Open Access Article
Open Access ArticleHydrogen-bond-assisted meta-nitrogen-doped graphyne enables real-time electrocatalytic NADH tracking in single cells
Jiale
Wei†
a,
Jing
Jiang†
a,
Chuqi
Li†
b,
Guo
Wang
a,
Jiayao
Wulan
a,
Huan
Yang
a,
Fangxu
Shen
a,
Daixin
Ye
c,
Kai
Li
*a,
Xianchan
Li
*b and
Yuqing
Lin
 *a
*a
aDepartment of Chemistry, Capital Normal University, Beijing 100048, China. E-mail: linyuqing@cnu.edu.cn; likai@cnu.edu.cn
bState Key Laboratory of Natural and Biomimetic Drugs and Department of Pharmaceutical Analysis, School of Pharmaceutical Sciences, Peking University, Beijing 100191, China. E-mail: xcli@pku.edu.cn
cDepartment of Chemistry & Institute for Sustainable Energy, College of Sciences, Shanghai University, Shanghai 200444, PR China
First published on 12th January 2026
Abstract
Nicotinamide adenine dinucleotide (NADH), a crucial biomarker for cellular redox homeostasis, is intrinsically linked to mitochondrial function and neurodegenerative diseases. However, electrochemical detection of intracellular NADH faces dual challenges: low abundance and biofouling, as well as high overpotential. Herein, we designed a hydrogen-bond-assisted meta-nitrogen/oxygen co-doped graphyne (3NGYO) nanoelectrode for high-performance NADH sensing. Precise tri-N doping in graphyne (3NGY) generates sp2-N atoms associated with hydrogen atoms. Specifically, pyrrolic N–H forms a 2.502 Å N–H⋯O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) P hydrogen bond with NADH's PO4 groups, boosting adsorption energy to −5.48 eV and reducing NADH oxidation potential to 0 V. This achieves a 59-fold higher catalytic current response compared to pristine graphyne. Subsequent acid oxidation introduces oxygen-containing functional groups (e.g., –COOH, C
P hydrogen bond with NADH's PO4 groups, boosting adsorption energy to −5.48 eV and reducing NADH oxidation potential to 0 V. This achieves a 59-fold higher catalytic current response compared to pristine graphyne. Subsequent acid oxidation introduces oxygen-containing functional groups (e.g., –COOH, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), increasing hydrophilicity (contact angle: 54.0°) and anti-fouling performance (80% current retention after 2 h BSA exposure). Electrodeposited 3NGYO nanotips attain a sensitivity of 0.419 pA µM−1 and a linear range of 0–20 µM at +0.2 V vs. Ag/AgCl. Real-time amperometry in SH-SY5Y cells demonstrates that 1-methyl-4-phenylpyridinium (MPP+)-induced mitochondrial dysfunction triggers NADH release. In contrast, hydrogen sulfide (H2S) pretreatment reduces NADH leakage by 95.4%, correlating with suppressed calcium ion (Ca2+) influx and reactive oxygen species (ROS) generation. This work provides a tool for studying mitochondrial dysfunction and establishes a new paradigm for in situ electrocatalytic biosensing.
O), increasing hydrophilicity (contact angle: 54.0°) and anti-fouling performance (80% current retention after 2 h BSA exposure). Electrodeposited 3NGYO nanotips attain a sensitivity of 0.419 pA µM−1 and a linear range of 0–20 µM at +0.2 V vs. Ag/AgCl. Real-time amperometry in SH-SY5Y cells demonstrates that 1-methyl-4-phenylpyridinium (MPP+)-induced mitochondrial dysfunction triggers NADH release. In contrast, hydrogen sulfide (H2S) pretreatment reduces NADH leakage by 95.4%, correlating with suppressed calcium ion (Ca2+) influx and reactive oxygen species (ROS) generation. This work provides a tool for studying mitochondrial dysfunction and establishes a new paradigm for in situ electrocatalytic biosensing.
Introduction
NADH, a pivotal coenzyme in cellular redox reactions,1,2 serves as a critical biomarker for mitochondrial function and energy metabolism.3,4 Its intracellular concentration dynamics are closely associated with neurodegenerative diseases such as Parkinson's disease (PD) and Alzheimer's disease (AD),5 cardiovascular disorders,6 and metabolic syndromes, where mitochondrial dysfunction characterized by impaired adenosine triphosphate (ATP) synthesis and excessive ROS production plays a central pathogenic role.7–9 For instance, NADH deficiency directly reduces neuronal viability in Parkinson's disease models, while its release through the mitochondrial permeability transition pore ultimately leads to cell death.10,11 Consequently, real-time monitoring of NADH dynamics in single living cells is essential for elucidating disease mechanisms and developing therapeutic strategies.12 However, the low intracellular concentration of NADH (sub-micromolar level), rapid signal transduction,13 and complex biochemical milieu of living cells impose stringent requirements on analytical tools.14,15To date, various methods have been developed for intracellular molecular detection, encompassing immunofluorescence labelling,16,17 DNA/RNA sequencing,18,19 mass spectrometry,20 electroporation-based surface-enhanced Raman scattering,21 and electrochemical techniques.22–24 Among existing techniques, nano electrochemical methods stand out due to their minimal invasiveness and high compatibility with single-cell analysis.25–29 For instance, Ewing's group used nanoelectrodes to develop vesicle impact electrochemical cytometry (VIEC) to detect neurotransmitters in single-cell vesicles.30,31 Long's group developed novel nanosensors for single-cell, single-molecule protein analysis, revealing the impact of cellular heterogeneity on protein function.32,33 Huang's group developed an advanced electrochemical sensor platform based on core–shell nanowires, enabling high-resolution molecular analysis at the single-cell and subcellular levels.34 Notably, the performance of these nanoelectrochemical sensing platforms is highly dependent on the choice of electrode materials.
Carbon-based nanomaterials, such as graphene and carbon nanotubes, have been widely explored for NADH detection.35–37 However, its performance is limited by high overpotential (>0.5 V),38 causing interference from intracellular coexisting electroactive substances such as ascorbic acid (AA) and uric acid (UA),39 while biomacromolecules (e.g., proteins and lipids) in the complex intracellular environment induce electrode surface fouling, thereby degrading signal sensitivity.40,41 Although nitrogen doping has been employed to enhance catalytic activity and hydrophilicity for example,42 pyridinic nitrogen-doped graphene reduces the overpo-tential for NADH oxidation43 and oxidized graphene improves anti-fouling through oxygen-rich functional groups current strategies often rely on empirical doping configurations,42 lacking atomic-level precision in modulating interactions with NADH. Moreover, integrating high sensitivity and robust anti-fouling properties into a single nanoelectrode platform remains challenging, severely hindering long-term intracellular monitoring.
In this study, we designed a hydrogen-bond-assisted meta-nitrogen/oxygen co-doped graphyne carbon fibernanoelectrode (3NGYO/CFNE) for real-time, in situ tracking of NADH dynamics in single cells (Scheme 1). By introducing three meta-positioned nitrogen atoms into the graphyne lattice, we created electron-deficient pockets that form a 2.502 Å N–H⋯O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) P hydrogen bond with NADH's phosphate groups, boosting the adsorption energy to −5.48 eV and reducing the oxidation potential to 0 V. Subsequent acid oxidation introduced oxygen-containing functional groups (e.g., –COOH and C
P hydrogen bond with NADH's phosphate groups, boosting the adsorption energy to −5.48 eV and reducing the oxidation potential to 0 V. Subsequent acid oxidation introduced oxygen-containing functional groups (e.g., –COOH and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), enhancing hydrophilicity (contact angle: 54.0°) and anti-fouling performance (80% current retention after 2 h of bovine serum albumin exposure). The optimized 3NGYO/CFNE achieved a sensitivity of 0.419 pA µM−1 and a linear range of 0–20 µM (at 0.2 V vs. Ag/AgCl), superior to traditional carbon-based electrodes. Real-time amperometric measurements in SH-SY5Y cells revealed that MPP+-induced mitochondrial dysfunction triggers NADH release, while H2S pretreatment reduces NADH leakage by 95.4%, correlating with suppressed Ca2+ influx and ROS generation. This work not only provides a powerful tool for investigating mitochondrial dysfunction in neurodegenerative diseases but also establishes a new paradigm for in situ electrocatalytic biosensing through atomically precise engineering of carbon materials.
