Open Access Article
Open Access ArticleA nuclear-targeted activity-based sensing probe for ratiometric imaging of formaldehyde reveals endogenous epigenetic contributors to the nuclear formaldehyde pool
Logan
Tenney
 ab,
Kuo-Kuang
Wen
c,
Seong-Su
Han
c,
Yatin M.
Vyas
c and
Christopher J.
Chang
ab,
Kuo-Kuang
Wen
c,
Seong-Su
Han
c,
Yatin M.
Vyas
c and
Christopher J.
Chang
 *ab
*ab
aDepartment of Chemistry, Princeton University, Princeton, NJ 08544, USA. E-mail: chrischang@princeton.edu
bDepartment of Chemistry and Molecular and Cell Biology, University of California, Berkeley, Berkeley, CA 94720, USA
cDepartment of Pediatrics, Division of Pediatric Hematology-Oncology, Penn State College of Medicine, Penn State Health Children's Hospital, Hershey, PA 17033, USA
First published on 1st December 2025
Abstract
Formaldehyde (FA) is both a one-carbon (1C) metabolite and a potent genotoxin in living cells. FA plays beneficial roles in endogenous catabolic processes and cellular signaling, but its potent electrophilicity necessitates strict regulation. This dichotomy is especially important in the nucleus, where endogenously produced FA has been shown to promote toxicity and disease by generating deleterious DNA adducts. More broadly, the sources and scavenging mechanisms of FA differ across subcellular compartments, underscoring the need for imaging sensors with subcellular spatial resolution to accurately probe contributions of FA to transient, local 1C pools. Here, we report NucRFAP-2, a nuclear-targeted, activity-based ratiometric probe for FA detection, and apply it to monitor dynamic changes in the nuclear FA pool. Using this first-generation reagent for nuclear FA imaging, we demonstrate that genetic perturbation of key FA clearance pathways alters nuclear FA levels by identifying alcohol dehydrogenase 5 (ADH5) as a principal regulator of nuclear FA homeostasis. Furthermore, NucRFAP-2 reveals elevated nuclear FA pools in patient-derived T and B lymphocytes deficient in Wiskott-Aldrich syndrome protein (WASp) and Fanconi anemia group D2 protein (FANCD2), suggesting that replication-associated epigenetic rewiring may contribute to aldehyde-associated pathologies. By demonstrating the ability of NucRFAP-2 to reveal an interplay between FA metabolism, genome integrity, and 1C homeostasis, we showcase this probe as a potentially powerful chemical tool to uncover novel mechanisms of nuclear FA biology.
Introduction
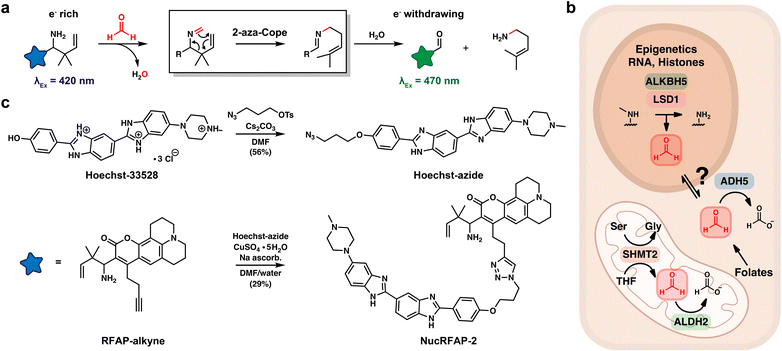
Formaldehyde (FA) is a highly reactive one-carbon (1C) metabolite generated by human metabolism. Traditionally recognized as an environmental toxin and carcinogen found in building materials, manufacturing processes, and wildfire smoke, a growing appreciation of FA as an endogenous metabolite and toxin has emerged in recent years.1,2 The intracellular FA pool has several contributors, including oxidative demethylation of histones,3,4 nucleic acids5,6 and other methylamines,7 and spontaneous decomposition of folates,8,9 such as 5,10-methylene-tetrahydrofolate, which is known as “active FA” generated by serine hydroxymethyltransferases (SHMT) in the cleavage of serine to produce glycine.10,11 Clearance of FA is regulated by several enzymes, including alcohol dehydrogenase 5 (ADH5, also known as ADH3 and GSNOR)12,13 and aldehyde dehydrogenase 2 (ALDH2).14,15 These systems are essential to organismal health16 as they both convert highly genotoxic FA to the less toxic and electrophilic 1C metabolite formate (Scheme 1).17,18 Indeed, one of the most common mutations in