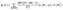Open Access Article
Open Access ArticleExplainable artificial intelligence for molecular design in pharmaceutical research
Alec
Lamens
 ab and
Jürgen
Bajorath
ab and
Jürgen
Bajorath
 *ab
*ab
aDepartment of Life Science Informatics and Data Science, B-IT, LIMES Program Unit Chemical Biology and Medicinal Chemistry, University of Bonn, Friedrich-Hirzebruch-Allee 5/6, D-53115 Bonn, Germany. E-mail: bajorath@bit.uni-bonn.de; Tel: +49-228-7369-100
bLamarr Institute for Machine Learning and Artificial Intelligence, University of Bonn, Friedrich-Hirzebruch-Allee 5/6, D-53115 Bonn, Germany
First published on 12th January 2026
Abstract
The rise of artificial intelligence (AI) has taken machine learning (ML) in molecular design to a new level. As ML increasingly relies on complex deep learning frameworks, the inability to understand predictions of black-box models has become a topical issue. Consequently, there is strong interest in the field of explainable AI (XAI) to bridge the gap between black-box models and the acceptance of their predictions, especially at interfaces with experimental disciplines. Therefore, XAI methods must go beyond extracting learning patterns from ML models and present explanations of predictions in a human-centered, transparent, and interpretable manner. In this Perspective, we examine current challenges and opportunities for XAI in molecular design and evaluate the benefits of incorporating domain-specific knowledge into XAI approaches for model refinement, experimental design, and hypothesis testing. In this context, we also discuss the current limitations in evaluating results from chemical language models that are increasingly used in molecular design and drug discovery.
Introduction
In the AI era, ML and deep generative modeling are increasingly applied in drug discovery and design.1,2 Typical applications include standard tasks such as physicochemical or biological property predictions for small molecules and quantitative structure–property relationship modeling or new tasks such as generative de novo design.1,2 Regardless of the applications, most ML and all deep learning models produce black-box predictions that are not understandable based on human reasoning.3–5 Incomprehensible predictions are not only scientifically unsatisfactory, but also limit the impact of ML in interdisciplinary research. This is the case because black-box predictions are rarely used to guide experimental work, which represents a substantial problem for applied ML3,4 and complex deep ML frameworks.5–7 As a consequence, the field of explainable AI (XAI) experiences increasing interest.8,9 XAI encompasses the development of computational concepts and practical methods to explain predictions and underlying data patterns, identify internal model bias, and determine applicability domains.8–11 Ultimately, XAI aims to increase the transparency of model decisions and enable human interpretation of predictions and causal reasoning. Beyond its scientific relevance, XAI has gained importance due to the need for transparency, trust, and interpretability in high-risk or high-cost scientific domains such as drug discovery.11 Here, explainability supports model validation and informed decision-making. In addition to predictive models used to identify or design new chemical matter, ML models are applied in regulatory and safety-related contexts, particularly for the evaluation of impurities, where explainability and interpretability support scientific justification, prioritization, and risk-based evaluation.12In the following, we present different XAI concepts and explore opportunities and limitations of XAI in molecular design. In addition, we discuss explanations that allow human interpretation and support causal reasoning. Using exemplary studies, we analyze benefits of combining existing XAI concepts with domain-specific knowledge for molecular design. Furthermore, we discuss attempts to explain transformer models for generative design tasks.
Evaluation criteria in the absence of fundamental truth
Given the conceptual diversity of XAI approaches and their different objectives, it is essential to define best practices for their use and identify specific limitations. Therefore, evaluation criteria have been proposed, often termed ‘desiderata’, to support the assessment and comparison of alternative XAI methods.13–16 An example is the evaluation of the accuracy (fidelity) of explanations for a given black-box model. This typically involves the comparison of explanations from alternative methods, the evaluation of the stability of explanations that are based on similar data points, or the evaluation of the robustness to data perturbation.13 Additional desiderata assess whether given explanations are concise, non-redundant, complete considering the underlying data patterns, or understandable within the applicability domain of a model.16 However, so far, desiderata have been rarely applied to explanations of ML predictions in molecular design. Notably, model explanations often lack a ground truth. An explanation depends on the chosen XAI approach, the ML model, parameter settings, and the calculation protocols. There is no ground truth for parameter or protocol variations. Therefore, it can often not be determined if an explanation is valid. Instead, one might need to formulate and test hypotheses or compare different XAI methods to assess the consistency of explanations.17 These aspects directly apply to the evaluation of model explanations in molecular design, as discussed below.From model explanation to interpretation
In the analysis of predictions, the terms explanation and interpretation are often interchangeably used. However, in XAI, a formal distinction is made between explainability, that is, the extraction of feature-based learning patterns from a model, and interpretability, that is, the translation of an explanation into terms understandable to humans.18–20 An explanation is therefore computational in nature (and thus falls into the AI domain), whereas interpretation requires human intelligence.From model interpretation to causality
In the life sciences including drug discovery, experimentally testable hypotheses and causal reasoning depend on explanations that can be directly related to chemically or biologically meaningful concepts, even when derived from complex or non-interpretable models.21 If ML predicts the outcome of a natural process, a causal relationship exists if features determining the prediction are directly responsible for the observed event.21 In such cases, establishing a causal relationship typically requires experimental follow-up. For example, one evaluates if chemical features determining activity predictions of small molecules are directly responsible for a specific biological activity. In addition, ML relying on causal inference (also termed causal ML) generally attempts to establish cause-and-effect relationships between different variables.22,23 Such relationships are often analyzed in medical diagnostics.23 They are statistically determined and also fall within the explanation-interpretation-causation framework of XAI. A misunderstanding of statistical relationships between different variables might lead to the assumption of false causal relationships.24 In ML, models producing predictions for other than apparent or assumed reasons are often referred to as ‘Clever Hans’ predictors.25,26 This term originated from psychology after a horse mistakenly believed to be capable of counting.25 In interdisciplinary research, Clever Hans predictors impair hypothesis-driven experimental design. In XAI, distinguishing between different types of explanations and understanding their foundations as well as limitations reduces the risk of over-interpretation.Potential biases in chemical data
Charted chemical space largely results from preferred synthetic reactions, and bioactive chemical space is strongly influenced by the nature of pharmaceutical targets and their compound binding characteristics. In addition, chemical optimization efforts produce series of analogues that influence and may distort compound distributions in bioactive chemical space. Therefore, corresponding molecular property distributions can show correlations that affect ML predictions and bias XAI methods relying on feature independence assumptions. As discussed below, this might lead to different explanations of predictions using closely related method variants.Methodological categories
XAI methods can be categorized based on algorithmic principles for generating explanations of predictions, as illustrated in Fig. 1. Feature attribution methods quantify the contribution of individual input features to model predictions.27 Locally interpretable model-agnostic explanations (LIME)28 is an exemplary model-agnostic feature attribution method. LIME derives a linear surrogate model in the feature space vicinity of a test instance to quantitatively approximate original feature contributions to its prediction.Rule-based methods define non-redundant (if-then) decision rules in feature space that consistently yield corresponding predictions.29 ‘Anchors’ is a popular rule-based approach that probes the feature space around a test instance to generate a set of rules that anchors a prediction locally, regardless of other feature values.30 Rules are evaluated using metrics such as precision (that is, the fraction of consistent instance-based predictions when the rule applies) or coverage (that is, the proportion of test instances covered by the rule).
