Open Access Article
Open Access ArticleSimultaneous occupancy of CuC and CuD in the ammonia monooxygenase active site
Frank J.
Tucci
 a,
Madeline B.
Ho
a,
Madeline B.
Ho
 a,
Aaron A. B.
Turner
a,
Aaron A. B.
Turner
 b,
Lisa Y.
Stein
b,
Lisa Y.
Stein
 b,
Brian M.
Hoffman
b,
Brian M.
Hoffman
 a and
Amy C.
Rosenzweig
a and
Amy C.
Rosenzweig
 *a
*a
aDepartments of Molecular Biosciences and of Chemistry, Northwestern University, Evanston, Illinois 60208, USA
bDepartment of Biological Sciences, University of Alberta, Edmonton, AB, Canada T6G 2E9. E-mail: amyr@northwestern.edu
First published on 18th December 2025
Abstract
Ammonia monooxygenase (AMO), a copper-dependent membrane enzyme, catalyzes the first and rate-limiting step of nitrification: the oxidation of ammonia to hydroxylamine. Despite its central role in the global nitrogen cycle and its biotechnological relevance, structural characterization of AMO has lagged behind that of its homolog, particulate methane monooxygenase (pMMO), due to the slow growth rates of ammonia-oxidizing bacteria and the instability of AMO upon purification. Recent cryoEM studies of Nitrosomonas europaea AMO and Methylococcus capsulatus (Bath) pMMO in native membranes revealed new structural features, including two adjacent copper-binding sites in the transmembrane region, CuC and CuD, believed to constitute the active site. Although multiple structures were determined under various conditions, simultaneous occupancy of CuC and CuD was never observed, leaving their potential functional interplay unresolved. Here we report the 2.6 Å resolution cryoEM structure of AMO from Nitrosospira briensis C-128 in isolated native membranes. This structure reveals the first instance of simultaneous copper occupancy of the CuC and CuD sites, along with occupancy of the periplasmic CuB site. Electron paramagnetic resonance (EPR) spectroscopic data indicate that the CuB site is primarily occupied by Cu(II), while CuC and CuD are primarily occupied by diamagnetic ions, presumably Cu(I). Notably, a lipid molecule is bound between the CuC and CuD sites, separating them by ∼8.0 Å. The results underscore the importance of studying these enzymes in their native environments across species to resolve conserved and divergent molecular features.
Introduction
Methane- and ammonia-oxidizing bacteria consume methane and release nitrous oxide, respectively, regulating atmospheric levels of these potent greenhouse gases.1 In the first steps of their metabolic pathways, methane oxidizers convert methane to methanol while ammonia oxidizers convert ammonia to hydroxylamine.2,3 These challenging chemical reactions are catalyzed by homologous copper-dependent, membrane-bound enzymes: particulate methane monooxygenase (pMMO) and ammonia monooxygenase (AMO).4–6 Understanding the molecular chemistry of these enzymes is of substantial interest for both environmental and biotechnological applications.7–10Initial crystal structures of pMMO, derived from inactive, detergent-solubilized samples, revealed an ∼300 kDa complex comprising three copies each of the PmoA, PmoB, and PmoC subunits.11 These structures also revealed three mononuclear copper-binding sites—the bis-His, CuB, and CuC sites.5 However, the crystal structures lacked parts of PmoA and PmoC, including a highly conserved portion of PmoC adjacent to CuC in the transmembrane region.12 This region was later resolved by reconstituting pMMO into lipid nanodiscs, discoidal lipid bilayers that partially restore pMMO activity, especially when prepared with native lipids extracted from whole cells of methane-oxidizing bacteria.13 High resolution cryoEM structures of these nanodisc samples revealed the missing portions of PmoA and PmoC, stabilized by interacting lipids, along with a previously unknown copper-binding site denoted CuD, located within 6 Å of CuC. Combined cryoEM and spectroscopic data then showed that a hydrophobic pocket adjacent to CuD binds the product analog trifluoroethanol, pinpointing this region as the likely active site.14
In multiple structures of active Methylococcus capsulatus (Bath) pMMO in lipid nanodiscs, CuD is occupied while CuC is empty. By contrast, structures of inactive Methylotuvimicrobium alcaliphilum 20Z and Methylocystis sp. Rockwell pMMOs in lipid nanodiscs contained occupied CuC sites with disordered CuD sites.13 These observations raised significant questions regarding the relationship between CuC and CuD, the possibility of simultaneous occupancy, and the role of lipids in pMMO function. In addition, the potential effects of purification and nanodisc reconstitution on the pMMO structure and copper sites remained unclear. To avoid structural distortions caused by purification, cryoEM single particle analysis (SPA) was then used to obtain structures of M. capsulatus (Bath) and M. sp. Rockwell pMMOs directly in native membranes.15 In both structures, CuD is occupied and CuC is vacant. Thus, CuD is present in pMMO from multiple species, supporting its identification as the active site.14
Attempts to characterize AMO had been hindered by its instability outside the membrane and the slow growth rates of ammonia-oxidizing bacteria, which precluded obtaining sufficient cell mass for structural studies.6,16 Recently, however, cryoEM SPA of native membranes also enabled structure determination of AMO from Nitrosomonas europaea (ATCC 19718).15 Like pMMO, N. europaea AMO is an α3β3γ3 trimer formed by the AmoA, AmoB, and AmoC subunits and contains CuB, CuC, and CuD binding sites. Notably, the N. europaea AMO structure revealed a previously unknown supernumerary helical subunit interacting with AmoC. In contrast to the native membrane pMMO structures, CuC is occupied while CuD is empty in the N. europaea AMO structure, deepening the mystery of these neighboring copper sites and underscoring the need for more data to establish a structural catalog of potential active-site copper occupancies and conformations.
