the technique aims to measure. Moreover, substantial individual variability in inflammatory intensity combined with the unpredictable duration of this pathological state undermines the reliability of long-term clinical monitoring.
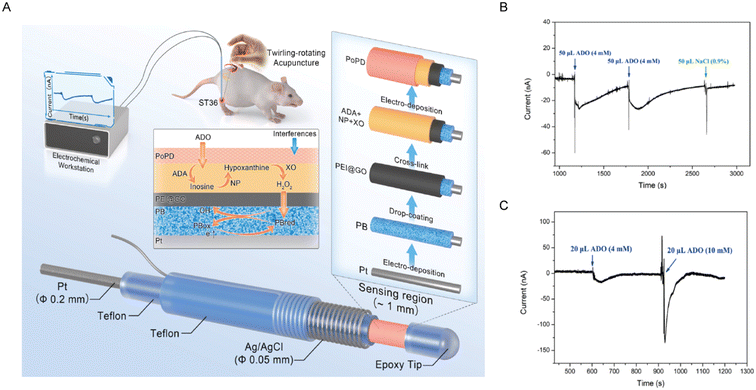 |
| | Fig. 3 (A) Schematic illustration of the fabrication and electrochemical application of adenosine microelectrode sensors. (B) Current response of implanted microsensors in the crus muscle following dual 4 mM adenosine or normal saline injections. (C) Current response of implanted microsensors in dorsal striatum following 4 mM or 10 mM adenosine injections. Reproduced with permission from ref. 52. Copyright 2023, Elsevier. | |
2.2 Optical detection methods
HPLC and electrochemical techniques for adenosine detection suffer from limitations, including limited spatiotemporal resolution and inherent invasiveness. There is an increasing demand for highly sensitive, real-time, and minimally invasive adenosine detection methods. Optical detection techniques offer several advantages, including noninvasiveness, superior spatiotemporal resolution, and excellent biocompatibility. These properties substantially reduce interference with the tissue microenvironment, enabling real-time monitoring and precise quantification of adenosine in complex biological environments. This section will focus on optical-based detection strategies, including fluorescence, colorimetry, chemiluminescence, and other approaches.
2.2.1 Fluorescence.
Fluorescence-based detection relies on the selective binding of fluorescent probes to adenosine for highly sensitive monitoring. The first detection of adenosine was achieved through fluorescence spectroscopy in 1979.57
Owing to its accuracy and rapid response, this approach is widely employed for measuring adenosine in biological samples. Multiple fluorescent probes based on diverse luminescence strategies have been engineered. For instance, a label-free sensor was constructed using a dimeric G-quadruplex and the fluorophore Thioflavin T. The displacement of Thioflavin T occurs in the presence of adenosine, resulting in a decreased fluorescence signal. This sensor exhibited a linear response to adenosine concentrations ranging from 0.5 to 120 μM, with a LOD of 245 nM.58 Besides, cascade amplification approaches have been used to further enhance the sensitivity of adenosine determination. As proof, Feng's team established two specific initiator chains that cooperatively triggered a hybridization chain reaction upon adenosine binding.59 This strategy effectively suppressed non-specific responses from the hairpin structures, improving the adenosine detection sensitivity to 2.0 × 10−7 M. Similarly, Quan et al. proposed a catalytic reaction-triggered hybridization chain reaction for adenosine detection.60 Through a catalytic cycle for recycling of intermediates, this strategy not only allowed real-time adenosine detection with a sensitivity down to 200 pM but demonstrated high reliability. Notably, Förster resonance energy transfer-based approaches have gained attention due to their unique “off–on” mechanism and excellent anti-interference capability. An example is a ratiometric fluorescent sensor based on the fluorescence quenching between CdS quantum dots and graphene oxide.61 In the absence of adenosine, graphene oxide significantly reduces the fluorescence of CdS quantum dots. Upon adenosine binding, fluorescence is recovered, enabling quantitative detection with a LOD of 1.3 nM. Cheng et al. further designed a ratiometric fluorescence sensor with dual-recognition capability.62 The sensor achieved highly selective adenosine recognition via the synergistic interaction of boronic acid and molecularly imprinted polymers. On the one hand, boronic acid possessed the ability for specific binding with the cis-diol groups in adenosine, providing the first recognition pathway. On the other hand, adenosine was incorporated as a template molecule during the preparation of molecularly imprinted polymers, creating imprinted cavities highly complementary to adenosine in shape and size. This dual-recognition strategy significantly suppressed non-specific interference and enhanced the selectivity and sensitivity, enabling adenosine detection at concentrations as low as 0.26 mg L−1. In addition, Chen et al. constructed a DNAzyme nanoreactor that enables precise detection of adenosine via conformational changes of nucleic acids, achieving a detection limit of 2.1 nM. This nanoreactor demonstrates excellent accuracy and specificity for adenosine assays. Significantly elevated adenosine levels detected in the urine of liver cancer patients compared to healthy controls suggest this nanoreactor's promise as a non-invasive, metabolic biomarker for early tumor screening and treatment surveillance.63 However, many adenosine detection strategies, which rely on designed DNA nanostructures or composite nanomaterials, are susceptible to enzymatic degradation, non-specific adsorption, and instability in complex clinical samples.64,65
As an in vivo neurorecording technique, fiber photometry offers sub-second temporal resolution and nanomolar sensitivity. This is crucial for capturing transient adenosine dynamics during neural activity. In 2020, Xu's group developed a genetically encoded fluorescent sensor based on activation of a G protein-coupled receptor (Fig. 4A).66 Using an adeno-associated virus vector, they constructed the GRABAdo sensor to monitor adenosine in specific brain regions. When combined with a fiber photometry system, this sensor enabled real-time tracking of fluorescence changes that report adenosine dynamics in live mice. GRABAdo has since been extensively utilized in neuroscience research. For example, to investigate the neural mechanisms of motor regulation, Zhong's group expressed GRABAdo in neurons of the mouse dorsolateral striatum and discovered a significant increase in adenosine levels during exercise (Fig. 4B), with the magnitude of this increase positively correlating with exercise intensity (Fig. 4C).67 Despite extensive research on adenosine function, its specific origins in the brain remain debated. To address this, Wu et al. expressed GRABAdo in neurons of brain regions, such as the hippocampus, and measured adenosine levels under different laser stimulation frequencies.68 Their findings showed that adenosine release induced by neural activity originates from the somatodendritic compartment, rather than the previously assumed axon terminals. This study provides a new perspective on adenosine release mechanisms and their role in neural signaling. It is important to note, however, that the antagonist ZM-241385 can inhibit GRABAdo, reducing its basal fluorescence and potentially leading to the erroneous results of endogenous adenosine levels. Building on this, Wei et al. recently developed an innovative genetically encoded fluorescent probe for monitoring intracellular adenosine.69 This probe integrated a circularly permuted enhanced green fluorescent protein with the adenosine deaminase from Plasmodium vivax, enabling rapid and specific detection of intracellular adenosine dynamics (Fig. 4D). In Drosophila, the researchers leveraged the probe to detect real-time intracellular adenosine dynamics (Fig. 4E). Furthermore, in acute brain slice experiments, the probe exhibited dose-dependent fluorescence responses to adenosine while showing minimal response to inosine, confirming its excellent selectivity and fast response kinetics (Fig. 4F). In summary, this strategy provides a robust approach for adenosine monitoring with high spatiotemporal resolution in vivo.
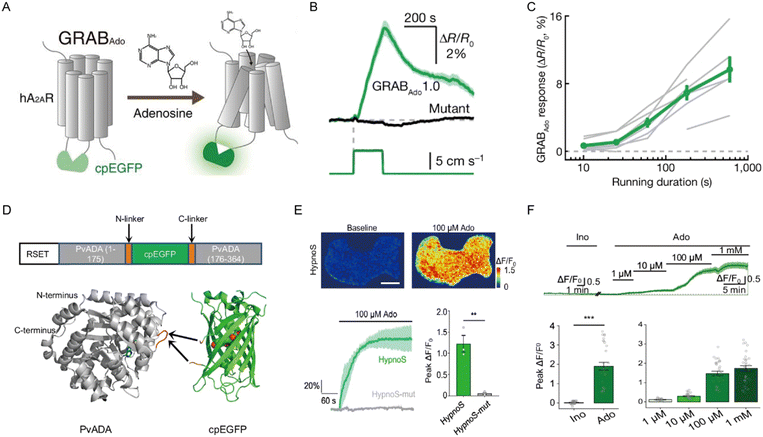 |
| | Fig. 4 (A) The working mechanism of the GRABAdo fluorescent sensor for adenosine detection. Reproduced with permission from ref. 66. Copyright 2020, The American Association for the Advancement of Science. (B) Averaged fluorescence responses of GRABAdo (green) and mutant (black) stimulated by running. (C) Correlation between adenosine signal responses and running duration. Reproduced with permission from ref. 67. Copyright 2022, Springer Nature. (D) Design principle of the genetically encoded sensor. (E) Representative images, fluorescence response curve, and aggregated data of ΔF/F0 in response to 100 μM adenosine. (F) Representative fluorescence response profiles of adenosine sensor to varying adenosine concentrations and 100 μM inosine in brain slices. Reproduced with permission from ref. 69. Copyright 2025, Springer Nature. | |