O), enhancing hydrophilicity (contact angle: 54.0°) and anti-fouling performance (80% current retention after 2 h of bovine serum albumin exposure). The optimized 3NGYO/CFNE achieved a sensitivity of 0.419 pA µM−1 and a linear range of 0–20 µM (at 0.2 V vs. Ag/AgCl), superior to traditional carbon-based electrodes. Real-time amperometric measurements in SH-SY5Y cells revealed that MPP+-induced mitochondrial dysfunction triggers NADH release, while H2S pretreatment reduces NADH leakage by 95.4%, correlating with suppressed Ca2+ influx and ROS generation. This work not only provides a powerful tool for investigating mitochondrial dysfunction in neurodegenerative diseases but also establishes a new paradigm for in situ electrocatalytic biosensing through atomically precise engineering of carbon materials.
Results and discussion
Structural characterization of 3NGY
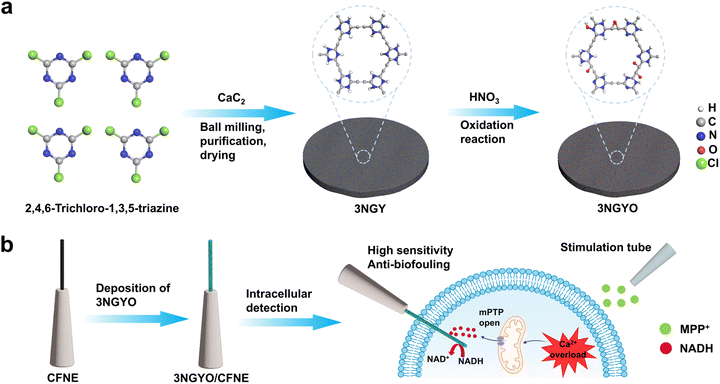
Graphynes, a novel carbon allotrope, consists of aromatic rings of sp2-hybridized carbon atoms connected by triple bond linkers of sp-hybridized carbon atoms, forming a one-atom-thick 2D network structure.44 This unique atomic arrangement endows graphynes with fascinating properties superior to those of conventional carbon materials, including intrinsic pores, a band gap, high conductivity,45,46 extensive conjugation, structural flexibility, and interesting chemical reactivity.47,48 Additional modifications and defect engineering can further enhance the reactivity of graphynes-based materials.49 Currently, graphynes and its derivatives have been extensively studied in the field of electrochemical sensing.50,51 In our team's previous work, we utilized the excellent adsorption capacity of graphdiyne (GDY) for Cu2+ to achieve highly selective detection of Cu2+.52 Hu et al. fabricated a high anti-interference electrocatalytic sensor based on N-doped graphdiyne (N-GDY) for the detection of dopamine (DA). The highly anti-interference ability was endowed by the abundant existing sp-N interactive sites on synthesized N-GDY surface that not only offered the very high-efficiency interactions between target DA molecules and developed interface but also effectively excluded other interferential component combinations.53 Qi et al. found that graphdiyne oxide serves as an exceptional substrate for the electroless deposition of ultrafine Pd clusters which exhibits remarkable catalytic performance in the reduction of 4-nitrophenol.54 However, reports on the electrocatalytic sensing of NADH by graphynes and its derivatives are very limited.We used CaC2 and 2,4,6-trichloro-1,3,5-triazine as the initial reactants and employed ball milling as a high-energy driving force for mechanochemical reactions to precisely synthesize 3NGY. Transmission electron microscopy (TEM), scanning electron microscopy (SEM), and atomic force microscopy (AFM) are among the most powerful methods for characterizing the morphology and thickness of carbon-based materials.55,56 Both GY and xNGY (x = 1, 2, 3) exhibit lamellar structures (Fig. 1a, b and S1), with 3NGY nanosheets having a thickness of 3.44 nm (Fig. 1c). The prepared GYs were examined using X-ray diffraction (XRD), and broad peaks around 23° were observed for the 1NGY, 2NGY, and 3NGY samples (Fig. 1d and S2). These peaks are characteristic of the interlayer spacing of typical two-dimensional carbon materials.57 The elemental configuration of GYs was determined using X-ray photoelectron spectroscopy (XPS) (Fig. S3 and S4). GY is composed solely of C and O elements, with no signals detected for other elements, indicating that GY is an all-carbon material.56 The presence of the O 1s peak is attributed to the oxygen-containing functional groups on GY. The total spectra of 1NGY, 2NGY, and 3NGY contain three main peaks (Fig. 1e and S3), corresponding to the binding energies of C 1s, N 1s, and O 1s, respectively, which confirm the presence of C, N and O elements in the material and the successful doping of N into GY, with N forming multiple covalent bonds with C. The substitution sites of N atoms can be identified by analyzing the N 1s spectra. For 1NGY, the N 1s spectrum (Fig. 1g) shows only one peak at approximately 399.8 eV, which can be attributed to pyridinic nitrogen. Since pyridinic nitrogen is formed by substituting nitrogen atoms for carbon atoms at planar defect sites or edges, we infer that N atoms have substituted the carbon atoms on the benzene ring. The N 1s spectrum of 2NGY (Fig. 1h) also indicates that the nitrogen exists solely in the form of pyridinic nitrogen (399.5 eV), suggesting that the doped N atoms are present as pyridinic nitrogen and that two nitrogen atoms are doped at opposite positions on a benzene ring. The three peaks in the total spectrum of 3NGY (Fig. 1e) correspond to the binding energies of C 1s, N 1s, and O 1s, indicating the successful doping of nitrogen. The C 1s peak (Fig. 1f) is mainly attributed to five types of bonds, corresponding to C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C at a binding energy of 284.6 eV, C
C at a binding energy of 284.6 eV, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N at 285.5 eV, C–N at 286.2 eV, C–O at 287.1 eV, and C
N at 285.5 eV, C–N at 286.2 eV, C–O at 287.1 eV, and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O at 288.9 eV.58 After nitrogen doping, the N 1s binding energy (Fig. 1i) is distinctly composed of two peaks: pyridinic nitrogen at 398.4 eV and pyrrolic nitrogen at 400.2 eV. The presence of C–N bonds may originate from cross-coupling reactions occurring under ambient N2 conditions in the presence of pyridinic nitrogen. Raman spectroscopy is an analytical method that uses the scattering spectrum to obtain information on molecular vibrations and rotations. It can be used to characterize the defect and bonding states of GYs materials. The intensity ratios of the D-band to the G-band (ID/IG) for GY, 1NGY, 2NGY, and 3NGY are 0.83, 1.52, 1.36, and 1.73, respectively, indicating that nitrogen doping introduces more structural defects (Fig. S5). Structural characterizations reveal that 3NGY features distinctive pyrrolic nitrogen moieties and high defect density, which are anticipated to provide abundant electrocatalytic activity. Subsequent electrochemical measurements systematically evaluate the structure–activity relationship between these structural advantages and the catalytic performance toward NADH oxidation.