Here we determined the 2.6 Å resolution cryoEM structure of AMO from Nitrosospira briensis C-128 (ref. 17) in native membranes, and assessed the Cu(II) occupancy of the metal-binding sites by electron paramagnetic resonance (EPR) spectroscopy. Surprisingly, the structure reveals a new active-site conformation with both CuC and CuD occupied, apparently with a phospholipid molecule wedged between them. This arrangement represents a dramatic deviation from those observed in prior AMO and pMMO structures, despite all being derived from native membranes with minimal biochemical manipulation. As such, it opens the possibility of catalytic involvement of both copper sites, which here are presumably occupied with Cu(I), and conceivably of lipids in the pMMO and AMO reactions. Further, this structure highlights the importance of studying pMMOs and AMOs across species to develop a comprehensive model of their molecular features.
Results and discussion
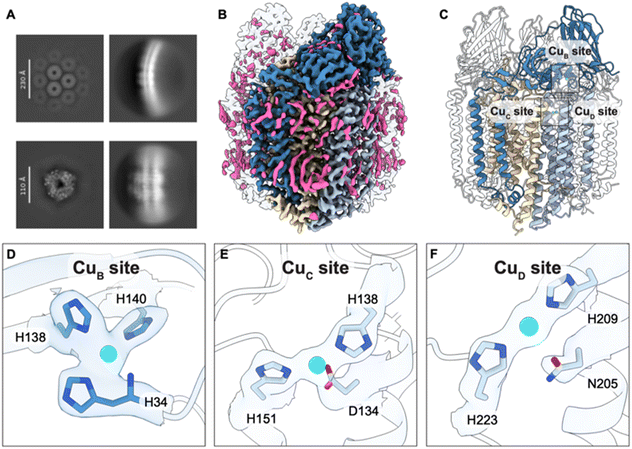
Membranes were isolated from ∼0.5 g of N. briensis cells. While both betaproteobacteria, N. europaea and N. briensis belong to divergent phyla,18 with 78, 85, and 88% identity of the AmoB, AmoA, and AmoC subunits, respectively. In addition, the two species have different intracytoplasmic membrane morphologies.19 The N. briensis membranes were applied to grids for data collection and SPA (Fig. S1). Like N. europaea AMO and the previously studied pMMOs,15,20N. briensis AMO forms hexagonal arrays in the membranes (Fig. 1A). The 2.6 Å resolution structure reveals a trimer, similar to prior pMMO and AMO structures, with numerous bound lipids encasing the transmembrane region (Fig. 1B and C). There is no density corresponding to supernumerary helices like those identified in N. europaea AMO (Fig. S2),15 suggesting that such subunits are not present in, or at least not strongly associated with, all AMOs.Inspection of the cryoEM map revealed three occupied metal-binding sites. First, the CuB site is occupied and exhibits the same coordination as observed in all pMMO structures5 and in the N. europaea AMO structure:15 three nitrogen atoms from histidine side chains and the amino terminal nitrogen of AmoB (Fig. 1D). The PmoB bis-His site5 is not present in AMOs, as one of the histidine ligands is replaced by glutamine. Second, the CuC site is clearly occupied by a metal ion coordinated by two histidines (His138, His151) and an aspartate residue (Asp134), consistent with previous pMMO and AMO structures.13,15 Surprisingly, the CuD site is also occupied, providing the first evidence for simultaneous occupancy of CuC and CuD as well as for the presence of CuD in AMO. The metal ion in the CuD site is coordinated by two histidine residues (His209, His223) in a linear, two-coordinate geometry (Fig. 1F). The side chain oxygen of the asparagine residue (Asn205) that typically coordinates CuD in pMMOs is >3 Å from the copper ion.
The occupancy of Cu(II) in these three sites was assessed by EPR spectroscopy of the same sample of N. briensis membranes used for cryoEM (Fig. 2A). Given that the coordination spheres of the three metal binding sites identified by cryoEM are essentially identical to those in pMMO, we assume that the spectra from a Cu(II) ion in each site should be characterized by the spin Hamiltonian parameters obtained previously for those sites in pMMO, with essentially identical parameters for CuC and CuD (g = [2.29, 2.07, 2.05] and 63Cu A‖ = 575 MHz), distinct from those for CuB (g = [2.241, 2.068, 2.035] and 63Cu A‖ = 575 MHz).21,22 The Cu hyperfine coupling in the g‖ region of the AMO spectrum is incompatible with a simulation that assumes the spectrum arises from the CuC/CuD sites (Fig. S3). Instead, the spectrum agrees with that of CuB, in particular matching well in the g‖ region. The simulated spectrum in the g⊥ region is improved by inclusion of a small contribution from a free-radical with g ∼ 2 (Fig. 2A, B and E). In addition, this region is further improved by assuming the AMO spectrum includes a minority contribution from the CuC/CuD sites (Fig. 2A), which together account for roughly ∼13% of the total Cu(II) signal (Fig. 2C and D).