2.2.2 Colorimetry.
Colorimetry quantifies analytes based on visual observation or spectrophotometric readout of color change and has been widely applied in detection. Since adenosine is colorless and lacks light-absorption properties at specific wavelengths, it cannot be directly monitored by absorbance measurements. Therefore, colorimetric detection of adenosine typically requires enzymatic or chemical reactions to convert adenosine into substances possessing color or light absorption characteristics. This method was applied to the determination of adenosine at the micromolar level in 1969.70 Metal nanoparticles have become an ideal strategy for colorimetric analysis because of their high molar extinction coefficients. Among them, gold nanoparticles are particularly suitable for the specific recognition of target molecules due to their unique surface plasmon resonance effect. Based on this principle, Liu and Lu exploited the synergistic action in which functionalized gold nanoparticles and linker DNA were used to monitor adenosine.71 In the absence of adenosine, nanoparticle aggregation mediated by base pairing via linker DNA yielded a purple solution (Fig. 5A). However, adenosine binding to the aptamer induces a conformational change, causing nanoparticle dissociation and a color shift from purple to red. Tracking the extinction ratio of the plasmon resonance peak revealed a rapid color transition only in adenosine-containing samples, validating its high selectivity for adenosine detection (Fig. 5B). Notably, the duration of the color transition correlated positively with adenosine concentration, enabling quantitative analysis (Fig. 5C). After that, Zhang et al. proposed a colorimetric assay based on the self-assembly of functionalized gold nanoparticles.72 Its core design leveraged aptamer-specific adenosine binding to initiate nanoparticle assembly, inducing a significant surface plasmon resonance shift (Fig. 5D). The researchers applied this method to quantify adenosine in serum at concentrations as low as 10 pM (Fig. 5E), providing a highly efficient and sensitive approach for the analysis of biological samples. With this strategy, the LOD for adenosine was 3.6 pM (Fig. 5F). Zhu's team engineered a dual-sensing system using gold nanoparticles.73 Specifically, adenosine triggered controlled gold nanoparticle aggregation, exploiting surface plasmon resonance effects to induce a color transition from blue to red. Through optimization of surface modification density and ionic strength, the system maintained over 90% specificity for adenosine recognition even with competing nucleotides present. Innovatively, Xu's team designed an enzymatic cleavage signal amplification mechanism.74 Within their dual-hairpin DNA system, adenosine binding initiated an Exonuclease III-mediated cyclic cleavage reaction. This process released guanine-rich sequences that formed G-quadruplex structures, catalyzing the oxidation of 3,3′,5,5′-tetramethylbenzidine to produce measurable color changes. With this strategy, a LOD as low as 17 nM for adenosine was achieved. Moreover, it avoided the use of precious metal nanomaterials, reducing detection costs by approximately 40%. Taken together, these innovative methodologies effectively circumvent the inherent limitations of direct colorimetric assays for adenosine detection—such as insufficient sensitivity and susceptibility to matrix interference—while maintaining operational simplicity and analytical robustness. The strategic integration of signal amplification mechanisms with cost-effective materials represents a significant advancement in the development of practical biosensing systems for routine adenosine monitoring. However, in hemolyzed samples, the release of hemoglobin introduces a strong background absorbance that overlaps with the typical detection wavelengths for adenosine. Similarly, the turbidity and high lipid content in lipemic sera can cause significant light scattering and nonspecific absorption, thereby distorting the accurate colorimetric readout.75,76 Overcoming this spectral interference barrier is pivotal for advancing colorimetric strategies toward more robust and routine clinical use.
 |
| | Fig. 5 (A) Design of a nanoparticle aggregation system based on adenosine-triggered disassembly for adenosine detection. (B) Kinetic profiles of adenosine aptamer-assembled nanoparticle aggregates for nucleoside discrimination. (C) Kinetic profiles of the concentration-dependent color changes in adenosine aptamer-assembled nanoparticle aggregates. Reproduced with permission from ref. 71. Copyright 2006, Springer Nature. (D) Design of gold nanoparticle-based colorimetric adenosine sensor. (E) UV-vis spectroscopic characterization of gold nanorod in serum samples spiked with varying adenosine concentrations. (F) The LOD of adenosine in the colorimetric assay. Reproduced with permission from ref. 72. Copyright 2019, Springer Nature. | |
2.2.3 Chemiluminescence.
Chemiluminescence has emerged as an analytical tool for adenosine detection, primarily owing to its rapid analysis and straightforward operation. The year 1987 saw the first application of this method for adenosine measurement.77 Recent years have witnessed growing efforts to boost the sensitivity of chemiluminescence sensors to meet the demand for detecting adenosine in complex samples. For instance, Asher's group devised an innovative sensor based on a DNA-crosslinked two-dimensional photonic crystal (2DPC) hydrogel for detecting adenosine.78 This sensor operates through the specific binding of an aptamer to adenosine that selectively cleaves DNA crosslinks and triggers expansion of the hydrogel. The expansion altered the particle spacing of the embedded 2DPC array, thereby causing a red-shift in the Bragg diffraction peak of the photonic crystal (Fig. 6A). Consequently, adenosine levels could be determined by measuring the change in Bragg diffraction. Importantly, the sensor exhibited minimal response to other nucleosides, such as cytidine, guanosine, and uridine (Fig. 6B). This sensor showed a LOD of 13.9 μM in adenosine-binding buffer (Fig. 6C). Combined with its rapid response time, these attributes rendered the 2DPC hydrogel platform an ideal solution for applications in low-resource environments. This approach not only improves sensitivity via multistage signal amplification but also exploits the environmentally responsive properties of DNA hydrogels to enhance their anti-interference capability. In short, the integration of DNA hydrogels with metal–organic frameworks offers a practical approach for adenosine detection in clinical application. As a notable example, an ultrasensitive sensor based on a target-responsive DNA hydrogel was developed by Lin et al.79 This sensor incorporated an acrylamide-modified adenosine aptamer, a single-stranded DNA linker, and a hemin aptamer, all encapsulated within a copper-based metal–organic framework possessing peroxidase-like activity. The catalytic performance and signal response were further optimized by incorporating gold nanoparticles into the cavities of HKUST-1, forming an Au@HKUST-1 composite. In the presence of hemin, the hemin aptamer formed a G-quadruplex/hemin DNAzyme complex, stabilizing the hydrogel structure. However, when adenosine was introduced, its specific binding to the acrylamide-modified adenosine aptamer triggered the dissociation of the hydrogel network, leading to hydrogel dissolution and subsequent release of the pre-encapsulated G-quadruplex/hemin complex and Au@HKUST-1. The released G-quadruplex/hemin complex then catalyzed the generation of a strong chemiluminescence signal in the luminol–H2O2 system (Fig. 6D). Furthermore, the Au@HKUST-1 synergistically catalyzed this reaction through its intrinsic peroxidase-like activity, achieving dual-signal amplification. With its outstanding sensitivity, this biosensor exhibited an exceptional LOD of 0.104 pM and demonstrated a robust linear response to adenosine (Fig. 6E). Validation studies in complex matrices confirmed the practicality and accuracy of this method, as evidenced by adenosine quantification in human urine. Nevertheless, the translation of this biosensing platform to serum-based clinical diagnostics presents substantially greater challenges. When deployed in undiluted human serum, the assay demonstrates heightened susceptibility to interference from both endogenous radical scavengers and structurally similar metabolites. These interfering species compete for binding sites and generate false-positive signals through non-specific interactions, ultimately compromising measurement reliability.80,81
 |
| | Fig. 6 (A) DNA sequences designed to form adenosine-responsive crosslinks within the hydrogel (up) and the design of adenosine sensing mechanism in DNA-crosslinked 2D photonic crystal (2DPC) hydrogels (down). (B) Selectivity response of 2DPC hydrogels to adenosine and interferents. (C) Concentration-dependent response of 2DPC hydrogel particle spacing to adenosine in buffer. Reproduced with permission from ref. 78. Copyright 2022, American Chemical Society. (D) Design of chemiluminescence biosensor for adenosine detection. (E) Standard curve of adenosine detection using the chemiluminescence biosensor. Reproduced with permission from ref. 79. Copyright 2019, Elsevier. | |
2.2.4 Other optical methods.
Beyond the aforementioned optical methods, surface plasmon resonance-based techniques, such as surface-enhanced Raman scattering (SERS), have garnered attention for their ability to facilitate highly sensitive, real-time and label-free detection of adenosine by monitoring optical signal changes at nanostructured composites. In 1998, Li et al. detected adenosine in separated solutions using the SERS method.82 Then, utilizing the Raman signal of graphene as a normalization reference across the entire Raman spectrum not only improves signal stability but also yields more reliable results for adenosine monitoring through synergistic enhancement effects. To enhance biocompatibility and improve the accuracy of adenosine detection, Qiu's group developed a graphene-gold nanoparticle composite substrate (Fig. 7A).83 They observed a strong linear correlation (Fig. 7C) between Raman intensity and adenosine concentration in both standard solutions (Fig. 7B) and diluted serum (Fig. 7D). The LOD for adenosine in serum samples reached 5 nM (Fig. 7E). Notably, the high reliability of the substrate was evidenced by its undegraded performance even after thermal oxidation treatment. To address the challenges posed by complex biological samples, SERS has been applied to diverse gold nanostructures that achieve direct detection in undiluted serum. This is accomplished through data augmentation and feature selection strategies, thereby overcoming the high dependence on sample pretreatment that is often required by non-optical methods.84 However, SERS is intrinsically susceptible to endogenous fluorescence and spectral overlap from proteins and metabolites, which can obscure or distort the adenosine signal. The ensuing data-processing further increases uncertainty, preventing the technique from truly tracking rapidly changing adenosine levels in a clinical setting.85 Although advanced computational algorithms can partially mitigate these effects, the requisite data processing introduce additional analytical uncertainty. Consequently, current SERS platforms still struggle to reliably track adenosine concentration in real-time clinical monitoring scenarios, particularly in critical care and neurosurgical settings where minute-to-minute variation monitoring is crucial.