O at 288.9 eV.58 After nitrogen doping, the N 1s binding energy (Fig. 1i) is distinctly composed of two peaks: pyridinic nitrogen at 398.4 eV and pyrrolic nitrogen at 400.2 eV. The presence of C–N bonds may originate from cross-coupling reactions occurring under ambient N2 conditions in the presence of pyridinic nitrogen. Raman spectroscopy is an analytical method that uses the scattering spectrum to obtain information on molecular vibrations and rotations. It can be used to characterize the defect and bonding states of GYs materials. The intensity ratios of the D-band to the G-band (ID/IG) for GY, 1NGY, 2NGY, and 3NGY are 0.83, 1.52, 1.36, and 1.73, respectively, indicating that nitrogen doping introduces more structural defects (Fig. S5). Structural characterizations reveal that 3NGY features distinctive pyrrolic nitrogen moieties and high defect density, which are anticipated to provide abundant electrocatalytic activity. Subsequent electrochemical measurements systematically evaluate the structure–activity relationship between these structural advantages and the catalytic performance toward NADH oxidation.
Enhanced electrocatalytic oxidation of NADH at 3NGY-Modified electrodes
The electrocatalytic oxidation performance of NADH by GY and xNGY catalysts was evaluated using cyclic voltammetry. Electrodes modified with 3NGY exhibited the highest catalytic activity, yielding 16.98 ± 0.84 µA at 0 V for 2 mM NADH (Fig. 2a, b and S6), representing a 59-fold increase compared to pristine graphyne (GY: 0.29 ± 0.10 µA). Furthermore, as shown by the electrochemically active surface area (ECSA) results in Fig. S7, 3NGY possesses a larger ECSA value compared with GY. While 3NGY does exhibit higher surface area, the 8-fold increase cannot account for the 59-fold current enhancement. Therefore, the remaining improvement mainly originate from the enhanced intrinsic catalytic activity of nitrogen-doped graphyne, indicating the doping strategy fundamentally improves the material's electrocatalytic properties beyond simple surface area effects. | ||
| Fig. 2 (a) Cyclic voltammograms (CV) of 3NGY-modified glassy carbon electrodes (GCE) in blank PBS and PBS containing 2 mM NADH vs. Ag/AgCl. (b) Catalytic oxidation currents of 2 mM NADH at 0 V for GY-, 1NGY-, 2NGY-, and 3NGY-modified GCE (n = 3). (c and d) Amperometric response and calibration curve of 3NGY/GCE to NADH (0–700 µM) at 0.05 V: I (µA) = 0.0074C (µM) − 0.0686, R2 = 0.998. (e) Comparison of 3NGYO/GCE with other conventional electrodes reported for the sensitivity of NADH. Here, electrodes 1–8 are modified with Co3O4 nano sheet/carbon ink, GC/MWCNTs/Flu, GCE/MWCNT/poly-FA, SPCE/MWCNT/AuNP/PNR, SPCE/RA, PCA24h@MWCNT, PGO/SPE and NPG/Os(bpy)2PVI/DIA, respectively.60–67 (f) Steady-state voltammograms of 3NGY/GCE in 1 mM NADH (0.05 M PBS, pH 7.4) at varying rotation speeds (scan rate: 50 mV s−1). (g) K–L plot at 0.5 V derived from (f). (h) Absorbance at 340 nm over time: 0.25 mM NADH electrolyzed at +0.05 V for 60 min (1); after adding 10 mM glucose and glucose dehydrogenase (2). | ||
Subsequently, we evaluated the sensitivity of the 3NGY-modified electrode and compared it with other traditional electrodes. The sensor based on 3NGY demonstrated a good linear relationship in detecting NADH within the range of 0–700 µM (Fig. 2c and d), with a sensitivity of 0.2357 µA µM−1 cm−2. The sensitivity and linear range of the 3NGY/GCE are superior to the values reported by several conventional sensors (Fig. 2e). Hydrodynamic voltammetry helps distinguish between kinetic-limited and mass-transport-limited currents in electrochemical reactions.59 To investigate the hydrodynamic characteristics of NADH oxidation, linear sweep voltammetry (LSV) was employed to measure the current response of 3NGY/GCE in the presence of 1 mM NADH within the potential range of −0.6 to 0.6 V at a scan rate of 50 mV s−1 and different rotation speeds. A steady-state increase in current with rotation speed was observed (Fig. 2f). Fig. 2g shows the Koutecký–Levich (K–L) plot at 0.5 V, where the linear correlation between I−1 and ω−1/2 can be used to calculate the kinetic parameters according to the following equation:
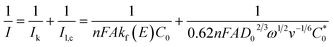 | (1) |
Here, n is the number of transferred electrons, C0 is the bulk concentration of NADH, D0 is the diffusion coefficient of NADH (2.4 × 10−6 cm2 s−1), ν is the kinematic viscosity of the solution (0.01 cm2 s−1), kf is the apparent catalytic rate constant, and A is the electrode area (7.065 × 10−2 cm2). Based on the slope of the K–L plot, the number of electrons transferred during the entire electrocatalytic oxidation process of NADH on 3NGY/GCE was calculated to be 2, indicating that the oxidation product of NADH catalyzed by 3NGY is NAD+.
To further verify that NAD+ is the product of NADH oxidation catalyzed by 3NGY, we employed ultraviolet-visible absorption spectroscopy (UV-Vis) to measure the absorbance of NADH before and after electrolysis, as well as the kinetics at 340 nm after adding 10 mM glucose and glucose dehydrogenase to the electrolyzed system. As shown in Fig. 2h, the absorbance at 340 nm of the solution system decreased after electrolyzing NADH for 60 min at +0.05 V, indicating that NADH had reacted and its concentration decreased. Subsequently, we added glucose and glucose dehydrogenase to the system and recorded the kinetic spectrum, observing that the absorbance at 340 nm gradually increased over time. This increase is attributed to the conversion of glucose to gluconolactone by glucose dehydrogenase, with NAD+ acting as the electron acceptor in the reaction. This experiment thus confirms that NADH is oxidized to NAD+ by the electrocatalytic action of 3NGY.
Theoretical calculation: tri-N doping in graphyne drives energetically favorable NADH adsorption via hydrogen bonding
We conducted a theoretical analysis of the catalytic activity of these four materials: GY and NGY (x = 1, 2, 3). We first performed density functional theory (DFT) calculations, in which the vacuum layer was set to be thicker than 15 Å so that possible interactions between image structures along the third dimension can be neglected. The PBE density functional and PAW basis in the VASP program68 were used. Weak interactions were treated through the DFT-D3(BJ) method. We speculate that π–π stacking interactions occurred between the GY and xNGY substrates and NADH molecules, and thus constructed plausible adsorption models (Fig. 3a–e). According to the formula:| Eads = ENADH–substrate − (ENADH + Esubstrate) |
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) P hydrogen bond with NADH's PO4 groups, enhancing substrate adsorption. The effective binding of NADH and 3NGY produces a more stable complex structure, which can effectively exclude the adsorption of other biomolecules on the surface of the 3NGY substrate, thereby further enhancing the catalytic performance for the target NADH. Molecular dynamics simulations69 further confirmed enhanced NADH accumulation on 3NGY surfaces (Fig. 3g and h), aligning with the observed electrocatalytic enhancement.
P hydrogen bond with NADH's PO4 groups, enhancing substrate adsorption. The effective binding of NADH and 3NGY produces a more stable complex structure, which can effectively exclude the adsorption of other biomolecules on the surface of the 3NGY substrate, thereby further enhancing the catalytic performance for the target NADH. Molecular dynamics simulations69 further confirmed enhanced NADH accumulation on 3NGY surfaces (Fig. 3g and h), aligning with the observed electrocatalytic enhancement.