humans, the ALDH2*2 mutation that affects approximately 8% of the world's population and 40% of East Asians, impairs the aldehyde-clearance capacity of the cell.19,20 Deficiency in aldehyde scavenging capacity has major potential health implications, particularly in contexts where endogenous aldehydes are elevated, such as after consumption of alcohol. Patel's laboratory and others have demonstrated that loss of either ADH5 or ALDH2 increases DNA-FA adduct formation, and simultaneous loss of both ADH5 and ALDH2 leads to bone marrow failure and leukemia in mice.16 In addition, disruption of DNA–protein crosslink (DPC) repair pathways, such as loss of Fanconi Anemia Complementation Group D2 (FANCD2) in the context of elevated endogenous FA, leads to toxic DNA damage, genome instability, the development of T-lymphoblastic leukaemia (T-ALL) and accelerated aging.21,22Beyond its well-known roles as a genotoxin and biomolecular crosslinker, our laboratory discovered that FA also functions as a molecular 1C signal, regulating endogenous SAM pools and the epigenetic status of the cell.23,24 By systematic profiling of FA reactivity with the cysteinome, we found that at relatively low concentrations FA does not react indiscriminately with all cysteines, but instead it selectively targets a subset of privileged, hyperreactive cysteine residues within specific proteins. In particular, FA modulates enzymes within 1C metabolic pathways through cysteine modification. Notably, we demonstrated that FA inhibits methionine adenosyltransferase 1A (MAT1A)—the enzyme responsible for the terminal step in S-adenosylmethionine (SAM) biosynthesis—leading to SAM depletion and changes in histone and DNA methylation patterns. These results establish a role for endogenous FA as a regulatory signal within 1C metabolism, warranting further investigation into its broader contributions to cell function.
Our prior work on FA imaging demonstrates that subcellular compartments respond differently to changes in 1C metabolism.25 For example, global cellular FA increases in ADH5 knockout cell lines, while mitochondrial FA levels are reduced in the same model—possibly reflecting segregation of the mitochondrial 1C pool, which is regulated by alternative machinery including ALDH2. Similarly, genetic depletion of SHMT1/2—where SHMT2 is a mitochondria-specific isoform—selectively decreases mitochondrial FA without affecting other compartments. Motivated by this observation, and the demonstrated importance of maintenance of the nuclear 1C pool, we sought to expand the repertoire of fluorescence-based tools for FA study by introducing a new probe to monitor fluctuations in the nuclear FA pool. The nucleus has several well-established sources of FA, including epigenetic demethylation reactions in which methyl groups on nucleic acids and histones are removed via enzymatic oxidation and subsequent hydrolysis.26 Given the high susceptibility of the genome to FA-induced genotoxic stress, stringent regulation of nuclear FA homeostasis is essential. Consequently, there is increasing interest in elucidating how human disease states characterized by epigenetic reprogramming or perturbations in 1C metabolism—such as dysregulated folate pathways9—may disrupt this nuclear FA pool, thereby contributing to pathological outcomes that could be broadly classified as aldehyde-induced toxicities. As such, the development of activity-based sensors capable of accurately monitoring fluctuations in nuclear FA would facilitate studies of FA biochemistry in healthy and disease states.