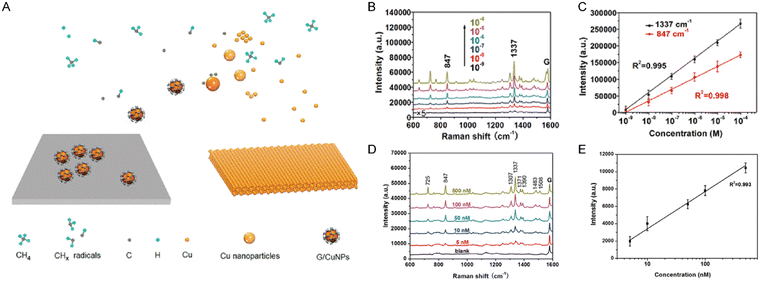 |
| | Fig. 7 (A) Design of the surface-enhanced SERS substrate for adenosine detection. Raman spectra of different adenosine concentrations in the samples (B) and the curves of linearity for adenosine peak at 847 and 1337 cm−1 (C). Raman spectra of different adenosine concentrations in diluted serum (D) and the curve of adenosine peak at 1337 cm−1 (E). Reproduced with permission from ref. 83. Copyright 2015, American Chemical Society. | |
2.3 Comparison of adenosine detection strategies
Each detection strategy exhibits significant differences in terms of analytical performance and application feasibility (Table 1). HPLC remains the clinical gold standard for quantitative accuracy and selectivity, but lacks real-time capability. Electrochemical sensors and genetically encoded fluorescent probes offer high temporal resolution and are suitable for monitoring transient adenosine fluctuations in the brain, but due to their invasiveness, their clinical compatibility is limited. Optical nanoprobes strike a balance between spatial resolution and invasiveness, demonstrating the potential for deep tissue imaging and future translation. In contrast, colorimetry and chemiluminescence, although having relatively lower temporal resolution, have excellent selectivity and clinical applicability and can be used for immediate diagnosis. In summary, these comparisons indicate that no single method is universally applicable; instead, the best choice depends on whether the research goal is real-time neurochemical dynamics or clinical quantitative requirements. The integration of multimodal sensing technologies may bridge this gap in future developments.
Table 1 Comparison of different adenosine detection technologies
| Method |
Invasiveness |
Selectivity |
Temporal resolution |
Key advantages |
Main limitations |
LOD |
Feasibility for in vivo application |
Potential for clinical translation |
Ref. |
| Legend: ✓ indicates the degree of feasibility or potential for clinical application (✓ < ✓✓ < ✓✓✓); ✗ denotes ex vivo application only or major barriers to clinical translation. |
| HPLC |
High |
High |
Minute |
Stable, matrix-tolerant |
Inadequate analyte retention; time-consuming |
6.25 pmol/sample |
✗ |
✓✓✓ |
42
|
| Electrochemistry |
High |
Moderate |
Sub-second |
Real-time, sensitive |
Invasive; poor long-term stability/selectivity |
20.32 fg mL−1 |
✓✓✓ |
✓ |
56
|
| Fluorescence |
Moderate |
High |
Millisecond-to-second |
High spatiotemporal resolution |
Viral inflammation; photobleaching |
1.3 nM |
✓✓ |
✗ |
62
|
| Colorimetry |
Low |
High |
Minute |
Visual, simple, cheap |
Color interference in hemolyzed/lipemic sera |
3.6 pM |
✗ |
✓ |
72
|
| Chemiluminescence |
Moderate |
High |
Minute |
Ultra-sensitive, dual amplification |
Reagent stability; matrix interference |
0.104 pM |
✗ |
✓✓✓ |
79
|
| Other methods |
Moderate |
High |
Second |
Label-free, serum-ready |
Protein/metabolite spectral overlap, complex processing |
5 nM |
✓ |
✓✓ |
83
|
3. Roles of adenosine detection technologies in neural circuit
Adenosine mediates diverse physiological and pathological processes ranging from sleep regulation to synaptic plasticity and dynamic neural networks through its actions on specific receptor subtypes.86 Determining adenosine concentrations in cells and tissues can offer valuable information on the states of health and disease. In this section, we will elaborate on the roles of adenosine, with a particular focus on its applications in sleep and memory, emotional modulation, and neurological disorders.
3.1 Roles in sleep and memory regulation
Adenosine serves as a key regulator in sleep homeostasis, with its levels dynamically fluctuating in response to wakefulness and sleep. Accumulating evidence has outlined that adenosine levels rise during wakefulness but decline markedly during sleep (Fig. 8D).87 This fluctuation primarily stems from activity-dependent metabolic processes in neurons, where sustained wakefulness leads to the accumulation of adenosine in the extracellular milieu, thereby increasing sleep pressure and triggering non-rapid eye movement (NREM) sleep.88 Peng et al. employed high-resolution sensors to detect extracellular adenosine in the murine basal forebrain, confirming state-dependent fluctuations with adenosine concentrations peaking during wakefulness and declining to baseline levels during NREM sleep (Fig. 8A).66 Crucially, the millisecond-scale resolution probe uncovered previously undetectable adenosine transients during brief rapid eye movement (REM) sleep, suggesting that adenosine regulates slow-wave activity primarily through the direct modulation of thalamocortical neurotransmission. To investigate the function of adenosine in the regulation of sleep–wake cycles, Zhu et al. injected the adenosine analog NECA into brain regions of freely moving mice, resulting in a large increase in wakefulness.89 Subsequent optogenetic activation of astrocytes indicated that endogenous adenosine induces arousal via adenosine A1 receptors (A1R). Strikingly, pharmacological blockade of A1R substantially reduced transition probabilities from both NREM and REM sleep to wakefulness, highlighting the critical role of A1R in the astrocyte-mediated wakefulness induction. Increased A1R availability and activation following sustained wakefulness have been observed in both mice and humans.90
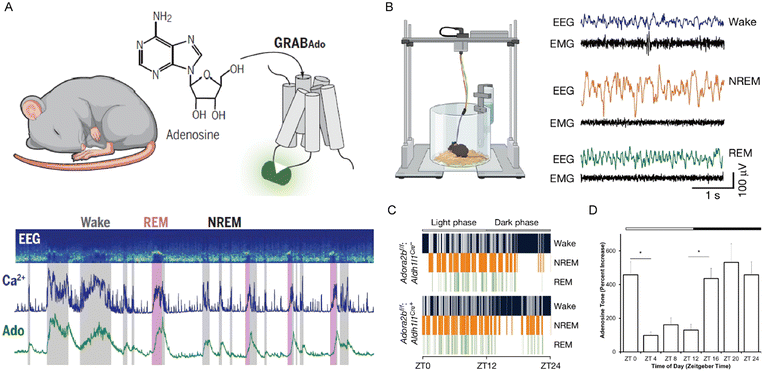 |
| | Fig. 8 (A) Schematic illustration of adenosine optical probes designed to record neural activity in the mouse basal forebrain during the sleep–wake cycles. Reproduced with permission from ref. 66. Copyright 2020, The American Association for the Advancement of Science. (B) Schematic representation of the electroencephalogram and electromyogram recording system and signals during the sleep–wake cycles. (C) Representative signals of sleep–wake stages recorded by electroencephalogram. Reproduced with permission from ref. 94. Copyright 2024, Springer Nature. (D) Adenosine dynamic changes in sleep–wake cycles. Reproduced with permission from ref. 87. Copyright 2012, Society for Neuroscience. | |
Adenosine dynamically regulates sleep via downstream signaling pathways of adenosine receptors.91 Early studies confirmed that A1R activation can maintain sleep homeostasis by suppressing cholinergic neurons that promote wakefulness in the basal forebrain and glutamatergic neurons in the preoptic area, thereby disinhibiting cortical slow-wave activity and forming a negative feedback loop.87 After that, neuroimaging studies in humans suggested that prolonged sleep deprivation increased A1R expression levels, which normalized following recovery sleep.90 The upregulation of A1R is a form of adaptation by the brain to this persistent high level of adenosine. This adaptive response aims to enhance the brain's sensitivity to “sleep pressure” signals, thereby reducing neuronal excitability and promoting the need for sleep. Meanwhile, adenosine A2A receptors (A2AR) signaling plays an important role in sleep regulation. The primary mechanism by which systemic caffeine induces arousal is antagonism of A2AR, not A1R.92 Gomez-Castro et al. treated CA1 pyramidal neurons with ADA and a CD73 inhibitor to reduce extracellular adenosine levels.93 Through methods such as super-resolution imaging and fluorescence labeling, they found that reduction of extracellular adenosine induces synaptic instability, yet selective activation of A2AR prevents this effect. In brief, the role of the A2AR in maintaining synaptic integrity was confirmed. Adenosine A2B receptors (A2BR) are another essential mediator of sleep homeostasis. Theparambil et al. employed the electroencephalogram and electromyogram recordings to demonstrate that genetic ablation of A2BR specifically in hippocampal astrocytes disrupted sleep behaviors and reduced NREM sleep (Fig. 8B and C).94 Therefore, these findings indicate that sleep–wake transitions are controlled by a molecular switch involving balanced adenosine receptors-mediated signaling. The coordinated action of the adenosine system has a profound influence on cognitive processes through its regulatory effects on synaptic plasticity and memory consolidation. These effects are particularly prominent during sleep, as physiological declines in adenosine levels create permissive conditions for memory consolidation.90 In contrast, under pathological conditions such as sleep deprivation, adenosine accumulates significantly in the striatum and cerebellum, impairing hippocampal memory and then suppressing cAMP/PKA signaling pathways through A1R overactivation.95 Moreover, accumulated sleep pressure from prolonged wakefulness can induce similar cognitive impairments.96 Studies have reported that astrocyte-derived adenosine can modulate both memory consolidation and cognitive flexibility through A1R signaling.97 Halassa et al. employed the novel object recognition test to investigate the role of astrocyte-derived adenosine in sleep deprivation-associated cognitive performance.98 Through electrophysiological techniques, they found that sleep deprivation impaired memory in wild-type mice, whereas mice with dominant-negative astrocytic SNARE were protected from these impairments. Importantly, the memory impairment in sleep-deprived wild-type mice could be rescued after pharmacological blockade of A1R. This result directly revealed the pathway of adenosine release from astrocytes and its action on A1R as a core mechanism regulating synaptic function in sleep homeostasis. Although A1R antagonists can temporarily reverse memory impairments caused by sleep deprivation, their non-selectivity may disrupt neuronal mechanisms for sleep pressure in the long term.99 This suggests that blockade of adenosine receptors may provide short-term benefits under specific conditions, yet chronic interventions perturb normal sleep–wake cycles and may produce adverse effects, hence this factor must be considered when using adenosine for sleep disorders. In conclusion, adenosine exerts multifaceted regulatory effects on sleep homeostasis and cognitive processes through its activation in neural circuits.100,101