Structural characterization of 3NGYO
To enhance the anti-fouling properties of the material, we subjected the relatively high-performance catalytic material 3NGY to acid treatment (Fig. S8a).70 The acid-treated material (3NGYO, exists four stable configurations) still exhibited a thin, flake-like structure (Fig. S8b and c). The XRD pattern of 3NGYO (Fig. S9a) showed a broad peak at 22°, indicating its containing amorphous structure. This open nanostructure significantly increased the material's surface area, facilitating better contact with the substrate.71 Raman spectroscopy was used to characterize 3NGYO, and the typical D and G bands could be easily distinguished from the Raman spectrum (Fig. S9b). The D band at 1340 cm−1 is closely related to disordered carbon atoms and structural defects such as edges,72 while the G band at 1585 cm−1 is associated with the first-order scattering of the E2g stretching vibration mode of sp2 carbon domains in aromatic rings.73 The ratio of the intensities of the ID/IG can be used to assess the defects and disorder in 3NGYO, with an ID/IG value of 1.16 indicating a high degree of structural defects. These defects, caused by N and O doping, provide a large number of active sites, which are beneficial for improving the sensing performance of sensors based on 3NGYO. Additionally, the molecular structure and atomic valence states of 3NGYO were investigated using XPS. The XPS survey spectrum of 3NGYO (Fig. S10a) contains three main peaks at 532 eV, 401 eV, and 285 eV, corresponding to the binding energies of C 1s, N 1s, and O 1s, respectively, which confirms the presence of C, N, and O elements in the material. Moreover, by comparing the XPS spectra with those of 3NGY, it is evident that the relative content of oxygen in 3NGYO has increased. This is due to the oxidation of 3NGY by concentrated nitric acid, leading to the formation of more oxygen-containing functional groups such as –OH, –COOH, and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O. In the C 1s spectrum of 3NGYO (Fig. S10b), four common sub-peaks are observed at 284.8 eV (C
O. In the C 1s spectrum of 3NGYO (Fig. S10b), four common sub-peaks are observed at 284.8 eV (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C), 287.2 eV (C–N), 285.9 eV (C
C), 287.2 eV (C–N), 285.9 eV (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), and 289.1 eV (C
N), and 289.1 eV (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). After O doping, 3NGYO exhibits a distinct N characteristic peak above 400 eV (Fig. S10c), located at approximately 400.2 eV, which can be attributed to pyrrolic nitrogen. The Fourier-transform infrared (FT-IR) spectrum of 3NGYO is recorded in Fig. S11 and the following functional groups were identified for sample characterization. The IR spectra show a strong peak at 1720 cm−1 corresponding to the stretching vibration of the carbonyl C
O). After O doping, 3NGYO exhibits a distinct N characteristic peak above 400 eV (Fig. S10c), located at approximately 400.2 eV, which can be attributed to pyrrolic nitrogen. The Fourier-transform infrared (FT-IR) spectrum of 3NGYO is recorded in Fig. S11 and the following functional groups were identified for sample characterization. The IR spectra show a strong peak at 1720 cm−1 corresponding to the stretching vibration of the carbonyl C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group, a peak at 3475 cm−1 caused by the –OH group, and a peak at 1210 cm−1 due to the stretching vibration of C–OH.74,75 All these findings indicate the presence of oxygen-containing functional groups after acid treatment.
O group, a peak at 3475 cm−1 caused by the –OH group, and a peak at 1210 cm−1 due to the stretching vibration of C–OH.74,75 All these findings indicate the presence of oxygen-containing functional groups after acid treatment.
3NGYO enables highly sensitive, selective, and anti-fouling electrochemical detection of NADH
Fig. S12 shows the electrochemical impedance spectra of different glassy carbon electrodes in a mixed solution of 5 mM [Fe(CN)6]3−/4− and 0.1 M KCl, with the inset displaying the corresponding equivalent circuit diagram. The charge transfer resistance (Rct value) of 3NGYO/GCE is lower than that of 3NGY/GCE and GCE, indicating that the acid-treated carbon material enhances the electron transfer ability at the electrode surface and electrolyte interface. The CV responses of 3NGYO/GCE in PBS solution with and without NADH (Fig. S13a) show that the oxidation current value at 0 V is 24.21 µA, which is significantly higher than that of 3NGY/GCE, demonstrating an enhanced current response of 3NGYO/GCE towards NADH. The CV responses of 3NGYO towards 1 mM NADH at different scan rates were investigated. As the scan rate increased, the redox peak currents also increased, and a linear regression equation between the scan rate and the anodic peak current was obtained (Fig. S13b and c), indicating that the oxidation of NADH is an adsorption-controlled process. Subsequently, we evaluated other sensing properties of 3NGYO/GCE, such as sensitivity, stability, selectivity, and anti-fouling ability. Fig. S13d shows the electrochemical response of 3NGYO/GCE to different concentrations of NADH at a constant range of 0–700 µM, with a sensitivity of 0.2866 µA µM−1 cm−2 voltage of 0.05 V (Fig. S13e). A good linear relationship was observed in the stability of 3NGYO catalyzing NADH oxidation was tested, and 3NGYO/GCE exhibited excellent stability in long-term testing, with almost no change in current (Fig. S13f). To investigate the selectivity of 3NGYO towards NADH, a series of common interfering substances, including hydrogen peroxide (H2O2), ATP, glucose (Glu), adenosine diphosphate (ADP), glutathione GSH, UA and nicotinamide adenine nucleotide phosphate (NADPH), were detected. Compared with NADH, most interfering substances showed negligible changes in current response, indicating that 3NGYO has excellent selectivity towards NADH (Fig. S13g and h).To investigate the performance of the 3NGYO-based sensor in complex biological fluids, we compared the CV curves of 3NGY/GCE and 3NGYO/GCE before and after soaking in phosphate-buffered saline (PBS) containing 5 mg per mL bovine serum albumin (BSA) for 2 hours (Fig. 4a and b). 3NGYO retained 80% of its initial peak current after 2 hours incubation in 5 mg per mL BSA solution (Fig. 4e). After soaking in cell culture medium (RPMI) containing BSA for 2 hours, the current response of 3NGYO/GCE was still higher than 85% of the initial response (Fig. 4c–e). In contrast, a larger current response decrease was observed on 3NGY/GCE (Fig. 4f, black curve). The antifouling ability of the 3NGYO against non-specific protein binding was visually examined by the binding of fluorescein isothiocyanate-tagged BSA (FITC-BSA) on the 3NGYO-coated carbon fiber microelectrode (CFE). As displayed in Fig. 4g–i, after immersing the 3NGYO-coated CFE in a solution of FITC-BSA (5 mg mL−1) for 2 h, almost no bright green color was observed. In contrast, the bare CFE appears bright green color. We further measured the water contact angle of the 3NGYO surface (54.0°), which is less than that of 3NGY (75.3°) and bare electrode (91.5°) (Fig. 4j–l). These results reveal that the 3NGYO has high ability of resisting nonspecific protein binding in complex biological fluids.