We now report the design, synthesis, and biological evaluation of Nuclear Ratiometric Formaldehyde Probe-2 (NucRFAP-2), a RFAP-Hoechst conjugate capable of reporting on the nuclear FA pool. We use the probe to identify accumulations of FA in the nucleus in response to exogenous additions of FA as well as genetic knockout of ADH5. Finally, we employed a NucRFAP-2 in human T and B cells under replication stress to demonstrate that deficiencies in WASp and FANCD2 lead to accumulation of genotoxic FA in the nucleus via aberrant epigenetic rewiring. Taken together, these results demonstrate the importance of activity-based sensors with precise spatiotemporal precision as tools to decipher the highly transient chemical species in the 1C metabolic cycle of the cell.
Results and discussion
Design, synthesis and in vitro characterization of a nuclear ratiometric FA probe, NucRFAP-2
Previously, our laboratory27–32 and others33–36 have developed selective activity-based sensing probes37–39 for FA imaging via 2-aza-Cope reactivity. This platform relies on a homoallyl amine trigger, which, upon condensation with FA, undergoes a rearrangement and subsequent hydrolysis to generate an aldehyde. The change from an electron-rich amine group to an electron-poor aldehyde group causes a change in the photophysical properties of a pendant fluorophore, enabling microscopy experiments that report on intracellular FA pools with preserved spatial resolution that is lost using traditional analytical methods.26,40–42 The reactive trigger is selective towards FA, as larger, more sterically encumbered aldehydes cannot readily achieve the six-member transition state required for the 2-aza-Cope rearrangement, providing a major advantage compared to hydrazine and hydroxylamine fluorescence-based probes that simply rely on condensation of aldehyde analytes and react with a broad range of metabolic species. Ratiometric FA Probes, or RFAPs, are a class of probes that undergo a change in excitation spectrum upon reaction with FA, where the amine has a maximum fluorescence emission at λex = 420 nm and product aldehyde at λex = 470 nm, this ratiometric readout helps reduce changes in probe signal due to probe loading and sample thickness.30 Recently our group has made improvements to the spatiotemporal resolution of RFAPs, via ligation to chemical agents that direct the localization of the probe. We reported MitoRFAP-2, a first-generation probe capable of reporting on ALDH2 scavenging of mitochondrial FA, that responds to both genetic knockout of ALDH2 and small molecule activation of ALDH2*2, with Alda-1.25To develop a new chemical tool for specific imaging of nuclear FA pools in living cells, we synthesized a fluorescent probe comprised of two parts: (i) a coumarin dye bearing a homoallylamine group that selectively reacts with FA through a 2-aza-Cope rearrangement and (ii) Hoechst, a widely used, non-toxic dye that binds chromatin with a dissociation constant (KD) of 1–10 nM.43,44 In a single step, we ligated a synthon previously reported by our group, RFAP-alkyne,25 to a Hoechst-azide conjugate using copper-catalyzed click chemistry to achieve the final probe, NucRFAP-2 (Scheme 1).