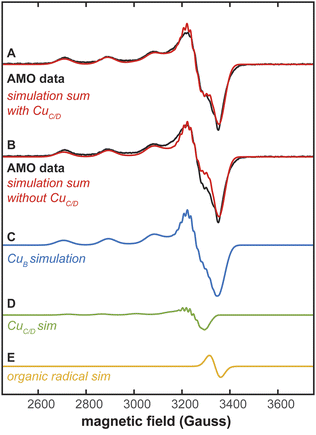 | ||
| Fig. 2 EPR spectroscopy of N. briensis AMO in native membranes. (A) EPR spectrum of N. briensis AMO in native membranes overlaid with a simulation that includes contributions from CuB, CuC/D, and an organic free radical with g ∼ 2. In this simulation, CuB comprises 87% of the total Cu(II) and CuC/D makes up 13% of the total Cu(II). (B) EPR spectrum of N. briensis AMO in native membranes overlaid with a simulation that includes only the contributions from CuB and the organic free radical. (C) The CuB component of the simulations in (A and B). (D) The CuC/CuD component of the simulation in (A). (E) The organic free radical component of simulations in (A and B). EPR conditions: 9.376 GHz microwave frequency, 200 µW microwave power, temperature 20 K, 320 ms time constant, 12.5 G modulation, 20 scans, 100 s per scan. EPR simulation parameters: g-tensors and hyperfine couplings adapted from ref. 21, CuBg = [2.243, 2.068, 2.035], A(63/65Cu) = [572, 15, 14] MHz, A(14Na) = [34.2, 38.5, 38.5] MHz, A(14Nb) = [37.8, 38.5, 38.5] MHz, A(14Nc) = [37.8, 38.5, 38.5] MHz, A(14Nd) = [37.8, 47.0, 47.0] MHz. CuC/Dg = [2.28, 2.07, 2.05], A(63/65Cu) = [440, 40, 20] MHz, A(14Na) = [34.2, 38.5, 38.5] MHz, A(14Nb) = [37.8, 47.0, 47.0]. The organic free radical was simulated as g = 2.0 with Hstrain = [60, 60, 60] MHz. Copper hyperfine couplings were simulated with natural abundance mixtures of 63Cu and 65Cu. The respective weights used in the summed simulation are 13% CuC/D and 87% CuB. | ||
These data suggest that the CuC/CuD sites are primarily occupied by EPR-silent Cu(I). The dominance of the AMO signal by CuB(II) is analogous to what is observed for pMMO,5,22 in which the CuC/CuD sites are present as Cu(I) in vivo and remain mostly reduced in native membranes, with oxidation occurring only upon solubilization in detergent and enzyme purification.21,22 Air oxidation of the AMO-containing sample did not lead to a significantly more intense CuC/CuD signal (Fig. S4), even upon bubbling with O2, suggesting that these membrane-bound sites may be more resistant to oxidation than the CuC/CuD sites in pMMO, or may be occupied in some part by non-copper, EPR-silent ions such as Zn(II). Since methane is also a substrate for AMOs,23 the sample was tested for methane oxidation activity using an established assay,24 but no methanol was detected. The absence of activity could be related to the difficulty oxidizing the CuC/CuD sites.
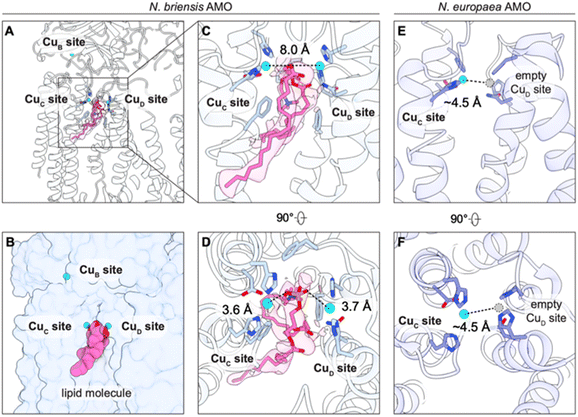
While both CuC and CuD are occupied by metal ions, presumably Cu(I) based on the EPR spectra and those of previous samples of pMMO and AMO, the two sites are significantly farther apart than predicted by previous pMMO and AMO cryoEM structures (Fig. 3A–D).13,15 In the previous structures, when either CuC or CuD is occupied, the ligands for the other site are oriented such that a second copper ion would be located ∼4.5–5.6 Å away (Fig. 3E, F and S5). By contrast, the two copper ions modeled in N. briensis AMO are separated by ∼8.0 Å (Fig. 3C). The active-site pocket is filled with a nonprotein density that appears to wedge the two copper sites apart (Fig. 3A–D). This density resembles a two-tailed phospholipid, showing clear densities for a headgroup and two acyl chains that extend into the transmembrane region, flanked by the three conserved phenylalanine residues that line the active site as identified by trifluoroethanol binding to pMMO (Fig. 3C and D).14 The density is well modeled as 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) (Fig. 3A–C). A lipid of this chain length is consistent with reported lipid compositions for ammonia oxidizers.25,26 The lipid does not appear to coordinate the copper centers, with the oxygen atoms of its phosphate group ∼3.6–3.7 Å from both CuC and CuD (Fig. 3C).