3.2 Involvement in depression and anxiety
Depressed mood is a core clinical symptom of depressive disorders. Adenosine accumulation and release in the brain have been implicated in the regulation of neural circuits underlying emotion. Yuan et al. recently introduced a NIR-activatable nanosensor that enables high-spatiotemporal imaging of adenosine in the intact mouse brain.102 This nanosensor remains optically silent in the absence of adenosine and is activated only in the presence of adenosine and NIR irradiation (Fig. 9A). In animals subjected to chronic unpredictable mild stress, cerebral fluorescence remained at baseline, whereas a marked, selective fluorescence increase was observed exclusively in the brains of depression-model mice, reflecting that adenosine levels were markedly higher in depressed mice than in normal controls. Pharmacological validation with the antidepressant fluoxetine reversed the elevated adenosine levels (Fig. 9B and C). Collectively, this work established a direct method for adenosine detection and demonstrated a relationship between depressive states and adenosine levels. Although the total adenosine levels show an upward trend in the depressed state, due to the significant differences in the expression distribution and signal transduction efficiency of A1R and A2AR in different brain regions and neuronal types, this global increase may lead to the disruption of the signal balance between the receptors. Furthermore, A1R and A2AR may have opposing roles in the progression of depressive disorders. Indeed, A1R activation exerts antidepressant effects and increases treatment sensitivity, although prolonged activation may lead to receptor desensitization.95 Conversely, A2AR activation has been shown to engender depression-like behaviors. These findings underscore the diverse involvement of adenosine receptor subtypes in the pathophysiology of depression. Moreover, Wang et al. created a chronic-stress mouse model to simulate depressive symptoms observed in humans.103 Their experiments identified that A2AR directly connected the lateral septum (LS) with both the dorsomedial hypothalamus (DMH) and the lateral habenula (LHb). Likewise, imaging studies revealed that A2AR in the LS is predominantly expressed in GABAergic neurons. A2AR upregulation in the LS enhanced the firing frequency of LS-A2AR neurons and hindered surrounding LS neurons. Importantly, this activation of A2AR in the LS subsequently activated the DMH and LHb, ultimately causing depression-like behaviors. Of particular note, local application of the selective A2AR antagonist KW6002 blocked this pathological process and reversed stress-induced depression-like behaviors (Fig. 9D and E). Under chronic stress, the persistently high adenosine levels may prompt an increase in adenosine release in the lateral septal nucleus (LS) region, which specifically upregulates the expression of A2AR within this region. This upregulation of A2AR, through the regulation of specific LS-DMH/LHb neural circuits, ultimately mediates the generation of depressive-like behaviors. Beyond depression, A2AR also significantly modulates anxiety-related behaviors. Emerging evidence suggests that an imbalance between A1R and A2AR signaling, rather than A2AR activation alone, contributes to anxiety pathogenesis.104,105 Hohoff et al. employed positron emission tomography to investigate the associations between both A1R and A2AR availability and anxiety-related brain regions in non-smokers.106 To sum up, these studies suggest that the coordinated interaction between A1R and A2AR signaling pathways plays a critical role in the neural modulation of stress-induced anxiety responses.
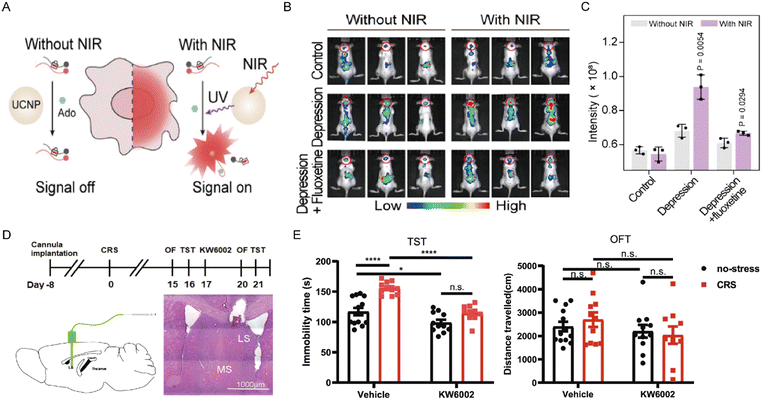 |
| | Fig. 9 (A) Schematic illustration of the nanosensor for adenosine detection. (B) Representative in vivo fluorescence images of adenosine detected by the nanosensor in different groups of mice. (C) Quantification of fluorescence intensity across brain regions. Reproduced with permission from ref. 102. Copyright 2025, American Chemical Society. Schematic diagram of A2AR antagonist KW6002 treatment in chronic restraint stress (CRS) – induced depression-like phenotypes (D) and the antidepressant effects of KW6002 in CRS mouse model (E). Reproduced with permission from ref. 103. Copyright 2023, Springer Nature. | |
3.3 Implications for neurological disorders
Adenosine signaling is an important modulator of both sleep–wake regulation and emotional processing, and it has been linked to the pathogenesis of many brain disorders, including Alzheimer's disease and epilepsy.107 These associations are mediated by diverse molecular mechanisms. The following sections will discuss the implications of adenosine in a spectrum of neurological disorders.
3.3.1 Alzheimer's disease (AD).
AD is pathologically hallmarked by the presence of extracellular β-amyloid plaques and tau-containing neurofibrillary tangles.108 In AD brains, adenosine signaling is dysregulated mainly through altered expression and function of its receptors.109,110 Zhou et al. used immunofluorescence imaging to identify that A1R activation can induce an astrocyte phenotype that promotes both neuroinflammation and synaptic dysfunction.111 Notably, neurons exposed to conditioned medium from A1R-overexpressing astrocytes exhibited marked dendritic atrophy and impaired long-term potentiation (Fig. 10B). Post-mortem human studies by Albasanz et al. further revealed increased A2AR binding affinity in the frontal cortex of AD patients versus controls.112 This observation was corroborated by electrophysiological recordings of hippocampal slices from wild-type mice, which showed that accumulation of adenosine generated from the enzymatic breakdown of ATP greatly activates A2AR, thereby triggering synaptic deterioration and memory impairment in murine models of early-stage AD.113 This finding establishes A2AR signaling as a potential therapeutic target for the prevention and reversal of associated neurodegenerative disorders. Orr et al. likewise reported up-regulated A2AR expression in glial cells from AD patients relative to non-demented controls.114 Moreover, to elucidate the cellular mechanism underlying the signal control behavior of A2AR, the study employed in vivo electrophysiological recordings, demonstrating that the activation of A2AR in hippocampal cells triggers impairments of spatial memory in AD.115 Moving beyond the vague understanding of A2AR function, this study precisely demonstrated its role within the globus pallidus neurons circuit. This work thereby establishes a critical experimental foundation for locally targeted therapies against neuropsychiatric diseases. Collectively, these findings suggest that adenosine receptor-mediated signaling may play a critical role in the regulation of synaptic dysfunction in AD.
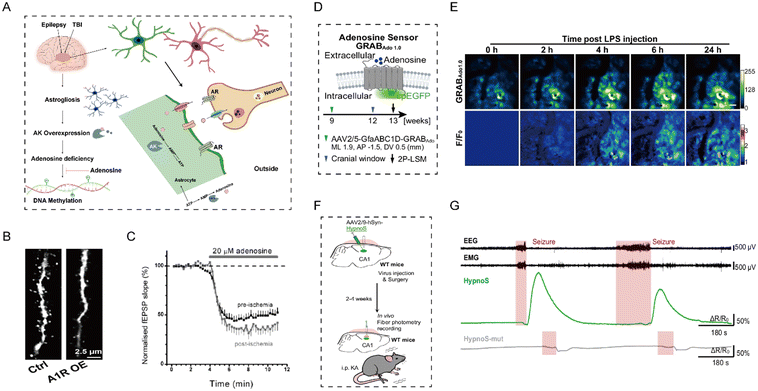 |
| | Fig. 10 (A) Regulatory role of local adenosine signaling pathways in epilepsy. (B) Synaptic impairment in neurons with A1R overexpression. Reproduced with permission from ref. 111. Copyright 2023, The American Association for the Advancement of Science. (C) Temporal profile of adenosine-induced fEPSP pre- (black) and post-ischemic state (grey). Reproduced with permission from ref. 125. Copyright 2006, John Wiley and Sons. (D) Schematic illustration of adenosine sensor mechanism (up) and LPS-induced adenosine dynamic imaging (down). (E) Fluorescence micrographs (up) and corresponding pseudo-color representations (down) of GRABAdo1.0 sensor signals following peripheral LPS administration. Reproduced with permission from ref. 131. Copyright 2024, Springer Nature. (F) Schematic diagram of adenosine fluorescence sensor monitoring epileptic activity in mice. (G) In vivo detection during seizure activity. Reproduced with permission from ref. 69. Copyright 2025, Springer Nature. | |