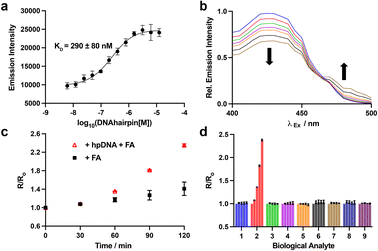
We first validated the capacity of NucRFAP-2 to bind DNA, as modifications to Hoechst's structure can affect its DNA-binding affinity, which could, in turn, influence its ability to localize to the nucleus. To our satisfaction, NucRFAP-2 retains a nanomolar affinity towards DNA (Fig. 1a); this assay relies on the change in fluorescence emission of Hoechst, which increases upon binding with DNA.45,46 Next, we evaluated the reactivity and selectivity of NucRFAP-2 towards FA in aqueous buffer. When incubated in the presence of FA (100 µM) and a hairpin DNA sequence in PBS at 37 °C for 2 hours, the probe exhibited the expected robust and facile fluorescence response – specifically, an increase in emission at λem = 510 nm when excited at λex = 470 nm and a corresponding decrease at a when excited at λex = 420 nm (Fig. 1b and SI 3–6). A unique observation, distinct from other RFAP probes previously reported by our laboratory, is that the FA reactivity of NucRFAP-2 is greater in the presence of DNA (Fig. 1c, SI 1, 7 and 9). We speculate that DNA prevents Hoescht-induced aggregation, leading to greater reactivity and more aldehyde product. Importantly, chromatin-dependent reactivity has been demonstrated with other reported Hoechst-conjugates, including those bearing cysteine reactive chemical functionalities.47 In addition to the high chromatin affinity that anchors the probe to the nucleus, this DNA-dependent FA response acts as a secondary spatial selectivity filter that ensures the probe is accurately reporting exclusively on the nuclear FA pool, as its reactivity will be impaired in organelles that lack DNA. As expected, NucRFAP-2 exhibits high selectivity for FA over several other biologically relevant aldehydes, including acetaldehyde and methylglyoxal, as well as other highly reactive metabolic species such as hydrogen peroxide (H2O2)48 and the cellular reductant glutathione, the latter of which is present at millimolar concentrations in cells (Fig. 1d).49
Validation of NucRFAP-2 nuclear localization and sensitivity to changes in nuclear FA pools with exogenous FA addition or endogenous FA accumulation
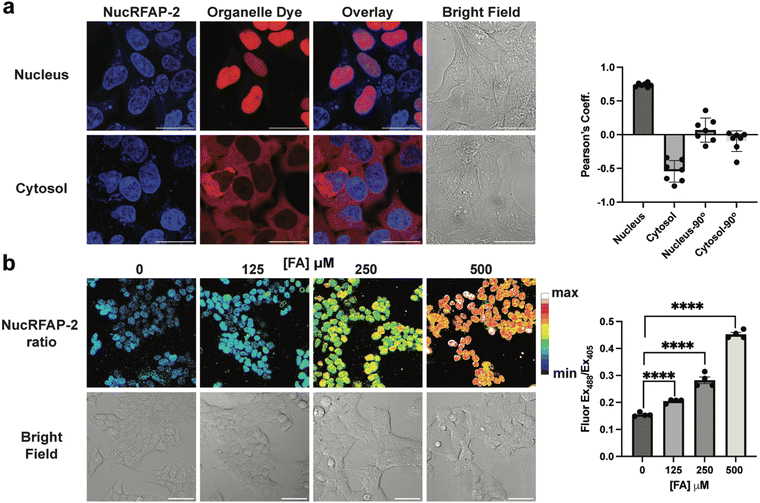
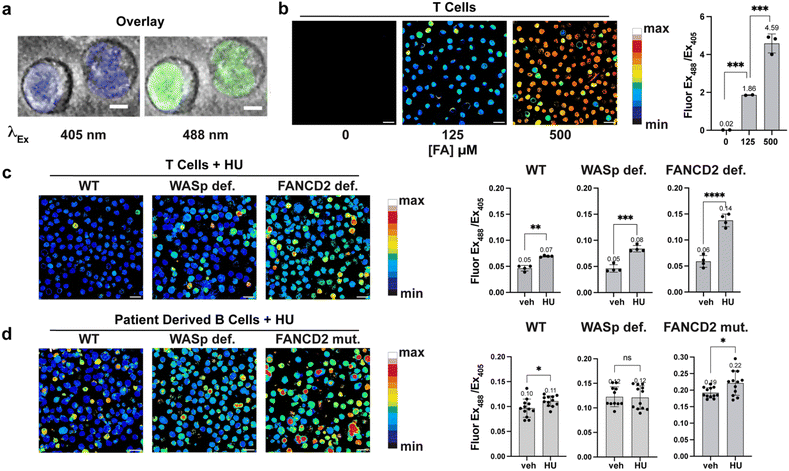
To demonstrate nuclear localization of NucRFAP-2 we stained HEK-293 FlpIn cells stably expressing Halo-SNAP protein (a protein capable of labeling with Halo- and SNAP-tag ligands) in two different subcellular localizations. We used Halo-SNAP-NLS for the nucleus and cyto-Halo-SNAP for the cytosol with the probe and a previously reported SiR-Halo probe that reacts with the Halo-tag protein.50–52 We then calculated the Pearson's correlation coefficient, a measure of the overlap between the two fluorescence signals, for NucRFAP-2 and SiR-Halo in each system. As expected, the Halo-SNAP-NLS cell line demonstrated strong overlap with NucRFAP-2 and the nucleus, with a positive average Pearson correlation coefficient of 0.74. In contrast, the cyto-Halo-SNAP showed a negative average value of −0.54, suggesting poor overlap between NucRFAP-2 and the cytosol. Also presented are 90° rotational controls that act as an important validation of the correlation coefficient calculations, where rotation of the image from either fluorescence channel leads to ablation of the correlation (Fig. 2a). We also noted the presence of a minimal amount of off-target punctate staining, likely attributable to lysosomal signal. We do not expect that this