Albeit algorithmically distinct, feature attribution and rule-based methods both attempt to answer the question ‘why was prediction P obtained?’. By contrast, example-based methods address the question “why was prediction P obtained but not Q?”. This question reflects a human tendency to gain insights through object comparisons.31 Therefore, example-based XAI methods generate hypothetical samples that closely resemble a given test instance but result in a different prediction. ‘Counterfactuals’ are a leading example-based approach.32,33 In molecular design, preferred exemplary counterfactuals are very similar molecules (structural analogues) that lead to opposite predictions. A related yet distinct approach termed ‘contrastive explanations’ addresses the question “why was prediction P obtained but not Q?” by identifying origins of different prediction outcomes and ‘contrasting’ them.33,34 Therefore, an expected prediction (P) is defined as the ‘fact’ and an alternative (unexpected) prediction (Q) as the ‘foil’. Then, one attempts to identify feature subsets that are essential for model decisions distinguishing the fact from the foil.33Table 1 compares different XAI approaches and summarizes advantages and disadvantages.
| Category | Algorithms | Advantages | Disadvantages |
|---|---|---|---|
| Feature attribution | LIME | Local fidelity; complete explanation with all features | Not generalizable; sensitive to small input changes |
| SHAP | Global importance assessment; axiomatic foundation | Stochastic approximation | |
| Context-dependent interpretation | |||
| Permutation feature importance | Importance linked to model error; concise global importance measure | No feature importance information for predictions; vulnerable to feature correlation | |
| Integrated gradients | Global importance estimates; axiomatic foundation | Explanations depend on chosen weight threshold; restricted to neural networks | |
| Attention quantification | Applicable to transformers; no additional calculations or surrogate models required | Limited correlation with other methods; prone to high variance | |
| Rule-based | Anchors | Succinct informative explanations; quantifiable local fidelity | Outcome depends on chosen precision/coverage thresholds; limited scalability with large feature sets |
| Example-based | Counterfactual explanations | Multiple equivalent explanations; immediate interpretability | Multiple plausible explanations; limited global insights |
| Contrastive explanations | Identifies highly differentiating features; mimics human reasoning | Explanations based on feature subsets; limited global insights |
Feature attribution using Shapley values and approximations
One of the currently most popular feature attribution approaches in many areas is based on the Shapley value concept originating from cooperative game theory.35 The Shapley values were introduced to divide the ‘payoff’ of a game among players of a team according to their individual contributions.35Here, T is the team including all players,
 an ordered subset of players (termed coalition) of players,
an ordered subset of players (termed coalition) of players,  the value of coalition
the value of coalition  , p is a player, and ϕf(ν) the Shapley value of player p.
, p is a player, and ϕf(ν) the Shapley value of player p.
For ML, the Shapley value concept is applied based on an analogy: Players are features, the game is the prediction of a given test instance, and the payoff is the prediction of the test instance after subtracting the mean value of all other test set predictions. The Shapley value of a feature is calculated as the mean marginal contribution to all possible coalitions. Accordingly, for n features, 2n coalitions must be accounted for. The Shapley value formalism makes it possible to quantify the importance of features that are either present or absent in test instances.
In ML, the number of features usually significantly exceeds the number of players in a team. Therefore, the calculation of exact Shapley over all possible coalitions is NP-hard and computationally infeasible for large feature sets. Furthermore, ML models require all features they were trained on to make predictions and cannot use different subsets. Therefore, to determine the contribution of a given coalition to a prediction, all features missing in the coalition must be randomized or sampled from a marginal distribution. These ML-specific challenges typically require the approximation of Shapley values, for which a variety of methods have been introduced.36 Among these, ‘Shapley additive explanations’ (SHAP)37 is the most popular approach overall and the currently most widely used explanation method in molecular design. SHAP relies on feature attribution and local approximations and represents an extension of LIME.37,38 It is model-agnostic and quantifies individual feature contributions that add up to the probability of a given class label prediction.
SHAP variants
The original implementation of the SHAP formalism is often referred to as KernelSHAP because it generates a local approximation model with a special kernel function to estimate Shapley values using linear regression.37 Various SHAP variants have been introduced that adjust the formalism for specific ML methods or molecular decomposition schemes.36 These variants include MolSHAP that decomposes a compound into its core structure and substituents (R-groups) and estimates the importance of each R-group for a prediction using SHAP values calculated during iterative masking of R-groups.39 In addition, SHAP variants have been introduced for decision tree methods (TreeSHAP)40 and deep neural networks (DeepSHAP).41 The SHAP variant for decision tree structures enables the calculation of exact Shapley values.40 For support vector machines with binary features (such as molecular fingerprints) and different kernels, exact Shapley values can also be computed using another (non-SHAP) methodology.42One should consider that approximation methods for Shapley values generally take the average model output as the expected value, given coalitions comprising a constant subset of features for which remaining features are sampled from their marginal distributions.36 This approach yields an approximation of feature contributions by implicitly treating features as independent entities. However, this assumption is problematic in the presence of data correlation and produces varying explanations. Alternatively, features not kept constant in coalitions can be sampled from the conditional (observational) distribution.36 In this case, feature relations are accounted for by restricting sampled coalitions to feature combinations observed in the data. However, sampling from the conditional distribution can assign importance to irrelevant features if they are correlated with relevant features. The choice of the marginal or conditional distribution depends on whether priority is given to model fidelity or data characteristics, but influences explanations of given predictions.