3.3.2 Schizophrenia.
Schizophrenia, a chronic brain disorder requiring long-term intervention, has been linked to the dysregulation of adenosine homeostasis in its pathological mechanism. Marta Valle-León et al. found that the A2AR-dopamine D2 receptor heteromer in the striatum of patients with schizophrenia was reduced, providing new evidence for the adenosine hypothesis of schizophrenia.116 Recent studies have revealed that adenosine signaling can affect the metabolic activities of astrocytes.117 Indeed, the A2AR regulates Na+/K+-ATPase activity in astrocytes and modulates the function of the glutamate transporter GLT-1, an essential component in maintaining glutamate homeostasis.118,119 Marco Matos et al. employed (3H) CGS 21680 radioligand binding and (11C) SCH 442416 PET imaging to reveal an upregulation of A2AR in brain regions of schizophrenia patients. Conversely, A2AR deletion in astrocytes has been shown to induce phenotypes of schizophrenia, accompanied by dysfunctional GLT-1.119 This contradictory finding suggests that balanced adenosine-A2AR signaling is important for maintaining glutamatergic stability; either excessive or insufficient levels of adenosine may disrupt this equilibrium. Notably, the associated disruption of astrocytic A2AR-glutamate homeostasis offers a valuable candidate target for diagnostic biomarkers, potentially reducing the high misdiagnosis rate in schizophrenia. Moreover, adenosine signaling has been involved in the regulation of synaptic plasticity, as affirmed by the research of Dang et al.120 Using electrophysiological recordings of synaptic transmission and plasticity in the hippocampus, they found that the deletion of the NLG3 gene in astrocytes impaired long-term potentiation in hippocampal slices, demonstrating adenosine signaling as the core pathway for restoring plasticity. Notably, exogenous adenosine and selective A2AR agonists were able to restore this deficit. Thus, these findings highlight the significance of adenosine signaling pathways in the regulation of synaptic plasticity in schizophrenia.
3.3.3 Traumatic brain injury (TBI).
In the context of global ischemia or TBI, local tissues rapidly initiate neuroprotective responses, which include the rapid depletion of intracellular ATP and a notable surge in adenosine generation and release (Fig. 10A).121,122 Locally released adenosine also mediates rapid, transient signaling of tissue injury and promotes pain resolution.123 Previous research has indicated that a critical response for cellular protection involved a several-fold increase in the regional concentration of adenosine during early ischemia. However, prolonged elevation of adenosine impedes the reconstruction of neural circuits, suppresses neurogenesis, and disrupts synaptic plasticity, thereby interfering with the recovery of higher cognitive functions such as learning and memory.124 Frenguelli and colleagues recorded neuronal electrical activity to investigate adenosine release during hippocampal ischemia in rats.125 They observed rapid adenosine efflux following ischemic stroke, which led to prolonged suppression of field excitatory postsynaptic potentials (Fig. 10C). Thus, adenosine produces sustained inhibition of excitatory transmission in the post-ischemic hippocampus. Importantly, even though exogenous adenosine was administered before or after ischemia, its effects on neural activity were not markedly altered, suggesting that adenosine signaling plays a sustained role in modulating neural activity rather than merely an acute response to ischemic stroke. Subsequent research by Jackson et al. measured the release of adenosine under conditions of glutamate excitotoxicity.126 Extracellular and intracellular adenosine levels increased by 2.9- and 1.7-fold, respectively. Under hypoxia and glucose deprivation conditions, these levels were elevated by 2.4- and 2.6-fold, respectively. These findings suggest that neurons can dynamically regulate the production and release of adenosine in response to neuronal homeostasis and function under pathological conditions.127 However, A2AR overactivation exacerbates neuronal dysfunction and excitotoxicity by promoting glutamate release from both neurons and astrocytes.121 Manuela Marcoli et al. found that in human brain tissue that there is a vesicle-like release mechanism for glutamate exocytosis during ischemia. Moreover, the A2AR antagonist can reduce glutamate accumulation by inhibiting this mechanism, providing a crucial experimental basis for neuroprotective treatment of cerebral ischemia.128,129 Meng et al. employed HPLC to reveal significantly reduced adenosine levels in the striatum of CD73 knockout mice compared to wild-type controls, highlighting the role of CD73-mediated adenosine in regulating microglial-mediated neuroinflammation. Conversely, neuroinflammation has been shown to elevate extracellular adenosine.130 Guo et al. used an adenosine sensor, GRABAdo1.0 to confirm that lipopolysaccharide (LPS)-induced systemic inflammation increases extracellular adenosine concentrations in the brain (Fig. 10D).131 In their study, fluorescence intensity began to rise two hours after LPS administration and reached a plateau at six hours, demonstrating that systemic inflammation can induce adenosine accumulation (Fig. 10E). On the whole, the generation and release of adenosine require precise regulation to maintain neuronal homeostasis.
3.3.4 Epilepsy.
Epilepsy is characterized by recurrent seizures arising from neuronal hyperexcitability. Maintaining adenosine homeostasis is thus critical for preventing seizure. Seizures are accompanied by explosive poly-spike-and-slow-wave discharges on electroencephalography.132 Wei et al. monitored real-time adenosine levels with a fluorescent probe following kainic-acid-induced hippocampal seizures in mice (Fig. 10F).69 Mice expressing the probe exhibited a larger fluorescence increase than controls. Strikingly, the enhancement of the fluorescent probe was temporally aligned with the termination of epileptic seizures, consistent with the role of adenosine signaling in seizure termination (Fig. 10G). Their work emphasized the importance of adenosine levels during epileptic seizures and underscored astrocytes as a crucial modulator in this mechanism. Besides, Shen et al. demonstrated that overexpression of adenosine kinase (ADK) resulted in a half reduction of adenosine in tissue.133 Li et al. proposed a novel astrocyte-based mechanism of epilepsy, laying the foundation for the ADK-mediated epilepsy hypothesis.134 ADK, a key enzyme regulating adenosine levels, is rapidly downregulated after epileptic seizures, leading to increased adenosine levels and exerting anti-seizure effects. However, inflammatory responses induce astrocytic proliferation during the latent period of epilepsy, resulting in ADK overexpression and an increased likelihood of seizures.135 This ADK-related dysregulation of adenosine concentrations suggests that adenosine dynamics act as an important driver of imbalances in neural circuits. Specifically, excessive ADK activity leads to decreased adenosine levels and insufficient A1R activation, resulting in the loss of a “braking” signal in neurons. This impairs their ability to suppress excitatory signals and exacerbates seizure severity.41 In contrast, blocking nucleoside transporters restores synaptic plasticity and reduces seizure severity.136 As another key receptor in the adenosine signaling pathway, A2AR regulates GLT-1 activity and directly impacts glutamate homeostasis. These studies are consistent with those of Boison's group, who validated that adenosine exerts inhibitory effects on seizures, suggesting its potential value in the treatment of epilepsy.137 Moreover, researchers have genetically modified fibroblasts to continuously release adenosine, encapsulating and transplanting them into a rat model of epilepsy, and significantly suppressed seizure-like activity induced by electrical stimulation.138 Taken together, these studies indicate that dysregulation of adenosine-based neuromodulation may contribute to epileptogenesis. The dynamic levels of adenosine in the cerebrospinal fluid or blood of epilepsy patients may potentially serve as an early warning indicator for seizures.69 The integration of a photoelectrochemical sensor into a microfluidic chip platform holds promise for providing a novel solution for at-home adenosine monitoring in epilepsy patients. Currently, the elevation of adenosine levels has emerged as a promising therapeutic approach in epilepsy.139
Conclusions
The neuromodulatory role of adenosine in the central nervous system has garnered widespread recognition over recent decades. Adenosine participates in the regulation of neuronal activity, and its dysregulation is closely associated with the pathological mechanisms of various neurological diseases. This recognition has prompted the development of adenosine detection methods, providing essential tools for elucidating adenosine regulatory mechanisms and understanding its contributions to neurological pathologies. In this review, we systematically discuss and categorize non-optical and optical-based approaches. By analyzing the applications of adenosine in sleep–wake regulation, mood disorders, and various neurological conditions, we emphasized the correlation between adenosine levels and neural activity. These advances have not only deepened our comprehension of adenosine's neuromodulatory roles but also laid the foundation for novel diagnostic tools and clinical therapeutic strategies.
At present, most adenosine-detection techniques remain at the experimental research, and only a few have entered pre-clinical or early clinical validation. In terms of technological development, although fiber-optic photometry enables high-spatial and temporal-resolution monitoring, implantable fibers cause tissue trauma and disrupt the microenvironment, prompting researchers to explore biodegradable implant designs that balance monitoring performance with biocompatibility. Regarding imaging, optical methods struggle to achieve high-signal-to-noise-ratio monitoring in deep brain regions because of limited tissue penetration and autofluorescence interference. Near-infrared-II imaging, which offers deeper penetration and lower background noise, now provides a new route for non-invasive monitoring of deep lesions. Notably, complex biological matrices (plasma proteins, lipids) and adenosine structural analogues in clinical samples severely compromise specificity. Multimodal data-analysis platforms that incorporate artificial intelligence (AI) can magnify subtle differences between adenosine and its analogues, markedly improving detection accuracy in complex matrices. Future efforts should concentrate on three synergistic advances: (i) developing endogenous optical pathways for minimally invasive intervention, (ii) leveraging NIR-II imaging to extend deep-tissue detection, and (iii) integrating biodegradable probes with AI analysis platforms, thereby accelerating the translation of adenosine detection from bench to bedside. In summary, technological advances will enable real-time, dynamic detection of brain adenosine, providing deeper insight into brain function and disease mechanisms.
Author contributions
Y. Q. Liao: writing – review and editing, investigation; J. Y. Jing: writing – original draft; W. K. Jin: visualization, investigation; X. D. Tian: writing – review and editing, supervision; T. H. Peng: writing – review and editing, supervision; L. Zhang: writing – review and editing, visualization; Q. Yuan: conceptualization, funding acquisition, resources, writing – review and editing. All authors have read and agreed to the published version of the manuscript.