signal would provide false FA-dependent positives, as we showed that probe reactivity and fluorescence response to FA treatment is much higher when bound to DNA, and reactivity is lower at lower pH compared to pH 7.4 (SI Fig. 1, 2, 7 and 9). Moreover, the fluorescence intensity profile of a single cell was also plotted to demonstrate the overlap of the dyes, which further validated a similar trend of colocalization (SI Fig. 12 and 13). Importantly, varying the FA concentration does not impact the localization of the dye, and NucRFAP-2 retains nuclear localization after reaction with FA (SI Fig. 15).To demonstrate the ability of NucRFAP-2 to report on fluctuations in nuclear FA pools in live-cell settings, we stained HEK-293T cells with the probe followed by treatment with biologically relevant exogenous doses of FA. As anticipated, NucRFAP-2 showed a FA-induced, dose-dependent increase in ratiometric fluorescence signal. Next, we sought to determine whether loss of one of the main endogenous scavengers of the FA pool, alcohol dehydrogenase 5 (ADH5), leads to nuclear accumulation of FA. ADH5 oxidizes the adduct of FA and glutathione, S-hydroxylmethylglutathione, to S-formylglutathione, and subsequent hydrolysis produces formate, a less electrophilic 1C metabolite. In the context of nuclear FA metabolism, it is worth noting that ADH5 is thought to be mainly a cytosolic enzyme, thus for this model, if nuclear accumulation is observed, it would suggest functional crosstalk and/or equilibrium between nuclear and cytoplasmic 1C pools. Indeed, we observe that genetic knockout of ADH5 lead to accumulation of nuclear FA in MEF cells relative to wild-type, where incubation times of approximately 3 h are required to observe appreciable and statistically significant difference in NucRFAP-2 signal (Fig. 3b and c). In addition, exogenous supplementation of a low dose of FA exacerbated the difference between wild-type and ADH5 knockout in MEF cell models (Fig. 3b). The collective results suggest the existence of some equilibrium between the nuclear and cytosolic FA pools, this observation is congruent with data that show increases in FA-dependent modifications of guanosine in ADH5 knockout mouse models.16 In contrast, other organelle FA pools, such as the pool within the mitochondria, show less sensitivity towards genetic ADH5 ablation and may have less, or alternative, crosstalk with the cytosol.25
Replication stress induces aberrant nuclear FA accumulation in WASp- and FANCD2-deficient lymphocytes
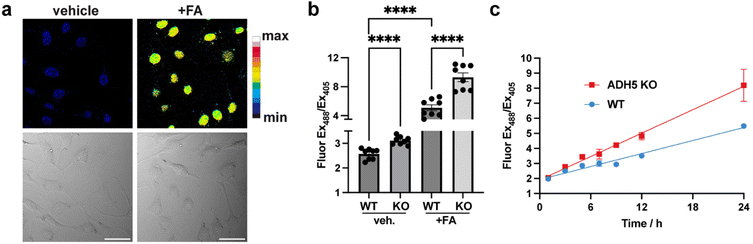
During hematopoietic cell differentiation, transcriptional reprogramming is expected to generate nuclear FA, in part through histone demethylation.4,53,54 To directly monitor nuclear FA dynamics in response to genetic alteration in proteins involved in histone methylation status and DNA–protein crosslink (DPC) repair, we employed NucRFAP-2 in human immune cells with genetically-induced deficiencies in genomic integrity. In wild-type human T cells, exogenous FA exposure produced a robust, dose-dependent increase in nuclear probe signal, validating the probe's sensitivity and specificity (Fig. 4a and b). Under hydroxyurea (HU)-induced replication stress, T cells deficient in WASp showed a more pronounced nuclear FA accumulation relative to wild-type controls (Fig. 4c). Moreover, FANCD2-deficient T and Fanconi anemia patient-derived B cells also exhibited significant, HU-induced nuclear FA accumulation, consistent with