SHAP analysis produces numerical explanations for molecular predictions. Such explanations require further analysis because they consist of importance values for each individual feature. Hence, they are neither concise nor immediately interpretable. To facilitate interpretation of explanations, chemical features are often projected on the structures of test compounds using atom-based feature mapping of Shapley values38 or heatmaps.43Fig. 2 shows examples that identify substructures with highest importance for correct predictions.
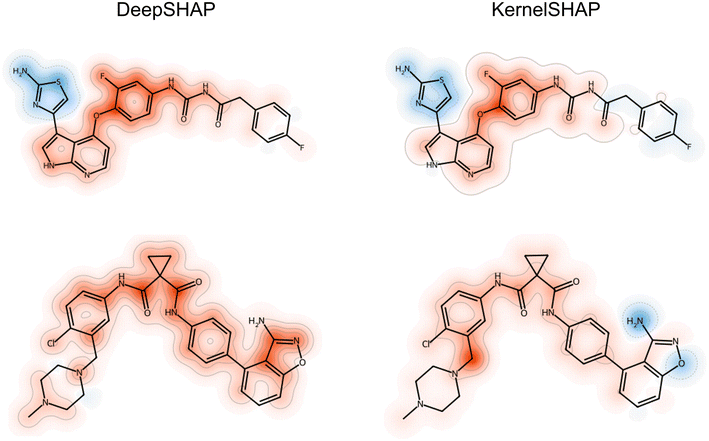 | ||
| Fig. 2 Feature mapping. A multi-layer perceptron was trained to distinguish between active and inactive compounds that were represented using atom environment (extended connectivity) fingerprints44 comprising atom sequence features. For two correctly predicted active compounds, feature importance values were calculated using model-agnostic KernelSHAP and the DeepSHAP variant for neural networks, respectively. The SHAP values for features present in these compounds were then projected on the structures by atom-based mapping. An atom might participate in multiple features and the corresponding SHAP values were summed. The atoms are color-coded in red (positive SHAP values, supporting the correct prediction) and blue (negative values opposing the prediction), with color intensity scaling with the size of the absolute values. | ||
Notably, such visualizations represent explanations incompletely because only features present in test compounds can be mapped, but not absent features. However, features absent in test compounds can also support or oppose predictions. Furthermore, the Shapley value concept requires that SHAP values add up to the difference between the expected value (average prediction of the model) and the prediction to be explained. As a result, identical importance values for different test instances have a different relative impact and should not be directly compared across multiple test instances without normalization.
Fig. 2 also illustrates that SHAP variants might generate different explanations for test compounds correctly predicted with a given ML model.17,42 For the compound at the top, the feature maps based on DeepSHAP and KernelSHAP explanations are similar, with the exception that the fluorophenyl ring weakly supports (DeepSHAP) or weakly opposes (KernelSHAP) the correct prediction. For the compound at the bottom, the magnitude of prediction support differs significantly for the explanations. For the aminoisoxazole ring, conflicting contributions are detected, with positive and negative values based on DeepSHAP and KernelSHAP, respectively. Notably, differences between explanations produced with different SHAP variants result from the stochastic nature of the underlying feature perturbation and sampling processes and ML method-specific algorithmic modifications. Importantly, such differences affect model explanation and interpretation.17 Therefore, it is advisable to consider alternative XAI concepts when explaining predictions.
While explanations based on feature attribution are primarily instance-based, cumulative (global) SHAP analysis can be carried out for multiple test compounds (or entire test sets) by aggregating explanations to quantify global feature importance.38 For example, global SHAP analysis was carried out to identify structural determinants of multi-target activity45,46 or metabolic stability47 of compounds. Global SHAP analysis also reveals general prediction characteristics of a given ML model. For example, in distinguishing between compounds with dual- and corresponding single-target activity using random forest models, correct predictions were mostly determined by features that were present in dual-target and absent in single-target compounds, as revealed using TreeSHAP.46 Thus, the models detected characteristic structural features in dual-target compounds and classified compounds in which these features were absent as single-target compounds.46
Structure-based explanations
Rationalizing predictions of new active compounds provides a basis for synthesis and experimental evaluation and the assessment of causality (see above). For numerical feature attribution, interpretation requires feature mapping or other follow-up analysis, as discussed above. However, by including domain-specific knowledge into XAI approaches, it is also possible to seamlessly integrate explanation and interpretation. Therefore, in molecular design, explanations of predictions in the applicability domain of a model should best be represented at the level of molecular structure.Anchors concept
The anchors concept, a representative rule-based XAI approach, has been supplemented with chemical knowledge, leading to the domain-specific MolAnchor methodology.48,49 In MolAnchor, test compounds are systematically decomposed by retrosynthetic substructure generation.50 Beginning with individual substructures, combinations are examined to derive rules of minimal structural composition that determine predictions. As an example, ‘if fragment X is present, then the compound is predicted to be active’. For high-dimensional feature representations such as molecular fingerprints, the combinatorial fragment exploration restricts the enormous search space of possible rules that the original anchors algorithm explores, leading to significant improvements in computational efficiency.48 Moreover, as illustrated in Fig. 3, MolAnchor analysis ensures that each explanation represents a chemically sound substructure (or combinations of substructures), rather than a feature combination from the global search space that might not be chemically understandable. By design, MolAnchor rules integrate explanation and chemical interpretation of predictions. In a recent application, MolAnchor analysis identified substructures in isoform-selective protein kinase inhibitors that consistently determined multi-task selectivity predictions and for which causal selectivity relationships were determined based on experimental data.49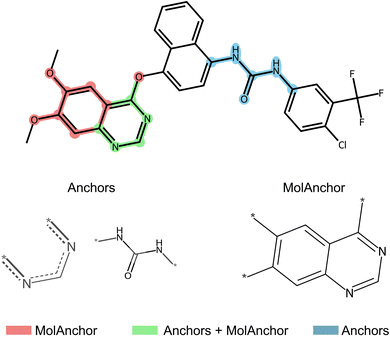 | ||
| Fig. 3 Structural explanations of predictions. For a correctly predicted active compound represented using an extended connectivity fingerprint,44 structural features representing decision rules are shown that were generated with the Anchors algorithm (blue), the MolAnchor variant (red), or both algorithms (green). The figure was taken from an open access publication by the authors48 and modified. Adapted with permission.48 Copyright 2024, Elsevier. | ||