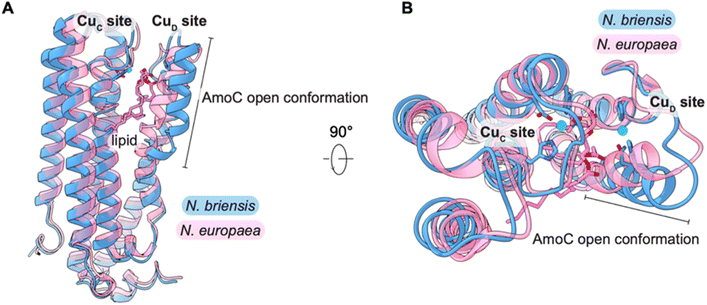
The presence of this lipid shifts the helix containing CuD ligands Asn205 and His209 markedly from its position in the N. europaea AMO structure (Fig. 4). This conformational change is facilitated by the presence of a conserved proline residue (Pro200)12 that introduces a kink in the helix, bending it toward the central opening of the AMO trimer (Fig. S6). The resultant open conformation of AmoC (spanning residues 194–217) is accommodated by a shift in residues 205–215 of the AmoA subunit. While this region of AmoC and PmoC is clearly flexible, as evidenced by its disorder in pMMO crystal structures,12 this conformation has not been observed previously. The lipid itself is partially exposed in the groove between protomers, suggesting it originates from this region (Fig. 3B).
Conclusions
The discovery of the pMMO CuD center13 raised critical questions regarding the active site, with the looming possibility that it could be dinuclear. Until now, simultaneous occupancy of CuC and CuD had not been observed, even computationally,27 precluding substantiated consideration of such a model. The present N. briensis AMO structure clearly shows density attributable to copper in both sites with an ∼8 Å separation imposed by an intervening lipid molecule. Importantly, the structure was determined directly from native membranes without biochemical manipulation. Thus, this lipid represents the first example of an endogenous molecule bound in the pMMO/AMO active site and could be relevant to function. Attempts to load the two sites in vitro with Cu(II), Cu(I), a mixture thereof, or other metal ions have not been successful, suggesting a potential scenario in which increased spatial separation facilitates dual copper loading in vivo. The structure may represent a loading conformation, in which CuC exists as a copper repository for a mononuclear CuD catalytic center.Alternatively, the two sites may both be occupied during catalysis, perhaps moving closer together, either to the predicted 4.5–5.6 Å separation or even closer, although there is no spectroscopic evidence for a coupled dicopper cluster. In general, occupancy of CuD correlates with activity, with CuD occupied and CuC empty in structures of active M. capsulatus (Bath) pMMO in lipid nanodiscs13 and native membranes15 as well as structures with the product analog and inhibitor trifluoroethanol.14 By contrast, only CuC is occupied in structures of inactive M. alcaliphilum 20Z and M. sp. Rockwell pMMOs in lipid nanodiscs13 as well as in all pMMO crystal structures, in which the CuD site and surrounding residues are not observed.12 However, only CuD is occupied in the structure of inactive M. sp. Rockwell pMMO in native membranes.15 In the case of AMO, CuC is occupied in N. europaea AMO, and both sites are occupied in the current N. briensis pMMO structure; neither AMO exhibited activity, but sample limitations precluded extensive activity studies. However, it remains possible that the copper centers in different pMMO and AMO membrane samples are differentially disrupted upon cryoEM grid preparation. Given these considerations, it is difficult to correlate structure with activity.
One possibility is that both CuC and CuD are involved in activity, but are more conformationally flexible that typical dicopper centers.28 Notably, recent studies of the dicopper enzymes dopamine β-hydroxylase and peptidylglycine monooxygenase suggest a more dynamic picture than envisioned previously, with Cu–Cu distances spanning 4–14 Å.29–33 Further, several new classes of enzymes containing two copper-binding sites have emerged recently, including the BURP-domain peptide cyclases34 and copper-dependent halogenases.35 In the BURP-domain cyclases, two binding sites each with two histidine ligands are predicted to be separated by 7 Å,34 and may also adopt multiple conformations.36 Whether pMMO and AMO exhibit similar dynamics, perhaps mediated by lipids, remains to be established.