known aldehyde hypersensitivity in Fanconi pathway defects (Fig. 4c and d).54,55 Importantly, the NucRFAP-2 probe provides the first direct evidence that FA accumulation in human T and B lymphocytes occurs within the nucleus. In contrast, WAS patient-derived B cells did not show a further HU-induced increase, despite having higher steady-state nuclear FA levels than wild-type B cells (Fig. 4d). We note that these cell lines are cultured in suspension rather than as adherent cells, which leads to images where some cells are slightly out of plane, with others aligning correctly on the imaging plane. This heterogeneity does not impact signal quantitation, as multiple Z planes are considered in the data workup. These findings suggest either cell-type-specific (T versus B) differences in the impact of replication stress on nuclear FA catabolism, or mutation-specific effects within the WAS genotypes that may vary even among B cells, and from one patient to another.Conclusions
In this work, we have reported the development and application of a first-generation, nuclear-targeted activity-based sensor for nuclear formaldehyde, NucRFAP-2. This probe leverages the chromatin dye Hoechst to confer nuclear localization through its selective high-affinity binding to DNA. Owing to its 2-aza-Cope trigger, NucRFAP-2 reports with high selectivity on fluctuations in FA over other metabolic aldehydes, including methylglyoxal and acetaldehyde, with a dose-dependent ratiometric fluorescence response.Using this probe, we observed a marked increase in nuclear FA following genetic disruption of ADH5, the principal FA detoxifying enzyme in mammalian cells. Notably, NucRFAP-2 shows a distinct optical response from the previously reported mitochondrial-targeting FA probe MitoRFAP-2, which did not show elevated levels of mitochondrial FA in ADH5-deficient cells. Indeed, these observations indicate that accurate measurements of labile FA – a highly reactive and transient metabolite – require detection in immediate proximity to its biochemical sources and sinks. In tandem, these subcellular-localized probes reveal distinct dynamics of different subcellular 1C pools regulated by ADH5, which affect nuclear FA pools but not mitochondrial FA ones.
Next, we applied NucRFAP-2 in WAS or FANCD2 KO (CRISPR/Cas9-mediated knock out) T cells, as well as B cells from a WAS patient and a Fanconi anemia patient. These experiments demonstrated impaired aldehyde catabolism in lymphocytes from both genetic disorders, which manifest in bone marrow failure and cancer predisposition. The ectopic accumulation of FA in the nucleus identifies a previously unrecognized cellular phenotype associated with WASp deficiency. These findings have important mechanistic and clinical implications for WAS pathophysiology, as Vyas and co-workers have previously demonstrated that WASp-deficient T cells exhibit markedly reduced histone H3K4me3 enrichment at the promoters of immune function genes.56 Because FA can deplete S-adenosylmethionine (SAM) and disrupt histone H3K4 methylation,23 we posit that persistent nuclear FA accumulation, as identified here, may contribute to the epigenetic dysregulation observed in WASp deficiency. Taken together with prior evidence that Fanconi pathway-deficient cells also accumulate nuclear FA, these findings point to a shared vulnerability to aldehyde stress across both WASp and Fanconi genome maintenance pathways, underscoring an unexpected functional alliance between them.