Counterfactuals and contrastive explanations
The XAI concepts of counterfactuals and contrastive explanations have also been adapted for chemistry and drug design. Molecular counterfactuals are designed to explore narrow chemical space around a test instance of interest. Therefore, different computational strategies based on chemical knowledge have been introduced. The first approach, termed molecular explanation generator (MEG), sampled compounds with high structural similarity to a test instance through reinforcement learning using graph neural networks.51 This study focused on toxicity and solubility predictions showing, for example, that minimal structural changes of a test instance such as the addition of a methyl group reversed a toxicity prediction.51 An alternative method termed model agnostic counterfactual explanations (MMACE)52 charted local chemical space around a test instance through algorithmic permutation of self-referencing embedded strings.53,54 Compared to MEG and MMACE, a simpler method was introduced to generate counterfactuals for an entire compound test set by structural decomposition followed by systematic recombination.55 Therefore, analogue series were extracted from the data sets, their core structures were isolated, and iteratively recombined with a library of substituents. Candidate compounds were systematically re-predicted to identify counterfactuals for test instances with isolated cores, typically producing large numbers of counterfactuals.55Fig. 4A shows a representative example from a case study on different protein kinase inhibitors.56 In this example, counterfactuals of a given test inhibitor with predicted activity against different kinases directly provide experimentally testable hypotheses for exploring causal relationships, that is, responsibility of the exchanged structural moieties for activity against different kinases. This illustrates the integration of the computational explanation and human interpretation of predictions through molecular counterfactuals.Different from counterfactuals, contrastive explanations including chemical domain knowledge have only recently been introduced. Contrastive explanations identify minimal feature sets required for opposite predictions, also known as pertinent positive or negative feature sets.57,58 Therefore, feature sets for model derivation are often systematically reduced or, alternatively, perturbed to analyze their relative impact on the predictions.57,58 Iterative feature perturbation is also used to determine increasingly contrasting predictions for the fact and the foil class.33,34 For classification models, this can be accomplished by monitoring the resulting probabilities of fact or foil predictions. The degree of contrast is formalized as contrastive behavior δcontr that corresponds to the normalized shift in the probability distribution of the foil relative to the fact.59
The principles of contrastive explanations were adapted for molecular design, leading to the molecular contrastive explanations (MolCE) methodology.60 Instead of randomly perturbing features, MolCE introduces molecular feature perturbation by exchanging cores or substituents of test compounds with structurally analogous fragments. The resulting virtual compounds represent foils for which the contrastive behavior is determined. This theoretical framework enables the generation of chemically intuitive contrastive explanations that are directly interpretable at the level of molecular structure, comparable to molecular counterfactuals.
In the proof-of-concept study, MolCE was applied to multi-class predictions distinguishing between selective and non-selective ligands for members of the D2-like dopamine receptor (D2R) family.60 Different prediction tasks were analyzed using the fact-foil formalism including the correct prediction of non-selective and incorrect prediction of selective compounds. Fig. 4B shows an example of correctly predicted non-selective compounds. By determining the contrast shifts of these compounds, it was possible to identify structural features guiding predictions of receptor isoform selectivity. Thus, MolCE also integrates explanation and interpretation of predictions, leading to immediate causal inferences.
Chemical language models and transformers
Language models have been adapted for molecular property prediction and generative compound design. These models operate on textual chemical representations such as molecular strings and are often termed chemical language models (CLMs).61–63 Their popularity is largely due to their versatility in tackling generative design tasks that are often impossible to attempt with other ML methods.63 First-generation CLMs were mostly recurrent neural networks that were then increasingly replaced by transformers with their hallmark attention/self-attention mechanism.64,65 Currently, transformers represent the preferred CLM architecture.63,66 In addition to task-specific transformer CLMs, large language models (LLMs) are also adopted for chemistry, for example, through additional training with large amounts of chemical information.67,68 A major attraction of such models is combining their interactive use in natural language with specific chemical knowledge and tasks. Depending on the scope of the domain-specific training, these models can be used as 'AI research assistants'.67,68 Another emerging trend is the use of such models as increasingly autonomous ‘AI agents’ systems.68,69Explaining transformer models
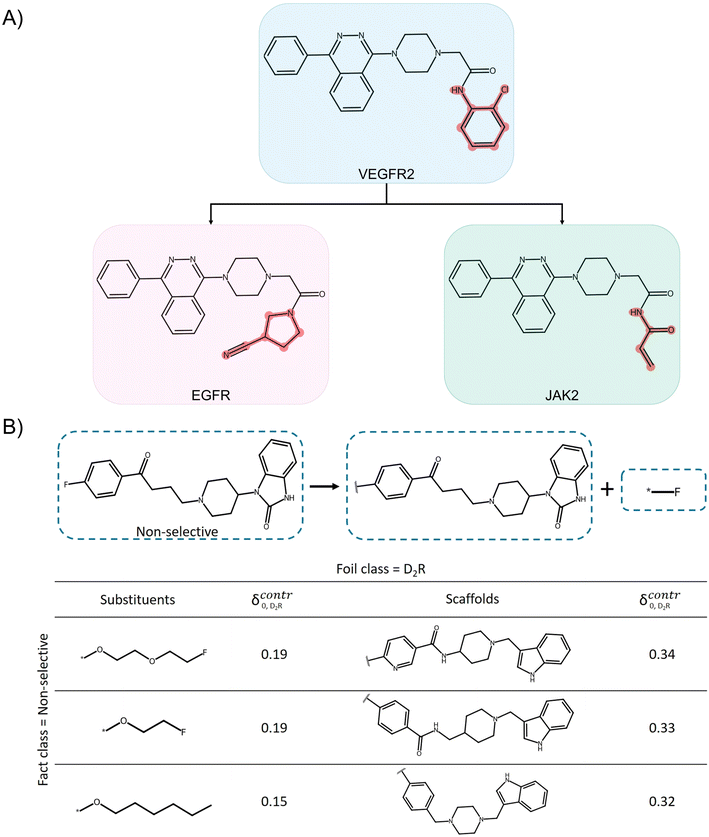