Materials and methods
Culturing of Nitrosospira briensis C-128
N. briensis C-128 was cultured in 1 L Wheaton bottles sealed with inlayed rubber butyl caps. HEPES-buffered HK medium (200 mL) containing 10 mM (NH4)2SO4 and 0.5% (v/v) phenol red was inoculated with 2% v/v late log phase culture.37 Cultures were incubated at 28 °C in the dark with shaking at 150 rpm and the pH was adjusted daily as needed with 5% (w/v) Na2CO3 to maintain pH 7.5–8.0. Growth was measured by NO2− production.38 To harvest biomass, cultures that achieved ca. 90% NH3 conversion to NO2− were centrifuged at 5000 × g for 45 min. Cell pellets were transferred to 15 mL sterile Falcon tubes and centrifuged at 2000 × g for 15 min to completely remove supernatant. Pellets were flash-frozen in a −70 °C ethanol bath and shipped on dry ice.Isolation of N. briensis membranes
Approximately 0.5 g of N. briensis cells were resuspended in buffer containing 25 mM PIPES (pH 7.3) and 250 mM NaCl with DNase (Sigma-Aldrich) and EDTA-free complete protease inhibitor cocktail (Sigma-Aldrich). The resuspended cells were lysed at 4 °C by sonication at 80% power (45 amplitude, 1 second on and 1 second off) for 10 min using a QSonica Q700 instrument. The lysate was clarified by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 45 min. The supernatant was preserved, and membranes were isolated from this supernatant by ultracentrifugation at 150
000 × g for 45 min. The supernatant was preserved, and membranes were isolated from this supernatant by ultracentrifugation at 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 1 h. The pellet was then harvested and resuspended in 200 µL of lysis buffer with a Dounce homogenizer.
000 × g for 1 h. The pellet was then harvested and resuspended in 200 µL of lysis buffer with a Dounce homogenizer.
CryoEM grid preparation and screening
Isolated membranes from N. briensis were diluted to 4 mg mL−1 of total protein as measured by the DC Lowry (Bio-Rad) assay. This sample was then blotted and plunge frozen in liquid ethane on C-Flat holey carbon copper grids with 1.2 µm holes, 1.3 µm spacing, and 400 mesh (Electron Microscopy Sciences) with an FEI Vitrobot. Prior to blotting, grids were plasma cleaned using a Solarus plasma cleaner (Gatan) for 10 s at a voltage of 10 W. 3 µL of sample were applied to the grids in the Vitrobot chamber set to 100% humidity and 4 °C and blotted for 4 s with a blot force of 5 and a wait time of 30 s followed by vitrification in liquid ethane.CryoEM data collection
Grids were screened in the Structural Biology Facility at Northwestern using a 200 kV Glacios (Thermo-Fisher) microscope. Suitable grids were identified for high resolution data collection at the National Center for CryoEM Access and Training (NCCAT) using a Titan Krios microscope (Thermo-Fisher) operating at 300 kV and controlled by Leginon. Details regarding the data collection parameters are provided in Table S1.CryoEM data processing and analysis
CryoEM data were processed using cryoSPARC v4.39 Movie frames were aligned using patch motion correction, and contrast transfer function (CTF) parameters were estimated using patch CTF estimation.40 Particle picking was performed first using template picking with the map of N. europaea AMO in native membranes, EMD-45663, as a preliminary template. Particles were curated extensively using 2D classification and heterogenous refinement to obtain clear top-down and side-on views of AMO, which were used to train a Topaz picking model.41,42 Topaz picking was then used to generate a stack of particles which were curated using multiple rounds of heterogenous refinement, using EMD-45663 as a reference volume with EMD-45663 lowpass filtered to 100 Å resolution as a decoy volume. These particles were then refined using non-uniform refinement43 at C1 and then C3 symmetry. Global and local CTF refinements were alternated with non-uniform refinements to improve map resolution and quality, followed by local motion correction and a final round of non-uniform refinement. Finally, local resolution estimation and local sharpening was used to sharpen the final volume. Resolution is reported based on gold-standard Fourier shell correlation with a cutoff of 0.143.44Model building, refinement, and visualization
An initial structural model was built into the final cryoEM map using ModelAngelo.45 Copper ions and lipids were manually added to the structure, inspected, and adjusted using Coot [version 0.9.8.93 EL (ccp4)].46 Further refinement of this model was performed using real-space refinement using the Phenix (version 1.21-5207)47 cryoEM suite along with automatic addition of waters, followed by manual inspection using Coot. Refined models were fitted into cryoEM maps, structures were visualized, maps were resampled to a uniform grid spacing, contour thresholds were chosen using sigma (σ) values, and figures were generated using ChimeraX-1.7.1.47EPR spectroscopy