Accordingly, in addition to its established role in homologous recombination-mediated DNA repair and preventing genome-destabilizing R loop accumulation,57–60 WASp appears critical for preventing aberrant nuclear aldehyde accumulation, thereby safeguarding DNA integrity against multiple endogenous genotoxic stressors. These data suggest that Wiskott-Aldrich syndrome belongs to an emerging class of hematologic aldehyde-dependent pathologies in which impaired detoxification (ADH5/ALDH2 enzyme deficiencies) and/or Fanconi-mediated DNA repair pathways fail to constrain endogenous FA-linked DNA damage, particularly under replication stress. While the specific nature of dysfunction in WAS, whether involving detoxification enzymes (ALDH2/ADH5)(Tier-1) or Fanconi D2-mediated DNA repair (Tier-2) mechanisms, remains to be formally investigated, this framework provides a rationale for developing (i) functional diagnostics that quantify nuclear FA levels in patient lymphocytes and correlate these with clinical severity-ranging from mild disease (X-linked thrombocytopenia) to more severe presentations involving autoimmunity and malignancy and (ii) FA detoxification-enhancing strategies, such as ALDH2 activation or glutathione (GSH) pool replenishment, as potential therapeutic interventions.
In summary, the development of NucRFAP-2 provides a powerful chemical tool for probing the spatial dynamics of nuclear FA biology, enabling precise detection of this transient metabolite at its sites of production and clearance within a compartment that requires stringent regulation of this established genotoxin. Application of this probe uncovered impaired aldehyde catabolism in Wiskott-Aldrich syndrome, linking WASp deficiency to persistent nuclear FA accumulation and downstream epigenetic dysregulation. By integrating activity-based sensing probe development and application to patient-derived, disease-relevant model systems, this work establishes both a framework for functional diagnostics based on nuclear FA measurement and a rationale for therapeutic strategies aimed at enhancing aldehyde detoxification capacity in patients. As such, activity-based sensing shows promise to contribute to basic biology and diagnostic medicine.
Author contributions
C. J. C. conceived and directed the project. L. T. designed the probe. L. T., K. K. W., S. S. H and Y. M. V. created CRISPR KO cell lines and designed the cell imaging assays. L. T. synthesized, purified, and characterized the compounds and performed spectroscopic assays. L. T. and K. K. W. performed cell imaging assays. L. T., C. J. C. and Y. M. V. wrote and edited the manuscript. All authors provided critical feedback and have approved the final version of the manuscript.Conflicts of interest
The authors declare no competing financial interest.Data availability
Additional synthetic, analytical, and biological data are available in the supplementary information (SI) of this article. Supplementary information is available. See DOI: https://doi.org/10.1039/d5sc07280h.Acknowledgements
We thank the NIH (ES 28096 and GM 139245 to C. J. C. and AI 146380 and AI 177269 to Y. M. V.) for support of this work. C. J. C. is a CIFAR Fellow. We thank Alison Killilea (UC Berkeley Tissue Culture Facility) for expert technical assistance and Hasan Celik, Alicia Lund, and UC Berkeley's NMR facility in the College of Chemistry (CoC-NMR) for spectroscopic assistance. Instruments in the CoC-NMR facility at UC Berkeley are supported in part by NIH S10OD024998. We thank Prof. Kai Johnsson for the Halo-SNAP cell lines. We thank Prof. K. J. Patel for sharing MEF wild-type and ADH5KO cell lines. We thank Prof. Alanna Schepartz for use of their laboratory's flow cytometer. This manuscript is the result of funding in whole or in part by the National Institutes of Health (NIH). It is subject to the NIH Public Access Policy. Through acceptance of this federal funding, NIH has been given a right to make this manuscript publicly available in PubMed Central upon the Official Date of Publication, as defined by NIH.Notes and references
- X. Tang, Y. Bai, A. Duong, M. T. Smith, L. Li and L. Zhang, Environ. Int., 2009, 35, 1210–1224 CrossRef CAS PubMed
.