After oxidation of 3NGY with concentrated nitric acid, there are four possible distributions of oxygen-containing functional groups (Fig. 5a). A two-dimensional model depicting the adsorption of NADH on 3NGYO was developed using theoretical calculations (Fig. 5b–e). The DFT calculation parameters employed for the 3NGYO system were identical to those used for the 3NGY system. Four stable configurations, each conducive to adsorption, were obtained. The respective adsorption energies were −0.02 eV, −1.47 eV, −1.53 eV, and −1.96 eV (Fig. S14) and hydrogen bonding still exists between 3NGYO and NADH.
 | ||
| Fig. 5 (a) Four stable configurations of 3NGYO. (b–e) Adsorption model of NADH on four stable configurations of 3NGYO (including top and front views). | ||
To investigate the hydrodynamic characteristics of NADH oxidation catalyzed by 3NGYO, LSV was employed within a potential range of −0.6 to 0.6 V at a scan rate of 50 mV s−1. The current response of 3NGYO/GCE in the presence of 1 mM NADH was measured at different rotation speeds. A steady increase in current with rotation speed was observed (Fig. S15a). Fig. S15b shows the K–L plot at 0.5 V, where the inverse current (I−1) is linearly correlated with the inverse square root of the rotation speed (ω−1/2). In the preceding text, we have already elaborated on the formulas and relevant parameters utilized for calculating the electron transfer number when 3NGY catalyzes NADH. Based on the same methodology and parameters, according to the slope of the Koutecký–Levich plot, the number of electrons transferred during the entire electrocatalytic oxidation of NADH on 3NGYO/GCE is calculated to be 2, indicating that the oxidation product of NADH catalyzed by 3NGY is NAD+. To further confirm that the product of NADH oxidation catalyzed by 3NGYO is NAD+, we added 10 mM glucose and glucose dehydrogenase to the system after the electrolysis of NADH and continuously monitored the absorbance at 340 nm. The gradual increase in absorbance over time indicated that the product of NADH electrocatalytic oxidation by 3NGYO is NAD+ (Fig. S15c). Additionally, mass spectrometry (MS) analysis showed the sodiated molecular ion (Fig. S16), with a peak at m/z = 686 corresponding to [MNAD+ + Na]+, further confirming that the product is NAD+.
We also tested the chemical catalytic effects of 3NGY and 3NGYO on NADH. After mixing the catalysts with NADH and allowing the reaction to proceed for 10 minutes, a significant decrease in the UV-Vis absorption of the system was observed for both catalysts (Fig. S17a). However, the kinetic spectra (Fig. S17b) showed that the concentration of NADH catalyzed by 3NGYO was lower, indicating that 3NGYO has a better catalytic effect than 3NGY, which is consistent with the conclusions drawn from the electrochemical methods. Additionally, we explored the superoxide dismutase (SOD)-like activity of 3NGYO. SOD is a classic antioxidant enzyme that can convert superoxide anion (O2˙−) into hydrogen peroxide and oxygen (O2). O2˙− can reduce nitroblue tetrazolium enables it to exert anti-inflammatory effects by modulating the levels of oxidative stress (Fig. S18).
Development of 3NGYO-Modified carbon fiber nanoelectrode for selective and sensitive detection of physiological NADH levels
To enable the modification of 3NGYO on nanoelectrodes, we first tested the zeta potential of the material, which was measured to be −26.69 mV (Fig. S19). Subsequently, a constant potential of +1.5 V was applied to electrodeposit 3NGYO onto CFNE, yielding 3NGYO/CFNE with nanotips of ∼400 nm in diameter and ∼30 µm in length (Fig. S20). First of all, we tested the catalytic performance of 3NGYO/CFNE in PBS containing NADH (Fig. 6a). A distinct oxidation peak of NADH was observed on 3NGYO/CFNE. To further demonstrate the successful modification of the material, we also tested the I–t curves. Based on the CV results, +0.2 V was selected as the working potential. At +0.2 V, the oxidation current on 3NGYO/CFNE increased with the addition of NADH (Fig. 6b), while the current on CFNE remained almost unchanged. Therefore, these results indicate that 3NGYO was successfully modified onto CFNE. The specificity and stability of the sensor are crucial for accuracy and reliability of electrochemical sensors in practical applications. Thus, we evaluated the specificity and long-term stability of the 3NGYO/CFNE-based sensor. The results showed that 3NGYO high selectivity for NADH over other biomolecules, with no significant interference from other biomolecules involved in mitochondrial metabolism (such as H2O2, Glu, GSH, ADP, ATP, and NADPH) (Fig. 6c). Additionally, 3NGYO/CFNE demonstrated excellent stability (Fig. 6d). We then tested the linear response of the 3NGYO/CFNE-based sensor. The sensor exhibited a good linear relationship in the range of 0 to 20 µM (Fig. 6e and f). In conclusion, the successful modification of 3NGYO onto nanoelectrodes has been realized, and it has exhibited remarkable sensing performance. These findings suggest that the 3NGYO/CFNE-based sensor is highly appropriate for the detection of intracellular NADH. Subsequently, we evaluated the cytocompatibility of 3NGYO using the CCK-8 assay. After exposure to 7.5 µg mL−1 of 3NGYO for 24 hours, the viability of SH-SY5Y cells remained above 85%, indicating that the material is safe for subsequent experiments (Fig. S21).Real-time tracking of mitochondrial NADH dynamics in single neuronal cells reveals MPP+-induced metabolic dysfunction and neuroprotective effects of H2S
Glu can promote the production of NADH through glycolysis, thereby increasing the intracellular level of NADH.76 To demonstrate that 3NGYO/CFNE can be used to monitor the dynamics of NADH in single cells, Glu was selected to treat the cells. First, the nanosensor was inserted into individual SH-SY5Y cells (Fig. 7a) and we compared the current responses of NADH on 3NGYO/CFNE and CFNE electrodes under glucose stimulation. As shown in Fig. 7b, no significant current response was observed on CFNE, while an increase in current was detected on 3NGYO/CFNE. When 3NGYO/CFNE was inserted into starved SH-SY5Y cells, glucose stimulation induced a pronounced increase in the amperometric signal (Fig. 7c, orange line), which was not observed in cells treated with PBS. These results demonstrate that 3NGYO/CFNE can detect changes in the intracellular concentration of NADH.MPP+ is a toxic metabolite that induces mitochondrial dysfunction,77 which in turn affects the intracellular levels of NADH.78 However, the underlying mechanisms by which MPP+ exerts these effects remain unclear. A deeper understanding of the cellular response mechanisms under MPP+ stimulation may provide new insights for the treatment of Parkinson's disease. Therefore, in the subsequent studies, cells were stimulated with MPP+Fig. 7d, after inserting 3NGYO/CFNE into SH-SY5Y cells, a pronounced amperometric signal was observed upon injection of 3 mM MPP+ around the cells, whereas no significant signal was detected when the cells were stimulated with buffer solution. This indicates that MPP+ can induce the release of NADH from mitochondria, thereby increasing the intracellular NADH content. As is well known, there are currently six major classes of drugs for the treatment of Parkinson's disease, including levodopa, dopamine receptor agonists, monoamine oxidase B inhibitors, catechol-O-methyltransferase inhibitors, anticholinergic drugs, and amantadine.79–81 We selected several of these drugs, such as L-DOPA, Rasagiline, H2S, and Resveratrol, to investigate their protective effects on cells. After incubation with different anti-Parkinson's drugs (L-DOPA, Rasagiline, H2S, and Resveratrol), SH-SY5Y cells exhibited varying degrees of NADH response currents under MPP+ stimulation (Fig. 7e–h). Among them, cells protected by H2S showed the lowest NADH response upon MPP+ stimulation (Fig. 7i), indicating that MPP+ did not cause significant damage to the cells and that H2S exerted a significant protective effect.