While the transformer architecture is highly flexible, its inherent complexity hinders model explanation.70 The black-box character is compounded by frequently observed hallucination behavior, especially for LLMs. Hallucination refers to the generation of seemingly plausible results without empirical support.71 So far, XAI research for transformers has mainly focused on quantifying the importance of individual tokens and token relationships using perturbation-, gradient-, and attention-based approaches.72–75 Perturbation-based approaches function analogously to those in feature attribution methods, as discussed above. Alternatively, gradient-based methods such as integrated gradients or layer-wise relevance propagation compute internal weight gradients based on back-propagation.75 Attention weights systematically quantify the importance of relationships between tokens within a sequence or across sequences for transformer modeling. For instance, the importance of input tokens for each output token can be determined. Attention weights are often displayed in so-called attention maps, which are heatmaps representing weights for pairwise token combinations. Fig. 5 shows an example for a CLM.While the attention map represents a visual explanation of a prediction, its chemical interpretability is limited. There is also a debate in the literature about whether attention weights can be used to explain transformer decisions, considering that their correlation with other feature importance values is typically weak.76,77 However, it was also shown that the application of feature attribution methods to transformer architectures and complex tokenization schemes is prone to high variance and errors.78 This raises the question of whether such comparisons should even be considered. Furthermore, in a notable study, feature attribution methods were applied in modified form to explain aqueous solubility predictions of an encoder-only transformer variant.79 Attention scores were calculated as a measure of token relevance and the SHAP formalism was extended using systematic masking of input tokens to compute SHAP values. The resulting explanations were not chemically interpretable, for instance, by analyzing functional groups and their influence on the predictions. Instead, the explanations appeared to be more related to molecular similarity relationships in latent space.79 These findings also raised the question of whether the transformer learned chemical information related to solubility during fine-tuning or largely relied on the similarity relationships of molecular embeddings encountered during the extensive pre-training procedure.
Given the difficulties in adapting or developing interpretable XAI approaches for transformer models, another recent study attempted to indirectly investigate learning characteristics of an encoder-decoder transformer CLM for a generative drug design application. Here, sequence-based compound design was chosen as a model system. Accordingly, the CLM was tasked with learning sequence-to-compound mappings to ultimately generate new compounds for given input protein sequences.80 This system made it possible to carry out many control calculations based on systematically modified sequences and sequence–compound pairs. The CLM successfully reproduced known active compounds for input proteins that were excluded from fine-tuning. However, the control calculations revealed that the model did not learn sequence information relevant for ligand binding. Instead, it heavily relied on sequence and molecular similarity relationships between training and test data and on compound memorization effects. Furthermore, the model tolerated sequence randomizations as long as ∼50–60% of a native sequence was retained, which was sufficient for learning. However, the distribution of retained sequence segments across native sequences had no influence on the results.80 Thus, sequence-based compound predictions were only statistically driven. The models relied on detectable compound and sequence similarity and memorization, but did not learn sequence motifs characteristic of protein families or implicated in ligand binding. These conclusions could be firmly drawn based on systematic control calculations, without the need to adopt XAI methods for transformer CLMs.
Conclusions
In molecular design, interpretable explanations of ML predictions play an important role. Candidate compounds are most likely not synthesized and tested if the predictions cannot be understood. This generally limits the impact of ML in drug discovery. In the AI era, the use of complex generative or property prediction models exacerbates this problem and leads to increasing interest in XAI. The inclusion of domain-specific knowledge helps to adapt XAI approaches for molecular design. For transformers, XAI approaches are still in their early stages. Attention maps are insufficient to generate chemically interpretable explanations. However, for CLMs, model-agnostic human-centered approaches such as counterfactuals are applicable. Such explanations offer immediate opportunities to formulate experimentally testable hypotheses for evaluating causal relationships. Notably, current CLMs rely entirely on statistical operations and the results may often not depend on learning chemically relevant information. In such cases, model overinterpretation likely leads to Clever Hans predictors. Purely statistically driven predictions may not benefit from structure-based explanations. Instead, concepts like uncertainty estimates or contrastive scoring of output instances might be more appropriate. Numerical estimates of feature importance can be visualized by feature mapping to identify and highlight regions in molecules that are primarily responsible for predictions. There are numerous opportunities for future research to develop new approaches for explaining black-box models in molecular design taking domain-specific knowledge into account.Author contributions
Conceptualization, J. B.; formal analysis, A. L. and J. B.; writing – original draft, A. L. and J. B.; writing – review & editing, A. L. and J. B.; supervision, J. B.Conflicts of interest
There are no conflicts to declare.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Acknowledgements
Support from the Lamarr Institute for research on explainable artificial intelligence is gratefully acknowledged.References
- J. Vamathevan, D. Clark, P. Czodrowski, I. Dunham, E. Ferran, G. Lee, B. Li, A. Madabhushi, P. Shah, M. Spitzer and S. Zhao, Applications of machine learning in drug discovery and development, Nat. Rev. Drug Discovery, 2019, 18, 463–477, DOI:10.1038/s41573-019-0024-5.
- R. S. K. Vijayan, J. Kihlberg, J. B. Cross and V. Poongavanam, Enhancing preclinical drug discovery with artificial intelligence, Drug Discovery Today, 2022, 27, 967–984, DOI:10.1016/j.drudis.2021.11.023.
- D. Castelvecchi, Can we open the black box of AI?, Nature, 2016, 538, 20–23, DOI:10.1038/538020a.
- C. Rudin, Why black box machine learning should be avoided for high-stakes decisions, in brief, Nat. Rev. Methods Primers, 2022, 2, 81, DOI:10.1038/s43586-022-00172-0.
- Y. Liang, S. Li, C. Yan, M. Li and C. Jiang, Explaining the black-box model: a survey of local interpretation methods for deep neural networks, Neurocomputing, 2021, 419, 168–182, DOI:10.1016/j.neucom.2020.08.011.