Conflicts of interest
There are no conflicts to declare.
Data availability
No primary research results, software or code have been included, and no new data were generated or analyzed as part of this review.
Acknowledgements
This work was supported by the National Key R&D Program of China (2023YFF1205900), the National Natural Science Foundation of China (52221001), the Hunan Provincial Key Research and Development Plan (2024JK2117), and the New Cornerstone Science Foundation through the XPLORER PRIZE.
Notes and references
- T. Kobayashi, K. Shimba, T. Narumi, T. Asahina, K. Kotani and Y. Jimbo, Nat. Commun., 2024, 15, 9547 CrossRef CAS PubMed.
- J. M. Brundege and T. V. Dunwiddie, Adv. Pharmacol., 1997, 39, 353–391 CrossRef CAS PubMed.
- A. N. Drury and A. Szent-Györgyi, J. Physiol., 1929, 68, 213–237 CrossRef CAS.
- N. Dale and B. G. Frenguelli, Curr. Neuropharmacol., 2009, 7, 160–179 Search PubMed.
- L. Antonioli, C. Blandizzi, B. Csóka, P. Pacher and G. Haskó, Nat. Rev. Endocrinol., 2015, 11, 228–241 CrossRef CAS PubMed.
- B. B. Fredholm, Cell Death Differ., 2007, 14, 1315–1323 CrossRef CAS PubMed.
- R. B. Dias, D. M. Rombo, J. A. Ribeiro, J. M. Henley and A. M. Sebastião, Trends Neurosci., 2013, 36, 248–257 CrossRef CAS PubMed.
- G. Liu, W. Zhang, J. Guo, F. Kong, S. Zhou, S. Chen, Z. Wang and D. Zang, Brain Res., 2018, 1700, 47–55 CrossRef CAS PubMed.
- B. B. Fredholm, A. P. Ijzerman, K. A. Jacobson, K.-N. Klotz and J. Linden, Pharmacol. Rev., 2001, 53, 527–552 CrossRef CAS PubMed.
- C.-P. Chang, K.-C. Wu, C.-Y. Lin and Y. Chern, J. Biomed. Sci., 2021, 28, 70 CrossRef CAS PubMed.
- G. Haskó, P. Pacher, E. Sylvester Vizi and P. Illes, Trends Pharmacol. Sci., 2005, 26, 511–516 CrossRef PubMed.
- F. Pedata, C. Corsi, A. Melani, F. Bordoni and S. Latini, Ann. N. Y. Acad. Sci., 2001, 939, 74–84 CrossRef CAS PubMed.
- A. M. Sebastião and J. A. Ribeiro, Brain Res., 2015, 1621, 102–113 CrossRef.
- D. Pereira-Figueiredo, A. A. Nascimento, M. C. Cunha-Rodrigues, R. Brito and K. C. Calaza, Cell. Mol. Neurobiol., 2022, 42, 1693–1725 CrossRef CAS.
- F. C. Tescarollo, D. M. Rombo, L. K. DeLiberto, D. E. Fedele, E. Alharfoush, Â. R. Tomé, R. A. Cunha, A. M. Sebastião and D. Boison, J. Caffeine Adenosine Res., 2020, 10, 45–60 CrossRef CAS.
- L. Cellai, K. Carvalho, E. Faivre, A. Deleau, D. Vieau, L. Buée, D. Blum, C. Mériaux and V. Gomez-Murcia, Front. Neurosci., 2018, 12, 520 CrossRef PubMed.
- L. V. Lopes, A. M. Sebastiao and J. A. Ribeiro, Curr. Top. Med. Chem., 2011, 11, 1087–1101 CrossRef CAS.
- H. K. Eltzschig, M. V. Sitkovsky and S. C. Robson, N. Engl. J. Med., 2012, 367, 2322–2333 CrossRef CAS.
- P. Alonso-Andrés, J. L. Albasanz, I. Ferrer and M. Martín, Brain Pathol., 2018, 28, 933–946 CrossRef PubMed.
- H. Liu, Y. Xiang, Y. Lu and R. M. Crooks, Angew. Chem., 2012, 124, 7031–7034 CrossRef.
- S. Zhang, J. Xia and X. Li, Anal. Chem., 2008, 80, 8382–8388 CrossRef CAS PubMed.
- L.-L. Li, P. Ge, P. R. Selvin and Y. Lu, Anal. Chem., 2012, 84, 7852–7856 CrossRef CAS.
- J.-W. Chen, X.-P. Liu, K.-J. Feng, Y. Liang, J.-H. Jiang, G.-L. Shen and R.-Q. Yu, Biosens. Bioelectron., 2008, 24, 66–71 CrossRef CAS PubMed.
- F. Aucella, V. Lauriola, G. Vecchione, G. L. Tiscia and E. Grandone, J. Pharm. Biomed. Anal., 2013, 86, 123–126 CrossRef CAS PubMed.
- Z. Gong, H. Zhao, Y. Mao, F. Zhou, Z. Shi and M. Lan, Bioelectrochemistry, 2025, 108906 CrossRef CAS PubMed.
- Z. Wu, D. Lin and Y. Li, Nat. Rev. Neurosci., 2022, 23, 257–274 CrossRef CAS.
- N. Dale, Purinergic Signalling, 2021, 17, 109–115 CrossRef CAS PubMed.
- A. S. Abdelfattah, S. Ahuja, T. Akkin, S. R. Allu, J. Brake, D. A. Boas, E. M. Buckley, R. E. Campbell, A. I. Chen and X. Cheng, Neurophotonics, 2022, 9, 013001 CAS.
- S. Ng, H. S. Lim, Q. Ma and Z. Gao, Theranostics, 2016, 6, 1683–1702 CrossRef CAS PubMed.
- B. Allard, D. Allard, L. Buisseret and J. Stagg, Nat. Rev. Clin. Oncol., 2020, 17, 611–629 CrossRef CAS PubMed.
- D. Vijayan, A. Young, M. W. L. Teng and M. J. Smyth, Nat. Rev. Cancer, 2017, 17, 709–724 CrossRef CAS PubMed.
- G. Haskó, J. Linden, B. Cronstein and P. Pacher, Nat. Rev. Drug Discovery, 2008, 7, 759–770 CrossRef.
- D. Boison and G. G. Yegutkin, Cancer Cell, 2019, 36, 582–596 CrossRef CAS PubMed.
- Z. Li, C. Wang, M. Zhang, S. Li, Z. Mao and Z. Liu, Nano Today, 2021, 39, 101239 CrossRef CAS.
- M. Hori and M. Kitakaze, Hypertension, 1991, 18, 565–574 CrossRef CAS.
- A. B. Reiss, D. Grossfeld, L. J. Kasselman, H. A. Renna, N. A. Vernice, W. Drewes, J. Konig, S. E. Carsons and J. DeLeon, Am. J. Cardiovasc. Drugs, 2019, 19, 449–464 CrossRef CAS.
- A. Bahreyni, S. S. Samani, F. Rahmani, R. Behnam-Rassouli, M. Khazaei, M. Ryzhikov, M. R. Parizadeh, A. Avan and S. M. Hassanian, J. Cell. Physiol., 2018, 233, 1836–1843 CrossRef CAS PubMed.
- Z. T. Kurt, D. Çimen, A. Denizli and N. Bereli, ACS Omega, 2023, 8, 18839–18850 CrossRef CAS PubMed.
- F. S. Anderson and R. C. Murphy, J. Chromatogr. A, 1976, 121, 251–262 CrossRef CAS PubMed.
- W. J. Wojcik and N. H. Neff, J. Neurochem., 1982, 39, 280–282 CrossRef CAS.
- D. Lovatt, Q. Xu, W. Liu, T. Takano, N. A. Smith, J. Schnermann, K. Tieu and M. Nedergaard, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 6265–6270 CrossRef CAS PubMed.
- R. M. Marin, K. G. Franchini and S. A. Rocco, J. Sep. Sci., 2007, 30, 2473–2479 CrossRef CAS.
- G. Cannazza, M. M. Carrozzo, A. S. Cazzato, I. M. Bretis, L. Troisi, C. Parenti, D. Braghiroli, S. Guiducci and M. Zoli, J. Pharm. Biomed. Anal., 2012, 71, 183–186 CrossRef CAS PubMed.
- L. Löfgren, S. Pehrsson, G. Hägglund, H. Tjellström and S. Nylander, PLoS One, 2018, 13, e0205707 CrossRef PubMed.
- E. Olesti, J. Rodríguez-Morató, A. Gomez-Gomez, J. G. Ramaekers, R. de la Torre and O. J. Pozo, Talanta, 2019, 192, 93–102 CrossRef CAS PubMed.
- C. Virgiliou, N. Fragakis, M. Sotiriadou, V. Vassilikos, S. Gerou, G. Theodoridis and H. Gika, Separations, 2021, 8, 222 CrossRef CAS.
- R. D. O'Neill, Neurosci. Lett., 1986, 63, 11–16 CrossRef.
- S. Cechova and B. J. Venton, J. Neurochem., 2008, 105, 1253–1263 CrossRef CAS PubMed.
- Y. Wang and B. J. Venton, J. Neurochem., 2017, 140, 13–23 CrossRef CAS.
- M. L. Pajski and B. J. Venton, Purinergic Signalling, 2013, 9, 167–174 CrossRef CAS PubMed.
- Y. Chang and B. J. Venton, ACS Chem. Neurosci., 2022, 13, 477–485 CrossRef CAS PubMed.
- H. Chang, M. Huo, Q. Zhang, M. Zhou, Y. Zhang, Y. Si, D. Zhang, Y. Guo and Y. Fang, Biosens. Bioelectron., 2023, 235, 115383 CrossRef CAS PubMed.
- K. K. Hussain, M. H. Akhtar, M.-H. Kim, D.-K. Jung and Y.-B. Shim, Biosens. Bioelectron., 2018, 109, 263–271 CrossRef CAS.