CW (continuous wave) X-band EPR measurements were performed on a Bruker ESP-300 spectrometer with an Oxford Instruments ESR-900 liquid helium flow cryostat. All spectra were background subtracted. Samples were prepared at ∼25 mg mL−1 protein (∼250 µM AMO protomer) in 25 mM PIPES (pH 7.3) containing 250 mM NaCl. EPR samples were prepared in parallel to cryoEM samples and loaded into custom quartz EPR tubes, flash frozen in liquid nitrogen, and stored in a liquid nitrogen dewar until analysis. EPR simulations were performed using EasySpin.48Methane oxidation activity assay
AMO membrane preparations (100 µL) were diluted to 0.5–2 mg mL−1 total protein and combined with 4 mg per mL NADH in 2 mL screw-cap vials (Agilent). After sealing, 1 mL of headspace was withdrawn and replaced with 1.5 mL of 13C-methane (Sigma-Aldrich). Three technical replicates were prepared for each independent sample, and control vials lacking methane and NADH were included. Reactions were shaken at 200 rpm in a 30 °C water bath for 5 min, then quenched by freezing at −20 °C. GC/MS analysis of 13C-methanol production was performed exactly as described previously.15Author contributions
F. J. T. and A. C. R. designed experiments, F. J. T. performed structural and biochemical analysis; F. J. T. and A. C. R. analyzed structural data; A. A. B. T. and L. Y. S. cultured N. briensis C-128; B. M. H. and M. B. H. performed EPR analysis; F. J. T. and A. C. R. wrote the paper with input from M. B. H., L. Y. S., and B. M. H.Conflicts of interest
The authors declare no competing financial interest.Data availability
The model and corresponding cryoEM map of N. briensis C-128 AMO have been deposited in the Protein Data Bank and Electron Microscopy Data Bank with accession codes PDB 9PXF and EMDB EMD-71966.Supplementary information (SI): Table S1 and Fig. S1–S6. See DOI: https://doi.org/10.1039/d5sc08447d.
Acknowledgements
This work was supported by Department of Energy grants DE-SC0016284 (A. C. R.) and DE-SC0019342 (B. M. H.), and NIH grants R35GM118035 (A. C. R.) and R01GM111097 (B. M. H.). L. Y. S. and A. A. B. T. were supported by NSERC-DG grant RGPIN-2025-04834. F. J. T. was supported by NIH grant F31ES034283. Some of this work was performed at the National Center for CryoEM Access and Training (NCCAT) and the Simons Electron Microscopy Center located at the New York Structural Biology Center, supported by the NIH Common Fund Transformative High-Resolution Cryo-Electron Microscopy program (U24 GM129539) and by Grants from the Simons Foundation (SF349247) and NY State Assembly.References
- L. Y. Stein and M. E. Lidstrom, Greenhouse gas mitigation requires caution, Science, 2024, 384, 1068–1069 Search PubMed.
- R. S. Hanson and T. E. Hanson, Methanotrophic bacteria, Microbiol. Rev., 1996, 60, 439–471 Search PubMed.
- D. J. Arp and L. Y. Stein, Metabolism of inorganic N compounds by ammonia-oxidizing bacteria, Crit. Rev. Biochem. Mol. Biol., 2003, 38, 471–495 CrossRef CAS.
- A. J. Holmes, A. Costello, M. E. Lidstrom and J. C. Murrell, Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related, FEMS Microbiol. Lett., 1995, 132, 203–208 Search PubMed.
- F. J. Tucci and A. C. Rosenzweig, Direct methane oxidation by copper- and iron-dependent methane monooxygenases, Chem. Rev., 2024, 124, 1288–1320 CrossRef CAS PubMed.
- K. M. Lancaster, J. D. Caranto, S. H. Majer and M. A. Smith, Alternative bioenergy: updates to and challenges in nitrification metalloenzymology, Joule, 2018, 2, 421–441 CrossRef CAS.
- N. K. Kang, T. H. T. Chau and E. Y. Lee, Engineered methane biocatalysis: strategies to assimilate methane for chemical production, Curr. Opin. Biotechnol., 2024, 85, 103031 CrossRef CAS.
- D. Sauvageau, L. Y. Stein, E. Arenas, S. Das, M. Iacobelli, M. Lawley, M. Lazic, F. L. Rondón and C. Weiblen, Industrializing methanotrophs and other methylotrophic bacteria: from bioengineering to product recovery, Curr. Opin. Biotechnol., 2024, 88, 103167 Search PubMed.
- S. Wendeborn, The chemistry, biology, and modulation of ammonium nitrification in soil, Angew. Chem., Int. Ed. Engl., 2020, 59, 2182–2202 CrossRef CAS.
- X. Wang, J. Bai, T. Xie, W. Wang, G. Zhang, S. Yin and D. Wang, Effects of biological nitrification inhibitors on nitrogen use efficiency and greenhouse gas emissions in agricultural soils: a review, Ecotoxicol. Environ. Saf., 2021, 220, 112338 CrossRef CAS.
- R. L. Lieberman and A. C. Rosenzweig, Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane, Nature, 2005, 434, 177–182 CrossRef CAS.
- C. W. Koo and A. C. Rosenzweig, Biochemistry of aerobic biological methane oxidation, Chem. Soc. Rev., 2021, 50, 3424–3436 RSC.
- C. W. Koo, F. J. Tucci, Y. He and A. C. Rosenzweig, Recovery of particulate methane monooxygenase activity in a lipid bilayer, Science, 2022, 375, 1287–1291 Search PubMed.