- K. Terayama, M. Sumita, R. Tamura and K. Tsuda, Black-box optimization for automated discovery, Acc. Chem. Res., 2021, 54, 1334–1346, DOI:10.1021/acs.accounts.0c00713.
- H. Askr, E. Elgeldawi, H. Aboul Ella, Y. A. M. M. Elshaier, M. M. Gomaa and A. E. Hassasien, Deep learning in drug discovery: an integrative review and future challenges, Artif. Intell. Rev., 2023, 56, 5975–6037, DOI:10.1007/s10462-022-10306-1.
- D. Gunning, M. Stefik, J. Choi, T. Miller, S. Stumpf and G. Z. Yang, XAI – explainable artificial intelligence, Sci. Robot., 2019, 4, eaay7120, DOI:10.1126/scirobotics.aay7120.
- G. Vilone and L. Longo, Notions of explainability and evaluation approaches for explainable artificial intelligence, Information Fusion, 2021, 76, 89–106, DOI:10.1016/j.inffus.2021.05.009.
- L. H. Gilpin, D. Bau, B. Z. Yuan, A. Bajwa, M. Specter and L. Kagal, Explaining explanations: an overview of interpretability of machine learning, IEEE 5th International Conference on Data Science and Advanced Analytics (DSAA), Turin, Italy, 2018, pp. 80–89, DOI:10.1109/DSAA.2018.00018.
- S. Ali, T. Abuhmed, S. El-Sappahg, K. Muhammad, J. M. Alonso-Moral, R. Confalonieri, R. Guidotti, J. Del Ser, N. Diaz-Rodriguez and F. Herrara, Explainable artificial intelligence (XAI): what we know and what is left to attain trustworthy artificial intelligence, Information Fusion, 2023, 99, 101805, DOI:10.1016/j.inffus.2023.101805.
- B. Goodman and S. Flaxman, European union regulations on algorithmic decision-making and a “right to explanation”, AI Magazine, 2017, 38, 50–57, DOI:10.1609/aimag.v38i3.2741.
- M. Velmurugan, C. Ouyang, Y. Xu, R. Sindhgatta, B. Wickramanayake and C. Moreira, Developing guidelines for functionally-grounded evaluation of explainable artificial intelligence using tabular data, Eng. Appl. Artif. Intell., 2025, 141, 109772, DOI:10.1016/j.engappai.2024.109772.
- D. V. Carvalho, E. M. Pereira and J. S. Cardoso, Machine learning interpretability: a survey on methods and metrics, Electronics, 2019, 8, 832, DOI:10.3390/electronics8080832.
- J. Zhou, A. H. Gandomi, F. Chen and A. Holzinger, Evaluating the quality of machine learning explanations: a survey on methods and metrics, Electronics, 2021, 10, 593, DOI:10.3390/electronics10050593.
- G. P. Wellawatte, H. A. Gandhi, A. Seshadri and A. D. White, A perspective on explanations of molecular prediction models, J. Chem. Theory Comput., 2023, 19, 2149–2160, DOI:10.1021/acs.jctc.2c01235.
- A. Lamens and J. Bajorath, Comparing explanations of molecular machine learning models generated with different methods for the calculation of Shapley values, Mol. Inform., 2025, 44, e202500067, DOI:10.1002/minf.202500067.
- W. J. Murdoch, C. Singh, K. Kumbier, R. Abbasi-Asl and B. Yu, Definitions, methods, and applications in interpretable machine learning, Proc. Natl. Acad. Sci. U. S. A., 2019, 44, 2071–2080, DOI:10.1073/pnas.1900654116.
- E. S. Ortigossa, T. Gonçalves and L. G. Nonato, Explainable artificial intelligence (XAI) – from theory to methods and applications, IEEE Access, 2024, 12, 80799–80846, DOI:10.1109/ACCESS.2024.3409843.
- A. Holzinger, G. Langs, H. Denk, K. Zatloukal and H. Müller, Causability and explainability of artificial intelligence in medicine, Wiley Interdiscip. Rev. Data Min. Knowl. Discov., 2019, 9, e1312, DOI:10.1002/widm.1312.
- J. Bajorath, From scientific theory to duality of predictive artificial intelligence models, Cell Rep. Phys. Sci., 2025, 6, 102516, DOI:10.1016/j.xcrp.2025.102516.
- J. Pearl, The seven tools of causal inference, with reflections on machine learning, Communications of the ACM, 2019, 62, 54–60, DOI:10.1145/324103.
- J. G. Richens, C. M. Lee and S. Johri, Improving the accuracy of medical diagnosis with causal machine learning, Nat. Commun., 2020, 11, 3923, DOI:10.1038/s41467-020-17419-7.
- J. Bajorath, Potential inconsistencies or artifacts in deriving and interpreting deep learning models and key criteria for scientifically sound applications in the life sciences, Artif. Intell. Life Sci., 2024, 5, 100093, DOI:10.1016/j.ailsci.2023.100093.
- O. Pfungst, Clever Hans (the horse of Mr Von Osten): contribution to experimental animal and human psychology, J. Philos. Psychol. Sci. Meth., 1911, 8, 663–666, DOI:10.2307/2012691.
- S. Lapuschkin, S. Wäldchen, A. Binder, G. Montavon, W. Samek and K. R. Müller, Unmasking Clever Hans predictors and assessing what machines really learn, Nat. Commun., 2019, 10, 1096, DOI:10.1038/s41467-019-08987-4.
- S. Kamolov, Feature attribution methods in machine learning: a state-of-the-art review, Annals Math Comp. Sci., 2025, 29, 104–111, DOI:10.56947/amcs.v29.635.
- M. T. Ribeiro, S. Singh and C. Guestrin, Why should I trust you? Explaining the predictions of any classifier, Proc. 22nd ACM SIGKDD Int. Conf. Knowl. Discov. Data Min., 2016, pp. 1135–1144. doi: DOI:10.1145/2939672.2939778.
- J. Van Der Waal, E. Nieuwburg, A. Cremers and M. Neerincx, Evaluating XAI: a comparison of rule-based and example-based, explanations, Artif. Intell., 2021, 291, 103404, DOI:10.1016/j.artint.2020.103404.