- P. Puthongkham, J. Rocha, J. R. Borgus, M. Ganesana, Y. Wang, Y. Chang, A. Gahlmann and B. J. Venton, Anal. Chem., 2020, 92, 10485–10494 CrossRef CAS.
- S. Cao, H. Zhao, K. Chen, F. Zhou and M. Lan, Anal. Chim. Acta, 2023, 1260, 341212 CrossRef CAS PubMed.
- K. Tian, Y. Ma, Y. Liu, M. Wang, C. Guo, L. He, Y. Song, Z. Zhang and M. Du, Sens. Actuators, B, 2020, 303, 127199 CrossRef CAS.
- D. G. Gardiner, Anal. Biochem., 1979, 95, 377–382 CrossRef CAS PubMed.
- S. Srinivasan, V. Ranganathan, M. C. DeRosa and B. M. Murari, Anal. Bioanal. Chem., 2019, 411, 1319–1330 CrossRef CAS PubMed.
- C. Feng, Z. Hou, W. Jiang, L. Sang and L. Wang, Biosens. Bioelectron., 2016, 86, 966–970 CrossRef CAS PubMed.
- K. Quan, J. Huang, X. Yang, Y. Yang, L. Ying, H. Wang, N. Xie, M. Ou and K. Wang, Anal. Chem., 2016, 88, 5857–5864 CrossRef CAS PubMed.
- P. Li, C. Luo, X. Chen and C. Huang, Spectrochim. Acta, Part A, 2024, 305, 123557 CrossRef CAS PubMed.
- P. Cheng, W. Guo, R. Li, Y. Yang and Q. Du, Microchem. J., 2023, 195, 109392 CrossRef CAS.
- X. Chen, X. Fu, Y. Wu, Y. Jin and W. Li, Anal. Methods, 2020, 12, 1579–1586 RSC.
- A. R. Chandrasekaran, Nat. Rev. Chem., 2021, 5, 225–239 CrossRef CAS PubMed.
- N. Stephanopoulos, ChemBioChem, 2019, 20, 2191–2197 CrossRef CAS PubMed.
- W. Peng, Z. Wu, K. Song, S. Zhang, Y. Li and M. Xu, Science, 2020, 369, eabb0556 CrossRef CAS PubMed.
- L. Ma, J. Day-Cooney, O. J. Benavides, M. A. Muniak, M. Qin, J. B. Ding, T. Mao and H. Zhong, Nature, 2022, 611, 762–768 CrossRef CAS PubMed.
- Z. Wu, Y. Cui, H. Wang, H. Wu, Y. Wan, B. Li, L. Wang, S. Pan, W. Peng, A. Dong, Z. Yuan, M. Jing, M. Xu, M. Luo and Y. Li, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2212387120 CrossRef CAS PubMed.
- Q. Wei, Z. Bai, L. Wang, J. Wang, Y. Wang, Y. Hu, S. Ding, Z. Ma, C. Li, Y. Li, Y. Zhuo, W. Li, F. Deng, B. Liu, P. Zhou, Y. Li, Z. Wu and J. Wang, Nat. Commun., 2025, 16, 4245 CrossRef CAS PubMed.
- H. Van Belle, Anal. Biochem., 1969, 32, 238–250 CrossRef CAS.
- J. Liu and Y. Lu, Nat. Protoc., 2006, 1, 246–252 CrossRef CAS PubMed.
- X. Zhang, C. Kong, Q. Liu, X. Zuo, K. Li and Z. Chen, Microchim. Acta, 2019, 186, 587 CrossRef PubMed.
- R. Zhu, J. Song, Y. Zhou, P. Lei, Z. Li, H.-W. Li, S. Shuang and C. Dong, Talanta, 2019, 204, 294–303 CrossRef CAS PubMed.
- L. Xu, X. Shen, B. Li, C. Zhu and X. Zhou, Anal. Chim. Acta, 2017, 980, 58–64 CrossRef CAS PubMed.
- S. ArulVijayaVani, P. S. Mohanraj and R. Reeta, J. Lab. Physicians, 2023, 15, 269–275 CrossRef PubMed.
- M. D. Krasowski, Acad. Pathol., 2019, 6, 2374289519888754 CrossRef PubMed.
- H. Kather, E. Wieland and W. Waas, Anal. Biochem., 1987, 163, 45–51 CrossRef CAS PubMed.
- K. Jang, J. H. Westbay and S. A. Asher, ACS Sens., 2022, 7, 1648–1656 CrossRef CAS PubMed.
- Y. Lin, X. Wang, Y. Sun, Y. Dai, W. Sun, X. Zhu, H. Liu, R. Han, D. Gao and C. Luo, Sens. Actuators, B, 2019, 289, 56–64 CrossRef CAS.
- C. M. Jabs, P. Neglen, B. Eklof and E. J. Thomas, Clin. Chem., 1990, 36, 81–87 CrossRef CAS.
- T. Fujita, E. K. Williams, T. K. Jensen, N. A. Smith, T. Takano, K. Tieu and M. Nedergaard, J. Cereb. Blood Flow Metab., 2012, 32, 1–7 CrossRef PubMed.
- H. Li, P. A. Walker III and M. D. Morris, J. Microcolumn Sep., 1998, 10, 449–453 CrossRef CAS.
- S. Xu, B. Man, S. Jiang, J. Wang, J. Wei, S. Xu, H. Liu, S. Gao, H. Liu, Z. Li, H. Li and H. Qiu, ACS Appl. Mater. Interfaces, 2015, 7, 10977–10987 CrossRef CAS PubMed.
- R. Nishitsuji, T. Nakashima, H. Hisamoto and T. Endo, Sensors, 2024, 24, 6648 CrossRef CAS PubMed.
- W. Lee, B.-H. Kang, H. Yang, M. Park, J. H. Kwak, T. Chung, Y. Jeong, B. K. Kim and K.-H. Jeong, Nat. Commun., 2021, 12, 159 CrossRef CAS PubMed.
- T. V. Dunwiddie and S. A. Masino, Annu. Rev. Neurosci., 2001, 24, 31–55 CrossRef CAS PubMed.
- L. I. Schmitt, R. E. Sims, N. Dale and P. G. Haydon, J. Neurosci., 2012, 32, 4417–4425 CrossRef CAS PubMed.
- T. Porkka-Heiskanen, R. E. Strecker, M. Thakkar, A. A. Bjørkum, R. W. Greene and R. W. McCarley, Science, 1997, 276, 1265–1268 CrossRef CAS PubMed.
- Y. Zhu, J. Ma, Y. Li, M. Gu, X. Feng, Y. Shao, L. Tan, H.-f. Lou, L. Sun, Y. Liu, L.-h. Zeng, Z. Qiu, X.-m. Li, S. Duan and Y.-q. Yu, Adv. Sci., 2024, 11, 2407706 CrossRef CAS PubMed.
- D. Elmenhorst, E.-M. Elmenhorst, E. Hennecke, T. Kroll, A. Matusch, D. Aeschbach and A. Bauer, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 4243–4248 CrossRef CAS PubMed.
- A. Jagannath, N. Varga, R. Dallmann, G. Rando, P. Gosselin, F. Ebrahimjee, L. Taylor, D. Mosneagu, J. Stefaniak, S. Walsh, T. Palumaa, S. Di Pretoro, H. Sanghani, Z. Wakaf, G. C. Churchill, A. Galione, S. N. Peirson, D. Boison, S. A. Brown, R. G. Foster and S. R. Vasudevan, Nat. Commun., 2021, 12, 2113 CrossRef CAS PubMed.
- Z.-L. Huang, W.-M. Qu, N. Eguchi, J.-F. Chen, M. A. Schwarzschild, B. B. Fredholm, Y. Urade and O. Hayaishi, Nat. Neurosci., 2005, 8, 858–859 CrossRef CAS PubMed.
- F. Gomez-Castro, S. Zappettini, J. C. Pressey, C. G. Silva, M. Russeau, N. Gervasi, M. Figueiredo, C. Montmasson, M. Renner, P. M. Canas, F. Q. Gonçalves, S. Alçada-Morais, E. Szabó, R. J. Rodrigues, P. Agostinho, A. R. Tomé, G. Caillol, O. Thoumine, X. Nicol, C. Leterrier, R. Lujan, S. K. Tyagarajan, R. A. Cunha, M. Esclapez, C. Bernard and S. Lévi, Science, 2021, 374, eabk2055 CrossRef CAS PubMed.
- S. M. Theparambil, O. Kopach, A. Braga, S. Nizari, P. S. Hosford, V. Sagi-Kiss, A. Hadjihambi, C. Konstantinou, N. Esteras, A. Gutierrez Del Arroyo, G. L. Ackland, A. G. Teschemacher, N. Dale, T. Eckle, P. Andrikopoulos, D. A. Rusakov, S. Kasparov and A. V. Gourine, Nature, 2024, 632, 139–146 CrossRef CAS PubMed.
- T. Serchov, H.-W. Clement, M. K. Schwarz, F. Iasevoli, D. K. Tosh, M. Idzko, K. A. Jacobson, A. de Bartolomeis, C. Normann, K. Biber and D. van Calker, Neuron, 2015, 87, 549–562 CrossRef CAS PubMed.
- K. P. Wright, C. A. Lowry and M. K. LeBourgeois, Front. Mol. Neurosci., 2012, 5, 50 Search PubMed.
- M. Santello, N. Toni and A. Volterra, Nat. Neurosci., 2019, 22, 154–166 CrossRef CAS PubMed.
- M. M. Halassa, C. Florian, T. Fellin, J. R. Munoz, S.-Y. Lee, T. Abel, P. G. Haydon and M. G. Frank, Neuron, 2009, 61, 213–219 CrossRef CAS PubMed.