- F. J. Tucci, R. J. Jodts, B. M. Hoffman and A. C. Rosenzweig, Product analog binding identifies the copper active site of particulate methane monooxygenase, Nat. Catal., 2023, 6, 1194–1204 Search PubMed.
- F. J. Tucci and A. C. Rosenzweig, Structures of methane and ammonia monooxygenases in native membranes, Proc. Natl. Acad. Sci. U. S. A., 2025, 122, e2417993121 CrossRef CAS.
- L. Y. Stein, Insights into the physiology of ammonia-oxidizing microorganisms, Curr. Opin. Chem. Biol., 2019, 49, 9–15 Search PubMed.
- M. C. Rice, J. M. Norton, F. Valois, A. Bollmann, P. J. Bottomley, M. G. Klotz, H. J. Laanbroek, Y. Suwa, L. Y. Stein, L. Sayavedra-Soto, T. Woyke, N. Shapiro, L. A. Goodwin, M. Huntemann, A. Clum, M. Pillay, N. Kyrpides, N. Varghese, N. Mikhailova, V. Markowitz, K. Palaniappan, N. Ivanova, D. Stamatis, T. B. K. Reddy, C. Y. Ngan and C. Daum, Complete genome of Nitrosospira briensis C-128, an ammonia-oxidizing bacterium from agricultural soil, Stand. Genomic Sci., 2016, 11, 46 Search PubMed.
- U. Purkhold, M. Wagner, G. Timmermann, A. Pommerening-Röser and H. P. Koops, 16S rRNA and amoA-based phylogeny of 12 novel betaproteobacterial ammonia-oxidizing isolates: extension of the dataset and proposal of a new lineage within the nitrosomonads, Int. J. Syst. Evol. Microbiol., 2003, 53, 1485–1494 CrossRef CAS PubMed.
- H.-P. Koops, U. Purkhold, A. Pommerening-Röser, G. Timmermann and M. Wagner, in The Prokaryotes: Volume 5: Proteobacteria: Alpha and Beta Subclasses, ed. M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer and E. Stackebrandt, Springer, New York, NY, 2006, pp. 778–811 Search PubMed.
- Y. A. Zhu, C. W. Koo, C. K. Cassidy, M. C. Spink, T. Ni, L. C. Zanetti-Domingues, B. Bateman, M. L. Martin-Fernandez, J. Shen, Y. W. Sheng, Y. Song, Z. Y. Yang, A. C. Rosenzweig and P. J. Zhang, Structure and activity of particulate methane monooxygenase arrays in methanotrophs, Nat. Commun., 2022, 13, 5221 Search PubMed.
- M. O. Ross, F. MacMillan, J. Wang, A. Nisthal, T. J. Lawton, B. D. Olafson, S. L. Mayo, A. C. Rosenzweig and B. M. Hoffman, Particulate methane monooxygenase contains only mononuclear copper centers, Science, 2019, 364, 566–570 Search PubMed.
- R. J. Jodts, M. O. Ross, C. W. Koo, P. E. Doan, A. C. Rosenzweig and B. M. Hoffman, Coordination of the copper centers in particulate methane monooxygenase: comparison between methanotrophs and characterization of the Cuc site by EPR and ENDOR spectroscopies, J. Am. Chem. Soc., 2021, 143, 15358–15368 CrossRef CAS PubMed.
- M. R. Hyman and P. M. Wood, Methane oxidation by Nitrosomonas europaea, Biochem. J., 1983, 212, 31–37 Search PubMed.
- S. Y. Ro, M. O. Ross, Y. W. Deng, S. Batelu, T. J. Lawton, J. D. Hurley, T. L. Stemmler, B. M. Hoffman and A. C. Rosenzweig, From micelles to bicelles: Effect of the membrane on particulate methane monooxygenase activity, J. Biol. Chem., 2018, 293, 10457–10465 CrossRef.
- M. Blumer, T. Chase and S. W. Watson, Fatty acids in the lipids of marine and terrestrial nitrifying bacteria, J. Bacteriol., 1969, 99, 366–370 Search PubMed.
- M. Kruse, S. Keuter, E. Bakker, E. Spieck, T. Eggers and A. Lipski, Relevance and diversity of Nitrospira populations in biofilters of brackish RAS, PLoS One, 2013, 8, e64737 Search PubMed.
- W. Peng, Z. Wang, Q. Zhang, S. Yan and B. Wang, Unraveling the valence state and reactivity of copper centers in membrane-bound particulate methane monooxygenase, J. Am. Chem. Soc., 2023, 145, 25304–25317 Search PubMed.
- I. Kipouros and E. I. Solomon, New mechanistic insights into coupled binuclear copper monooxygenases from the recent elucidation of the ternary intermediate of tyrosinase, FEBS Lett., 2023, 597, 65–78 Search PubMed.