- M. T. Ribeiro, S. Singh and C. Guestrin, Anchors: high-precision model-agnostic explanations, Proc. AAAI Conf. Artif. Intell., 2018, 32, 1727–1735, DOI:10.1609/aaai.v32i1.11491.
- T. Miller, Explanation in artificial intelligence: insights from the social sciences, Artif. Intell., 2019, 267, 1–38, DOI:10.1016/j.artint.2018.07.007.
- S. Dandl, C. Molnar, M. Binder and B. Bischl, Multi-objective counterfactual Explanations, Proc. Int. Conf. Parallel Problem Solving from Nature (PPSN XVI), 2020, 448–469, DOI:10.1007/978-3-030-58112-1_31.
- I. Stepin, J. M. Alonso, A. Catala and M. Pereira-Fariña, Survey of contrastive and counterfactual explanation generation methods for explainable artificial intelligence, IEEE Access, 2021, 9, 11974–12001, DOI:10.1109/ACCESS.2021.3051315.
- O. Lipton, Contrastive explanation, Royal Institute of Philosophy Supplements, 1990, 27, 247–266, DOI:10.1017/S1358246100005130.
- L. S. Shapley, A value for n-person games, in Contributions to the Theory of Games, Princeton University Press, 1953, vol. II, pp. 307–318 Search PubMed.
- H. Chen, I. C. Covert, S. M. Lundberg and S. Lee, Algorithms to estimate Shapley value feature attributions, Nat. Mach. Intell., 2023, 5, 590–601, DOI:10.1038/s42256-023-00657-x.
- S. M. Lundberg and S. Lee, A unified approach to interpreting model predictions, Adv. Neur. Inf. Proc. Sys., 2017, 30, 4765–4774 Search PubMed.
- R. Rodriguez-Perez and J. Bajorath, Interpretation of compound activity predictions from complex machine learning models using local approximations and Shapley values, J. Med. Chem., 2019, 63, 8761–8777, DOI:10.1021/acs.jmedchem.9b01101.
- T. Tian, S. Li, M. Fang, D. Zhao and J. Zeng, MolSHAP: interpreting quantitative structure-activity relationships using Shapley values of R-groups, J. Chem. Inf. Model., 2023, 63, 2236–2249, DOI:10.1021/acs.jcim.3c00465.
- S. M. Lundberg, G. Erion, H. Chen, A. DeGrave, J. M. Prutkin, B. Nair, R. Katz, J. Himmelfarb, N. Bansal and S. Lee, From local explanations to global understanding with explainable AI for trees, Nat. Mach. Intell., 2020, 2, 56–67, DOI:10.1038/s42256-019-0138-9.
- H. Chen, S. M. Lundberg and S. Lee, Explaining a series of models by propagating Shapley values, Nat. Commun., 2023, 13, 4512, DOI:10.1038/s41467-022-31384-3.
- J. P. Roth and J. Bajorath, Machine learning models with distinct Shapley value explanations decouple feature attribution and interpretation for chemical compound predictions, Cell Rep. Phys. Sci., 2024, 5, 102110, DOI:10.1016/j.xcrp.2024.102110.
- T. Harren, H. Matter, G. Hessler, M. Rarey and C. Grebner, Interpretation of structure–activity relationships in real-world drug design data sets using explainable artificial intelligence, J. Chem. Inf. Model., 2022, 62, 447–462, DOI:10.1021/acs.jcim.1c01263.
- D. Rogers and M. Hahn, Extended-connectivity fingerprints, J. Chem. Inf. Model., 2010, 50, 742–754, DOI:10.1021/ci100050t.
- R. Rodriguez-Perez and J. Bajorath, Interpretation of machine learning models using Shapley values: application to compound potency and multi-target activity predictions, J. Comput.-Aided Mol. Des., 2020, 34, 1013–1026, DOI:10.1007/s10822-020-00314-0.
- C. Feldmann, M. Philipps and J. Bajorath, Explainable machine learning predictions of dual-target compounds reveal characteristic structural features, Sci. Rep., 2021, 11, 21594, DOI:10.1038/s41598-021-01099-4.
- A. Wojtuch, R. Jankowski and S. Podlewska, How can SHAP values help to shape metabolic stability of chemical compounds?, J. Cheminform., 2021, 13, 74, DOI:10.1186/s13321-021-00542-y.
- A. Lamens and J. Bajorath, MolAnchor method for explaining compound predictions based on substructures, Eur. J. Med. Chem. Rep, 2024, 12, 100230, DOI:10.1016/j.ejmcr.2024.100230.
- A. Lamens and J. Bajorath, Rationalizing predictions of isoform-selective phosphoinositide 3-kinase inhibitors using MolAnchor analysis, J. Chem. Inf. Model., 2025, 65, 1357–1366, DOI:10.1021/acs.jcim.4c02153.
- J. Degen, C. Wegscheid-Gerlach, A. Zaliani and M. Rarey, On the art of compiling and using ‘drug-like’ chemical fragment spaces, ChemMedChem, 2008, 3, 1503–1507, DOI:10.1002/cmdc.200800178.
- D. Numeroso and D. Bacciu, MEG: generating molecular counterfactual explanations for deep graph networks, in 2021 International Joint Conference on Neural Networks (IJCNN), IEEE, 2021, pp. 1–8, DOI:10.1109/IJCNN52387.2021.9534266.
- G. P. Wellawatte, A. Seshadri and A. D. White, Model-agnostic generation of counterfactual explanations for molecules, Chem. Sci., 2022, 13, 3697–3705, 10.1039/d1sc05259d.
- M. Krenn, F. Häse, A. K. Nigam, P. Friederich and A. Aspuru-Guzik, Self-referencing embedded strings (SELFIES): a 100% robust molecular string representation, Mach. Learn. Sci. Technol., 2020, 1, 045024, DOI:10.1088/2632-2153/aba947.
- A. Nigam, R. Pollice, M. Krenn, G. dos Passos Gomes and A. Aspuru-Guzik, Beyond generative models: superfast traversal, optimization, novelty, exploration and discovery (STONED) algorithm for molecules using SELFIES, Chem. Sci., 2021, 12, 7079–7090, 10.1039/d1sc00231g.