H2S mitigates MPP+-induced mitochondrial NADH leakage by suppressing pathological Ca2+ influx
The release of NADH from mitochondria is associated with the opening of the mPTP. Ca2+ act as an agonist for the opening of the mPTP.11,82 To explore the mechanisms underlying the increase in intracellular NADH levels induced by MPP+ and the protective effects of H2S, we first assessed the levels of the abnormal protein α-synuclein in SH-SY5Y cells using the ThT fluorescence probe. The fluorescence intensity in the MPP+ group was significantly higher, which was associated with the abnormal aggregation of α-synuclein. Cells pretreated with H2S prior to MPP+ exposure (H2S-MPP+ group) still exhibited partial abnormal α-synuclein aggregation (Fig. 7j and m). Subsequently, we performed fluorescence imaging of Ca2+ in SH-SY5Y cells using the Fluo-4 AM fluorescent probe. The results showed that, compared with the control group, the level of Ca2+ was significantly increased in MPP+-treated SH-SY5Y cells. This may be attributed to the fact that MPP+ induces the aggregation of α-synuclein, leading to ERS,83–85 which in turn triggers the store-operated calcium entry (SOCE) mechanism and promotes the influx of intracellular calcium ions.86 However, the fluorescence in the H2S-MPP+ group was significantly reduced (Fig. 7k and n), indicating that H2S effectively blocked Ca2+ influx. Cellular ROS levels were measured using the fluorescent probe (2′,7′-dichlorodihydrofluorescein diacetate, DCFH-DA). Compared with the control group, the fluorescence in MPP+-treated SH-SY5Y cells was significantly increased, indicating that MPP+ induced substantial ROS production, which led to mitochondrial dysfunction. Conversely, the fluorescence in the H2S-MPP+ group was significantly lower than that in the MPP+ group (Fig. 7l and o). Therefore, based on these findings, we speculate that MPP+ induces α-synuclein aggregation in cells, triggering ERS, which leads to a substantial influx of Ca2+ into the mitochondria. These changes further cause mitochondrial dysfunction, induce the opening of the mPTP, and ultimately result in the release of NADH from the mitochondria into the cytosol, thereby increasing the intracellular NADH level. However, pretreatment with hydrogen H2S revealed that, although α-syn aggregation still occurred, Ca2+ influx was significantly blocked, and the release of NADH was significantly reduced. These results suggest that H2S can protect cells by blocking Ca2+ influx, thereby alleviating mitochondrial dysfunction and mitigating MPP+-induced cellular damage (Fig. 7p).Conclusions
In summary, this study introduces a groundbreaking hydrogen-bond-assisted 3NGYO/CFNE for real-time, in situ tracking of NADH dynamics in single cells. By leveraging the unique electronic structure and precise tri-N doping of graphyne, our electrocatalytic sensitivity has been enhanced compared with pristine graphyne, and it operates within a wide linear range of 0–700 µM. The integration of oxygen-containing functional groups significantly enhanced the hydrophilicity and anti-fouling performance, ensuring stable operation even in complex biological environments. Real-time amperometry in SH-SY5Y cells demonstrated the sensor's capability to monitor MPP+-induced mitochondrial dysfunction and the protective effects of H2S, providing valuable insights into neurodegenerative disease mechanisms. This work not only establishes a new paradigm for in situ electrocatalytic biosensing through atomically precise carbon material engineering but also offers a powerful tool for investigating mitochondrial dysfunction and developing therapeutic strategies for neurodegenerative diseases. While the 3NGYO/CFNE demonstrates excellent performance in vitro, translating this technology from in vitro to in vivo applications may encounter challenges such as non-specific interference from the complex in vivo physiological environment and the requirement for long-term biocompatibility of the electrode. Nevertheless, with further optimization of electrode materials and structural design, this sensing platform holds great promise for in vivo NADH monitoring in the study of neurodegenerative diseases.Author contributions
Y. Q. Lin and K. Li designed and supervised the research. J. L. Wei and J. Jiang conducted material characterization, performance testing and biological application exploration. D. X. Ye assisted in material synthesis. G. Wang and J. Y. Wulan conducted theoretical calculations. H. Yang assisted in drawing the material model diagram. F. X. Shen assisted in the electrochemical testing. X. C. Li and C. Q. Li assisted in the intracellular electrochemical testing. J. L. Wei wrote the manuscript. Y. Q. Lin and K. Li revised the manuscript. All authors approved the final version of the manuscript.Conflicts of interest
The authors declare no conflict of interest.Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.Supplementary information (SI) is available. See DOI: https://doi.org/10.1039/d5sc08242k.
Acknowledgements
We gratefully acknowledge financial support from the National Natural Science Foundation (22074095 and 22374103 for Prof. Y. L.), Beijing Natural Science Foundation (Z230022), National Natural Science Foundation of China (22374005), National Key Research and Development Program of China (2022YFA1206101), and Recruitment Program of Global Experts (Youth).References
- H. Chen, J. Yu, X. Men, J. Zhang, Z. Ding, Y. Jiang, C. Wu and D. T. Chiu, Angew. Chem., Int. Ed., 2021, 60, 12007–12012 CrossRef CAS.
- P. D. Giang, D. Niks, S. Hakopian, R. Hille and P. V. Bernhardt, Chem. Sci., 2025, 16, 6035–6049 RSC.
- Y.-L. Ying, Y.-X. Hu, R. Gao, R.-J. Yu, Z. Gu, L. P. Lee and Y.-T. Long, J. Am. Chem. Soc., 2018, 140, 5385–5392 CrossRef CAS PubMed.
- X. T. Zheng and C. M. Li, Chem. Soc. Rev., 2012, 41, 2061–2071 RSC.
- Y. Liu, L. Cao, J. Zhou, C. Li, J. Du, Y. Xue, X. Li and L. Mao, J. Am. Chem. Soc., 2024, 147, 149–160 Search PubMed.
- M. Bonora, M. R. Wieckowski, D. A. Sinclair, G. Kroemer, P. Pinton and L. Galluzzi, Nat. Rev. Cardiol., 2019, 16, 33–55 CrossRef CAS PubMed.
- R. Coukos and D. Krainc, Nat. Rev. Neurosci., 2024, 25, 393–413 Search PubMed.
- M. T. Lin and M. F. Beal, Nature, 2006, 443, 787–795 Search PubMed.
- X. Xu, Y. Pang and X. Fan, Signal Transduction Targeted Ther., 2025, 10, 190 CrossRef.
- M. Vranas, D. Wohlwend, D. Qiu, S. Gerhardt, C. Trncik, M. Pervaiz, K. Ritter, S. Steimle, A. Randazzo, O. Einsle, S. Günther, H. J. Jessen, A. Kotlyar and T. Friedrich, Angew. Chem., Int. Ed., 2021, 60, 27277–27281 CrossRef CAS.
- H. Jiang, Y.-T. Qi, W.-T. Wu, M.-Y. Wen, Y.-L. Liu and W.-H. Huang, Chem. Sci., 2020, 11, 8771–8778 Search PubMed.
- D. V. Titov, V. Cracan, R. P. Goodman, J. Peng, Z. Grabarek and V. K. Mootha, Science, 2016, 352, 231–235 CrossRef CAS.
- K. Hu, Y. Li, S. A. Rotenberg, C. Amatore and M. V. Mirkin, J. Am. Chem. Soc., 2019, 141, 4564–4568 CrossRef CAS.
- L. Ren, A. Oleinick, I. Svir, C. Amatore and A. G. Ewing, Angew. Chem., Int. Ed., 2020, 59, 3083–3087 CrossRef CAS.
- Y.-T. Qi, F.-L. Zhang, S.-Y. Tian, H.-Q. Wu, Y. Zhao, X.-W. Zhang, Y.-L. Liu, P. Fu, C. Amatore and W.-H. Huang, Nat. Nanotechnol., 2024, 19, 524–533 Search PubMed.
- F. Shamsi, M. Piper, L.-L. Ho, T. L. Huang, A. Gupta, A. Streets, M. D. Lynes and Y.-H. Tseng, Nat. Metab., 2021, 3, 485–495 Search PubMed.