- C. Florian, C. G. Vecsey, M. M. Halassa, P. G. Haydon and T. Abel, J. Neurosci., 2011, 31, 6956–6962 CrossRef CAS PubMed.
- R. A. Cunha, J. Neurochem., 2016, 139, 1019–1055 CrossRef CAS PubMed.
- F. F. Ribeiro and A. M. Sebastião, Neural Regener. Res., 2016, 11, 706–708 CrossRef CAS.
- M. Chu, L. Zhang, X. Yang, C. Ji, W. Tan, J. Tan and Q. Yuan, J. Am. Chem. Soc., 2025, 147, 32447–32457 CrossRef CAS PubMed.
- M. Wang, P. Li, Z. Li, B. S. da Silva, W. Zheng, Z. Xiang, Y. He, T. Xu, C. Cordeiro and L. Deng, Nat. Commun., 2023, 14, 1880 CrossRef CAS PubMed.
- A. P. Simões, N. J. Machado, N. Gonçalves, M. P. Kaster, A. T. Simões, A. Nunes, L. Pereira de Almeida, K. A. Goosens, D. Rial and R. A. Cunha, Neuropsychopharmacology, 2016, 41, 2862–2871 CrossRef PubMed.
- C. Hohoff, T. Kroll, B. Zhao, N. Kerkenberg, I. Lang, K. Schwarte, D. Elmenhorst, E.-M. Elmenhorst, D. Aeschbach and W. Zhang, Transl. Psychiatry, 2020, 10, 406 CrossRef CAS PubMed.
- C. Hohoff, V. Garibotto, D. Elmenhorst, A. Baffa, T. Kroll, A. Hoffmann, K. Schwarte, W. Zhang, V. Arolt and J. Deckert, Neuropsychopharmacology, 2014, 39, 2989–2999 CrossRef CAS PubMed.
- L. Weltha, J. Reemmer and D. Boison, Brain Res. Bull., 2019, 151, 46–54 CrossRef CAS PubMed.
- M. A. Busche and B. T. Hyman, Nat. Neurosci., 2020, 23, 1183–1193 CrossRef CAS PubMed.
- E. Angulo, V. Casadó, J. Mallol, E. I. Canela, F. Viñals, I. Ferrer, C. Lluis and R. Franco, Brain Pathol., 2003, 13, 440–451 CrossRef CAS.
- L. T. Woods, D. Ajit, J. M. Camden, L. Erb and G. A. Weisman, Neuropharmacology, 2016, 104, 169–179 CrossRef CAS PubMed.
- L.-T. Zhou, D. Liu, H.-C. Kang, L. Lu, H.-Z. Huang, W.-Q. Ai, Y. Zhou, M.-F. Deng, H. Li, Z.-Q. Liu, W.-F. Zhang, Y.-Z. Hu, Z.-T. Han, H.-H. Zhang, J.-J. Jia, A. K. Sarkar, S. Sharaydeh, J. Wang, H.-Y. Man, M. Schilling, L. Bertram, Y. Lu, Z. Guo and L.-Q. Zhu, Sci. Adv., 2023, 9, eabq7105 CrossRef CAS PubMed.
- J. L. Albasanz, S. Perez, M. Barrachina, I. Ferrer and M. Martín, Brain Pathol., 2008, 18, 211–219 CrossRef CAS PubMed.
- F. Q. Gonçalves, J. P. Lopes, H. B. Silva, C. Lemos, A. C. Silva, N. Gonçalves, Â. R. Tomé, S. G. Ferreira, P. M. Canas and D. Rial, Neurobiol. Dis., 2019, 132, 104570 CrossRef PubMed.
- A. G. Orr, E. C. Hsiao, M. M. Wang, K. Ho, D. H. Kim, X. Wang, W. Guo, J. Kang, G.-Q. Yu, A. Adame, N. Devidze, D. B. Dubal, E. Masliah, B. R. Conklin and L. Mucke, Nat. Neurosci., 2015, 18, 423–434 CrossRef CAS PubMed.
- P. Li, D. Rial, P. M. Canas, J.-H. Yoo, W. Li, X. Zhou, Y. Wang, G. J. van Westen, M.-P. Payen and E. Augusto, Mol. Psychiatry, 2015, 20, 1339–1349 CrossRef CAS PubMed.
- M. Valle-León, L. F. Callado, E. Aso, M. M. Cajiao-Manrique, K. Sahlholm, M. López-Cano, C. Soler, X. Altafaj, M. Watanabe, S. Ferré, V. Fernández-Dueñas, J. M. Menchón and F. Ciruela, Neuropsychopharmacology, 2021, 46, 665–672 CrossRef PubMed.
- C. Lemos, B. S. Pinheiro, R. O. Beleza, J. M. Marques, R. J. Rodrigues, R. A. Cunha, D. Rial and A. Köfalvi, Purinergic Signalling, 2015, 11, 561–569 CrossRef CAS PubMed.
- M. Matos, E. Augusto, P. Agostinho, R. A. Cunha and J.-F. Chen, J. Neurosci., 2013, 33, 18492–18502 CrossRef CAS PubMed.
- M. Matos, H.-Y. Shen, E. Augusto, Y. Wang, C. J. Wei, Y. T. Wang, P. Agostinho, D. Boison, R. A. Cunha and J.-F. Chen, Biol. Psychiatry, 2015, 78, 763–774 CrossRef CAS PubMed.
- R. Dang, A. Liu, Y. Zhou, X. Li, M. Wu, K. Cao, Y. Meng, H. Zhang, G. Gan, W. Xie and Z. Jia, Nat. Commun., 2024, 15, 8639 CrossRef CAS.
- S. Latini, F. Bordoni, R. Corradetti, G. Pepeu and F. Pedata, Brain Res., 1998, 794, 325–328 CrossRef CAS PubMed.
- J. Ribeiro, Curr. Drug Targets: CNS Neurol. Disord., 2005, 4, 325–329 CAS.
- B. B. Fredholm, A. P. IJzerman, K. A. Jacobson, J. Linden and C. E. Müller, Pharmacol. Rev., 2011, 63, 1–34 CrossRef CAS PubMed.
- R. L. Williams-Karnesky and M. P. Stenzel-Poore, Curr. Neuropharmacol., 2009, 7, 217–227 CrossRef CAS PubMed.
- T. Pearson, K. Damian, R. E. Lynas and B. G. Frenguelli, J. Neurochem., 2006, 97, 1357–1368 CrossRef CAS PubMed.
- E. K. Jackson, S. E. Kotermanski, E. V. Menshikova, R. K. Dubey, T. C. Jackson and P. M. Kochanek, J. Neurochem., 2017, 141, 676–693 CrossRef CAS PubMed.
- F. Bartoli, G. Burnstock, C. Crocamo and G. Carrà, Brain Sci., 2020, 10, 160 CrossRef CAS PubMed.
- M. Marcoli, L. Raiteri, A. Bonfanti, A. Monopoli, E. Ongini, M. Raiteri and G. Maura, Neuropharmacology, 2003, 45, 201–210 CrossRef CAS PubMed.
- M. Marcoli, A. Bonfanti, P. Roccatagliata, G. Chiaramonte, E. Ongini, M. Raiteri and G. Maura, Neuropharmacology, 2004, 47, 884–891 CrossRef CAS PubMed.
- F. Meng, Z. Guo, Y. Hu, W. Mai, Z. Zhang, B. Zhang, Q. Ge, H. Lou, F. Guo and J. Chen, Brain, 2019, 142, 700–718 CrossRef PubMed.
- Q. Guo, D. Gobbo, N. Zhao, H. Zhang, N.-O. Awuku, Q. Liu, L.-P. Fang, T. M. Gampfer, M. R. Meyer, R. Zhao, X. Bai, S. Bian, A. Scheller, F. Kirchhoff and W. Huang, Nat. Commun., 2024, 15, 6340 CrossRef CAS PubMed.
- Y.-J. Liu, J. Chen, X. Li, X. Zhou, Y.-M. Hu, S.-F. Chu, Y. Peng and N.-H. Chen, CNS Neurosci. Ther., 2019, 25, 899–910 CrossRef PubMed.
- H.-Y. Shen, T. A. Lusardi, R. L. Williams-Karnesky, J.-Q. Lan, D. J. Poulsen and D. Boison, J. Cereb. Blood Flow Metab., 2011, 31, 1648–1659 CrossRef CAS PubMed.
- T. Li, G. Ren, T. Lusardi, A. Wilz, J. Q. Lan, T. Iwasato, S. Itohara, R. P. Simon and D. Boison, J. Clin. Invest., 2008, 118, 571–582 CrossRef CAS PubMed.
- A. Huber, M. Güttinger, H. Möhler and D. Boison, Neurosci. Lett., 2002, 329, 289–292 CrossRef CAS PubMed.
- S.-Y. Ho, I.-C. Chen, K.-C. Chang, H.-R. Lin, C.-W. Tsai, C.-J. Lin and H.-H. Liou, Front. Neurosci., 2020, 14, 610898 CrossRef PubMed.
- G. Ren, T. Li, J. Q. Lan, A. Wilz, R. P. Simon and D. Boison, Exp. Neurol., 2007, 208, 26–37 Search PubMed.
- A. Huber, V. Padrun, N. Déglon, P. Aebischer, H. Möhler and D. Boison, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 7611–7616 CrossRef CAS PubMed.
- D. Boison, Glia, 2012, 60, 1234–1243 Search PubMed.
Footnote |
| † These authors are equally contributed. |
|
| This journal is © The Royal Society of Chemistry 2026 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article d,
Tianhuan
Peng
*c,
Lei
Zhang
*b and
Quan
Yuan
d,
Tianhuan
Peng
*c,
Lei
Zhang
*b and
Quan
Yuan
 *bc
*bc

