- S. Maheshwari, C. Shimokawa, K. Rudzka, C. D. Kline, B. A. Eipper, R. E. Mains, S. B. Gabelli, N. Blackburn and L. M. Amzel, Effects of copper occupancy on the conformational landscape of peptidylglycine α-hydroxylating monooxygenase, Commun. Biol., 2018, 1, 74 Search PubMed.
- T. V. Vendelboe, P. Harris, Y. Zhao, T. S. Walter, K. Harlos, K. El Omari and H. E. Christensen, The crystal structure of human dopamine β-hydroxylase at 2.9 Å resolution, Sci. Adv., 2016, 2, e1500980 Search PubMed.
- E. F. Welch, K. W. Rush, R. J. Arias and N. J. Blackburn, Copper monooxygenase reactivity: Do consensus mechanisms accurately reflect experimental observations?, J. Inorg. Biochem., 2022, 231, 111780 CrossRef CAS.
- R. J. Arias, E. F. Welch and N. J. Blackburn, New structures reveal flexible dynamics between the subdomains of peptidylglycine monooxygenase. Implications for an open to closed mechanism, Protein Sci., 2023, 32, e4615 CrossRef CAS.
- E. F. Welch, K. W. Rush, K. A. S. Eastman, V. Bandarian and N. J. Blackburn, The binuclear copper state of peptidylglycine monooxygenase visualized through a selenium-substituted peptidyl-homocysteine complex, Dalton Trans., 2025, 54, 4941–4955 Search PubMed.
- L. S. Mydy, J. Hungerford, D. N. Chigumba, J. R. Konwerski, S. C. Jantzi, D. Wang, J. L. Smith and R. D. Kersten, An intramolecular macrocyclase in plant ribosomal peptide biosynthesis, Nat. Chem. Biol., 2024, 20, 530–540 CrossRef CAS.
- C. Y. Chiang, M. Ohashi, J. Le, P. P. Chen, Q. Zhou, S. Qu, U. Bat-Erdene, S. Hematian, J. A. Rodriguez, K. N. Houk, Y. Guo, J. A. Loo and Y. Tang, Copper-dependent halogenase catalyses unactivated C–H bond functionalization, Nature, 2025, 638, 126–132 Search PubMed.
- N. J. Blackburn, Metal-mediated peptide processing. How copper and iron catalyze diverse peptide modifications such as amidation and crosslinking, RSC Chem. Biol., 2025, 6, 1048–1067 Search PubMed.
- A. Krümmel and H. Harms, Effect of organic matter on growth and cell yield of ammonia-oxidizing bacteria, Arch. Microbiol., 1982, 133, 50–54 CrossRef.
- A. Bollmann, E. French and H. J. Laanbroek, Isolation, cultivation, and characterization of ammonia-oxidizing bacteria and archaea adapted to low ammonium concentrations, Methods Enzymol., 2011, 486, 55–88 CAS.
- A. Punjani, J. L. Rubinstein, D. J. Fleet and M. A. Brubaker, cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination, Nat. Methods, 2017, 14, 290–296 CrossRef CAS.
- K. Zhang, Gctf: real-time CTF determination and correction, J. Struct. Biol., 2016, 193, 1–12 Search PubMed.
- T. Bepler, A. Morin, M. Rapp, J. Brasch, L. Shapiro, A. J. Noble and B. Berger, Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs, Nat. Methods, 2019, 16, 1153–1160 CrossRef CAS PubMed.
- T. Bepler, K. Kelley, A. J. Noble and B. Berger, Topaz-Denoise: general deep denoising models for cryoEM and cryoET, Nat. Commun., 2020, 11, 5208 CrossRef CAS.
- A. Punjani, H. Zhang and D. J. Fleet, Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction, Nat. Methods, 2020, 17, 1214–1221 CrossRef CAS.
- S. H. Scheres and S. Chen, Prevention of overfitting in cryo-EM structure determination, Nat. Methods, 2012, 9, 853–854 CrossRef CAS.
- K. Jamali, L. Käll, R. Zhang, A. Brown, D. Kimanius and S. H. W. Scheres, Automated model building and protein identification in cryo-EM maps, Nature, 2024, 628, 450–457 Search PubMed.
- P. Emsley and K. Cowtan, Coot: model-building tools for molecular graphics, Acta Crystallogr., Sect. D:Biol. Crystallogr., 2004, 60, 2126–2132 CrossRef PubMed.
- P. D. Adams, P. V. Afonine, G. Bunkoczi, V. B. Chen, I. W. Davis, N. Echols, J. J. Headd, L. W. Hung, G. J. Kapral, R. W. Grosse-Kunstleve, A. J. McCoy, N. W. Moriarty, R. Oeffner, R. J. Read, D. C. Richardson, J. S. Richardson, T. C. Terwilliger and P. H. Zwart, PHENIX: a comprehensive Python-based system for macromolecular structure solution, Acta Crystallogr., Sect. D:Biol. Crystallogr., 2010, 66, 213–221 Search PubMed.
- S. Stoll and A. Schweiger, EasySpin, a comprehensive software package for spectral simulation and analysis in EPR, J. Magn. Reson., 2006, 178, 42–55 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2026 |