- D. Wu, D. L. Harrison, T. Szasz, C.-F. Yeh, T.-P. Shentu, A. Meliton, R.-T. Huang, Z. Zhou, G. M. Mutlu, J. Huang and Y. Fang, Nat. Metab., 2021, 3, 714–727 Search PubMed.
- S. J. Clark, R. Argelaguet, C.-A. Kapourani, T. M. Stubbs, H. J. Lee, C. Alda-Catalinas, F. Krueger, G. Sanguinetti, G. Kelsey, J. C. Marioni, O. Stegle and W. Reik, Nat. Commun., 2018, 9, 781 Search PubMed.
- M. Fasolino, G. W. Schwartz, A. R. Patil, A. Mongia, M. L. Golson, Y. J. Wang, A. Morgan, C. Liu, J. Schug, J. Liu, M. Wu, D. Traum, A. Kondo, C. L. May, N. Goldman, W. Wang, M. Feldman, J. H. Moore, A. S. Japp, M. R. Betts, M. Fasolino, G. W. Schwartz, A. R. Patil, A. Mongia, M. L. Golson, Y. J. Wang, A. Morgan, C. Liu, J. Schug, J. Liu, M. Wu, D. Traum, A. Kondo, C. L. May, N. Goldman, W. Wang, M. Feldman, J. H. Moore, A. S. Japp, M. R. Betts, R. B. Faryabi, A. Naji, K. H. Kaestner, G. Vahedi, R. B. Faryabi, A. Naji, K. H. Kaestner, G. Vahedi and H. C. the, Nat. Metab., 2022, 4, 284–299 Search PubMed.
- L. Zhang, T. Xu, J. Zhang, S. C. C. Wong, M. Ritchie, H. W. Hou and Y. Wang, Anal. Chem., 2021, 93, 10462–10468 CrossRef CAS PubMed.
- S. Zhang, S. Chen and R. Zhu, ACS Sens., 2023, 8, 555–564 CrossRef CAS.
- K. Hu, T. D. K. Nguyen, S. Rabasco, P. E. Oomen and A. G. Ewing, Anal. Chem., 2021, 93, 41–71 Search PubMed.
- C. Xu, D. Yang, Y. Wang, R. Liu, F. Wang, Z. Tian and K. Hu, Nano Res., 2024, 17, 196–206 CrossRef CAS.
- W. Ma, D.-W. Li, T. C. Sutherland, Y. Li, Y.-T. Long and H.-Y. Chen, J. Am. Chem. Soc., 2011, 133, 12366–12369 Search PubMed.
- Y. Liu, L. Cao, J. Zhou, C. Li, J. Du, Y. Xue, X. Li and L. Mao, J. Am. Chem. Soc., 2025, 147, 149–160 Search PubMed.
- Y.-T. Qi, H. Jiang, W.-T. Wu, F.-L. Zhang, S.-Y. Tian, W.-T. Fan, Y.-L. Liu, C. Amatore and W.-H. Huang, J. Am. Chem. Soc., 2022, 144, 9723–9733 CrossRef CAS PubMed.
- A. Schulte and W. Schuhmann, Angew. Chem., Int. Ed., 2007, 46, 8760–8777 Search PubMed.
- F.-L. Zhang, X.-K. Yang, Y.-T. Qi, S.-Y. Tian and W.-H. Huang, Chem. Sci., 2024, 15, 7651–7658 RSC.
- J. Liu, Y. Jiang, R. Liu, J. Jin, S. Wei, W. Ji, X. He, F. Wu, P. Yu and L. Mao, J. Am. Chem. Soc., 2024, 146, 17747–17756 Search PubMed.
- K. Hu, K. L. Le Vo, A. Hatamie and A. G. Ewing, Angew. Chem., Int. Ed., 2022, 61, e202113406 Search PubMed.
- X.-W. Zhang, A. Hatamie and A. G. Ewing, J. Am. Chem. Soc., 2020, 142, 4093–4097 Search PubMed.
- Z.-Q. Zhou, S.-C. Liu, J. Wang, K.-L. Chen, B.-K. Xie, Y.-L. Ying and Y.-T. Long, Chem. Sci., 2025, 16, 8501–8508 RSC.
- K. L. Chen, R. J. Yu, C. B. Zhong, Z. Wang, B. K. Xie, H. Ma, M. Ao, P. Zheng, A. G. Ewing and Y. T. Long, Angew. Chem., Int. Ed., 2024, 63, e202406677 CrossRef CAS PubMed.
- Y.-T. Qi, X.-K. Yang, X.-W. Zhang and W.-H. Huang, Acc. Chem. Res., 2025, 58, 3294–3305 CrossRef CAS PubMed.
- F. Poletti, A. Scidà, B. Zanfrognini, A. Kovtun, V. Parkula, L. Favaretto, M. Melucci, V. Palermo, E. Treossi and C. Zanardi, Adv. Funct. Mater., 2022, 32, 2107941 CrossRef CAS.
- Z. Xiong, G. Fang, R. K. Mondal, Y. Liao, N. Nie, Y.-C. Chen and M. Kim, Nano Lett., 2024, 24, 14993–15000 CrossRef CAS PubMed.
- X. Zhao, Z. Liu, Y. Qiu, Q. Zhang, Y. Chen, D. Wang, Z. Zhu, L. Meng and H. Zheng, Anal. Chem., 2024, 96, 12120–12128 CrossRef CAS PubMed.
- S. Immanuel and R. Sivasubramanian, Mater. Chem. Phys., 2020, 249, 123015 CrossRef CAS.
- S. Deng, X. Men, M. Hu, X. Liang, Y. Dai, Z. Zhan, Z. Huang, H. Chen and Z. Dong, Biosens. Bioelectron., 2024, 250, 116082 CrossRef CAS PubMed.
- T. Feng, W. Ji, Y. Zhang, F. Wu, Q. Tang, H. Wei, L. Mao and M. Zhang, Angew. Chem., Int. Ed., 2020, 59, 23445–23449 Search PubMed.
- F. Liu, C. Ding, S. Tian, S.-M. Lu, C. Feng, D. Tu, Y. Liu, W. Wang and C. Li, Chem. Sci., 2022, 13, 13361–13367 RSC.
- X. Zhao, F. Li, R. Wang, J.-M. Seo, H.-J. Choi, S.-M. Jung, J. Mahmood, I.-Y. Jeon and J.-B. Baek, Adv. Funct. Mater., 2017, 27, 1605717 CrossRef.
- J. Balamurugan, T. D. Thanh, N. H. Kim and J. H. Lee, Biosens. Bioelectron., 2016, 83, 68–76 CrossRef CAS PubMed.
- X. Gao, J. Tian, S. Cheng, Z. Zuo, R. Wen, F. He and Y. Li, Angew. Chem., Int. Ed., 2023, 62, e202304491 CrossRef CAS PubMed.
- Z. Xie, B. Zhang, Y. Ge, Y. Zhu, G. Nie, Y. Song, C.-K. Lim, H. Zhang and P. N. Prasad, Chem. Rev., 2021, 122, 1127–1207 CrossRef PubMed.
- Y. Fang, Y. Liu, L. Qi, Y. Xue and Y. Li, Chem. Soc. Rev., 2022, 51, 2681–2709 RSC.
- Z. Zheng, Y. Xue and Y. Li, Trends Chem., 2022, 4, 754–768 CrossRef CAS.
- K. Khan, A. K. Tareen, M. Iqbal, Z. Shi, H. Zhang and Z. Guo, Nano Today, 2021, 39, 101207 CrossRef CAS.
- M. Wang, J. Pu, Y. Hu, Y. Zi, Z.-G. Wu and W. Huang, Adv. Funct. Mater., 2024, 34, 2308601 CrossRef CAS.