- A. Lamens and J. Bajorath, Generation of molecular counterfactuals for explainable machine learning based on core–substituent recombination, ChemMedChem, 2024, 19, e202300586, DOI:10.1002/cmdc.202300586.
- A. Lamens and J. Bajorath, Systematic generation and analysis of counterfactuals for compound activity predictions using multi-task models, RSC Med. Chem., 2024, 15, 1547–1555, 10.1039/d4md00128a.
- T. Miller, Contrastive explanation: a structural-model approach, Knowl. Eng. Rev., 2021, 36, e14, DOI:10.1017/S0269888921000102.
- A. Dhurandhar, T. Pedapati, A. Balakrishnan, P. Y. Chen, K. Shanmugam and A. Puri, Model-agnostic contrastive explanations for classification models, IEEE J. Emerg. Sel. Top. Circuits Syst., 2024, 14, 789–798, DOI:10.1109/JETCAS.2024.3486114.
- A. Jacovi, S. Swayamdipta, S. Ravfogel, Y. Elazar, Y. Choi and Y. Goldberg, Contrastive explanations for model interpretability, arXiv, preprint, arXiv:2103.01378, DOI:10.48550/arXiv.2103.01378.
- A. Lamens and J. Bajorath, Contrastive explanations for machine learning predictions in chemistry, J. Cheminform., 2025, 17, 143, DOI:10.1186/s13321-025-01100-6.
- M. A. Skinnider, R. G. Stacey, D. S. Wishart and L. J. Foster, Chemical language models enable navigation in sparsely populated chemical space, Nat. Mach. Intell., 2021, 3, 759–770, DOI:10.1038/s42256-021-00368-1.
- F. Grisoni, Chemical language models for de novo drug design: challenges and opportunities, Curr. Opin. Struct. Biol., 2023, 79, 102527, DOI:10.1016/j.sbi.2023.102527.
- J. Bajorath, Chemical language models for molecular design, Mol. Inform., 2024, 43, e202300288, DOI:10.1002/minf.202300288.
- A. Vaswani, N. Shazeer, N. Parmar, J. Uszkoreit, L. Jones, A. N. Gomez, L. Kaiser and I. Polosukhin, Attention is all you need, Adv. Neural Inf. Process. Syst., 2017, 30, 5998–6008 Search PubMed.
- Z. Niu, G. Zhong and H. Yu, A review on the attention mechanism of deep learning, Neurocomputing, 2021, 452, 48–62, DOI:10.1016/j.neucom.2021.03.091.
- K. D. Luong and A. Singh, Application of transformers in cheminformatics, J. Chem. Inf. Model., 2024, 64, 4392–4409, DOI:10.1021/acs.jcim.3c02070.
- Z. Zhao, D. Ma, L. Chen, L. Sun, Z. Li, Y. Xia, B. Chen, H. Xu, Z. Zhu, S. Zhu, S. Fan, G. Shen, K. Yu and X. Chen, Developing ChemDFM as a large language foundation model for chemistry, Cell Rep. Phys. Sci., 2025, 6, 102523, DOI:10.1016/j.xcrp.2025.102523.
- M. C. Ramos, C. J. Collison and A. D. White, A review of large language models and autonomous agents in chemistry, Chem. Sci., 2025, 16, 2514–2572, 10.1039/d4sc03921a.
- D. A. Boiko, R. MacKnight, B. Kline and G. dos Passos Gomes, Autonomous chemical research with large language models, Nature, 2023, 624, 570–578, DOI:10.1038/s41586-023-06792-0.
- C. Singh, J. P. Inala, M. Galley, R. Caruana and J. Gao, Rethinking interpretability in the era of large language models, arXiv, 2024, preprint, arXiv:2402.01761, DOI:10.48550/arXiv.2402.01761.
- L. Huang, W. Yu, W. Ma, W. Zhong, Z. Feng, H. Wang, Q. Chen, W. Peng, X. Feng, B. Qin and T. Liu, A survey on hallucination in large language models: principles, taxonomy, challenges, and open questions, ACM Trans. Inf. Syst., 2025, 43, 1–55, DOI:10.1145/3703155.
- S. Liu, F. Le, S. Chakraborty and T. Abdelzaher, On exploring attention-based explanation for transformer models in text classification, Proc. IEEE Int. Conf. Big Data, 2021, 1193–1203, DOI:10.1109/BigData52589.2021.9671639.
- S. Kitada and H. Iyatomi, Attention meets perturbations: robust and interpretable attention with adversarial training, IEEE Access, 2021, 9, 92974–92985, DOI:10.1109/ACCESS.2021.3093456.
- S. Abnar and W. Zuidema, Quantifying attention flow in transformers, arXiv, 2020, preprint, arXiv:2005.00928, DOI:10.48550/arXiv.2005.00928.
- M. Ribeiro, B. Malcorra, N. B. Mota, R. Wilkens, A. Villavicencio, L. C. Hubner and C. Rennó-Costa, A methodology for explainable large language models with integrated gradients and linguistic analysis in text classification, arXiv, 2024, preprint, arXiv:2410.00250, DOI:10.48550/arXiv.2410.00250.
- S. Jain and B. C. Wallace, Attention is not explanation, arXiv, 2019, preprint, arXiv:1902.10186, DOI:10.48550/arXiv.1902.10186.
- S. Wiegreffe and Y. Pinter, Attention is not not explanation, arXiv, 2019, preprint, arXiv:1908.04626, DOI:10.48550/arXiv.1908.04626.
- P. B. R. Hartog, F. Krüger, S. Genheden and I. V. Tetko, Using test-time augmentation to investigate explainable AI: inconsistencies between method, model and human intuition, J. Cheminform., 2024, 16, 39, DOI:10.1186/s13321-024-00824-1.
- S. Hödl, T. Kachman, Y. Bachrach, W. T. S. Huck and W. E. Robinson, What can attribution methods show us about chemical language models?, Digit. Discov., 2024, 3, 1738–1748, 10.1039/d4dd00084f.
- J. P. Roth and J. Bajorath, Unraveling learning characteristics of transformer models for molecular design, Patterns, 2025, 6, 101392, DOI:10.1016/j.patter.2025.101392.
| This journal is © The Royal Society of Chemistry 2026 |