- K. Niu, Z. Zuo, X. Lu, L. Zou and J. Chen, Biosens. Bioelectron., 2022, 205, 114111 CrossRef CAS PubMed.
- D. Bahari, B. Babamiri, K. Moradi, A. Salimi and R. Hallaj, Biosens. Bioelectron., 2022, 195, 113657 Search PubMed.
- J. Jiang, X. Wang, Y. Bao, F. Shen, G. Wang, K. Li and Y. Lin, ACS Sens., 2024, 9, 2317–2324 Search PubMed.
- M. Cui, P. Xin, Z. Che, M. Zou, M. Zhang, X. Sun, Y. Yuan, Z. Zou, G. Lv, S. Wang and W. Hu, Chem. Eng. J., 2023, 464, 142629 CrossRef CAS.
- H. Qi, P. Yu, Y. Wang, G. Han, H. Liu, Y. Yi, Y. Li and L. Mao, J. Am. Chem. Soc., 2015, 137, 5260–5263 Search PubMed.
- S. R. Falsafi, H. Rostamabadi, E. Assadpour and S. M. Jafari, Adv. Colloid Interface Sci., 2020, 280, 102166 CrossRef CAS.
- X. Shi, W. Qing, T. Marhaba and W. Zhang, Electrochim. Acta, 2020, 332, 135472 Search PubMed.
- H. Shang, Z. Zuo, H. Zheng, K. Li, Z. Tu, Y. Yi, H. Liu, Y. Li and Y. Li, Nano Energy, 2018, 44, 144–154 CrossRef CAS.
- X. Zheng, H. Wu, Y. Gao, S. Chen, Y. Xue and Y. Li, Angew. Chem., Int. Ed., 2024, 63, e202316723 Search PubMed.
- N. B. Watkins, Z. J. Schiffer, Y. Lai, C. B. Musgrave III, H. A. Atwater, W. A. Goddard III, T. Agapie, J. C. Peters and J. M. Gregoire, ACS Energy Lett., 2023, 8, 2185–2192 CrossRef CAS.
- M. Bilgi, E. M. Sahin and E. Ayranci, J. Electroanal. Chem., 2018, 813, 67–74 Search PubMed.
- C.-H. Chen, Y.-C. Chen and M.-S. Lin, Biosens. Bioelectron., 2013, 42, 379–384 CrossRef CAS PubMed.
- L. V. da Silva, C. B. Lopes, W. C. da Silva, Y. G. de Paiva, F. A. dos Santos Silva, P. R. Lima, L. T. Kubota and M. O. F. Goulart, Microchem. J., 2017, 133, 460–467 CrossRef.
- M. Elancheziyan, K. Theyagarajan, V. K. Ponnusamy, K. Thenmozhi and S. Senthilkumar, Micro Nano Eng., 2022, 15, 100133 CrossRef CAS.
- F. Otero, T. Mandal, D. Leech and E. Magner, Sens. Actuators Rep., 2022, 4, 100117 CrossRef.
- T. Rębiś, M. Kuznowicz, A. Jędrzak, G. Milczarek and T. Jesionowski, Electrochim. Acta, 2021, 386, 138384 CrossRef.
- M. Sahin and E. Ayranci, Electrochim. Acta, 2015, 166, 261–270 Search PubMed.
- A. Sobczak, T. Rębiś and G. Milczarek, Bioelectrochemistry, 2015, 106, 308–315 Search PubMed.
- G. Kresse and J. Furthmüller, Phys. Rev. B: Condens. Matter Mater. Phys., 1996, 54, 11169–11186 CrossRef CAS PubMed.
- X. F. Cheng, J. H. He, H. Q. Ji, H. Y. Zhang, Q. Cao, W. J. Sun, C. L. Yan and J. M. Lu, Adv. Mater., 2022, 34, 2205767 CrossRef CAS PubMed.
- C. Xu, W. Ma, H. Wang, L. Shao, W. Li, P. Yu and L. Mao, Nano Res., 2023, 17, 1123–1131 CrossRef.
- Y. Guo, R. Zhang, S. Zhang, H. Hong, P. Li, Y. Zhao, Z. Huang and C. Zhi, Angew. Chem., Int. Ed., 2024, 63, e202401501 CrossRef CAS PubMed.
- N. Gao, G. Ren, M. Zhang and L. Mao, J. Am. Chem. Soc., 2024, 146, 3836–3843 Search PubMed.
- K. Niu, J. Gao, L. Wu, X. Lu and J. Chen, Anal. Chem., 2021, 93, 8656–8662 CrossRef CAS PubMed.
- H. Li, C. Pan, X. Peng, B. Zhang, S. Song, Z. Xu, X. Qiu, Y. Liu, J. Wang and Y. Guo, Nat. Commun., 2025, 16, 2439 CrossRef CAS PubMed.
- W. Li, C. Xu, T. Xiong, Y. Jiang, W. Ma, P. Yu and L. Mao, Angew. Chem., Int. Ed., 2023, 62, e202216530 CrossRef CAS PubMed.
- S. J. Kierans and C. T. Taylor, J. Biol. Chem., 2024, 300, 107906 CrossRef CAS.
- E. Martín-Montañez, N. Valverde, D. Ladrón de Guevara-Miranda, E. Lara, Y. S. Romero-Zerbo, C. Millon, F. Boraldi, F. Ávila-Gámiz, A. M. Pérez-Cano, P. Garrido-Gil, J. L. Labandeira-Garcia, L. J. Santin, J. Pavia and M. Garcia-Fernandez, Redox Biol., 2021, 46, 102095 CrossRef PubMed.
- H. B. Cho, H.-R. Kim, S. Lee, C. W. Cho, J.-I. Park, S. Youn, G. So, S. Kang, H. J. Kim and K.-H. Park, Adv. Sci., 2025, 12, 2415303 CrossRef CAS.
- D. Charvin, R. Medori, R. A. Hauser and O. Rascol, Nat. Rev. Drug Discovery, 2018, 17, 804–822 CrossRef CAS.
- R. M. A. de Bie, C. E. Clarke, A. J. Espay, S. H. Fox and A. E. Lang, Lancet Neurol., 2020, 19, 452–461 CrossRef CAS PubMed.
- D. R. Hill, A. D. Huters, T. B. Towne, R. E. Reddy, J. L. Fogle, E. A. Voight and P. R. Kym, Chem. Rev., 2023, 123, 13693–13712 CrossRef CAS PubMed.
- R. Behera, V. Sharma, A. K. Grewal, A. Kumar, B. Arora, A. Najda, G. M. Albadrani, A. E. Altyar, M. M. Abdel-Daim and T. G. Singh, Biomed. Pharmacother., 2023, 162, 114599 CrossRef CAS PubMed.
- W. Han, M. Wei, F. Xu and Z. Niu, Chem. Commun., 2024, 60, 6581–6590 RSC.
- N. Siwecka, K. Saramowicz, G. Galita, W. Rozpędek-Kamińska and I. Majsterek, Pharmaceutics, 2023, 15, 2051 CrossRef CAS.
- J. Wei, G. Meisl, A. Dear, M. Oosterhuis, R. Melki, C. Emanuelsson, S. Linse and T. P. J. Knowles, Chem. Sci., 2024, 15, 8430–8442 RSC.
- B. Manicassamy, F. Diao, C. Jiang, Y. Sun, Y. Gao, J. Bai, H. Nauwynck, X. Wang, Y. Yang, P. Jiang and X. Liu, PLoS Pathog., 2023, 19, e1011295 Search PubMed.
Footnote |
| † These authors have contributed equally. |
| This journal is © The Royal Society of Chemistry 2026 |