- O. US EPA, Facts About Formaldehyde, https://www.epa.gov/formaldehyde/facts-about-formaldehyde, accessed October 18, 2023.
- Y. Shi and J. R. Whetstine, Mol. Cell, 2007, 25, 1–14 CrossRef CAS PubMed
.
- Y. Shi, F. Lan, C. Matson, P. Mulligan, J. R. Whetstine, P. A. Cole, R. A. Casero and Y. Shi, Cell, 2004, 119, 941–953 CrossRef CAS PubMed
.
- J. D. W. Toh, S. W. M. Crossley, K. J. Bruemmer, E. J. Ge, D. He, D. A. Iovan and C. J. Chang, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 25284–25292 CrossRef CAS PubMed
.
- T. Gerken, C. A. Girard, Y.-C. L. Tung, C. J. Webby, V. Saudek, K. S. Hewitson, G. S. H. Yeo, M. A. McDonough, S. Cunliffe, L. A. McNeill, J. Galvanovskis, P. Rorsman, P. Robins, X. Prieur, A. P. Coll, M. Ma, Z. Jovanovic, I. S. Farooqi, B. Sedgwick, I. Barroso, T. Lindahl, C. P. Ponting, F. M. Ashcroft, S. O'Rahilly and C. J. Schofield, Science, 2007, 318, 1469–1472 CrossRef CAS PubMed
.
- D. Leys, J. Basran and N. S. Scrutton, EMBO J., 2003, 22, 4038–4048 CrossRef CAS PubMed
.
- G. Burgos-Barragan, N. Wit, J. Meiser, F. A. Dingler, M. Pietzke, L. Mulderrig, L. B. Pontel, I. V. Rosado, T. F. Brewer, R. L. Cordell, P. S. Monks, C. J. Chang, A. Vazquez and K. J. Patel, Nature, 2017, 548, 549–554 CrossRef CAS PubMed
.
- C. Mellor, E. A. Larson and M. Wang, J. Cell Biol., 2025, 224, e202502205 CrossRef CAS PubMed
.
- Y. Xu, H. Meng, J. Ren and A.-P. Zeng, J. Biol. Eng., 2020, 14, 15 CrossRef CAS PubMed
.
- U. Vekariya, M. Toma, M. Nieborowska-Skorska, B. V. Le, M.-C. Caron, A.-M. Kukuyan, K. Sullivan-Reed, P. Podszywalow-Bartnicka, K. N. Chitrala, J. Atkins, M. Drzewiecka, W. Feng, J. Chan, S. Chatla, K. Golovine, J. Jelinek, T. Sliwinski, J. Ghosh, K. Matlawska-Wasowska, G. Chandramouly, R. Nejati, M. Wasik, S. M. Sykes, K. Piwocka, E. Hadzijusufovic, P. Valent, R. T. Pomerantz, G. Morton, W. Childers, H. Zhao, E. M. Paietta, R. L. Levine, M. S. Tallman, H. F. Fernandez, M. R. Litzow, G. P. Gupta, J.-Y. Masson and T. Skorski, Blood, 2023, 141, 2372–2389 CAS
.
- R. J. Hopkinson and C. J. Schofield, Biochemistry, 2018, 57, 904–906 CrossRef CAS PubMed
.
- C. A. Staab, M. Hellgren and J.-O. Höög, Cell. Mol. Life Sci., 2008, 65, 3950 CrossRef CAS PubMed
.
- B. Jackson, C. Brocker, D. C. Thompson, W. Black, K. Vasiliou, D. W. Nebert and V. Vasiliou, Hum. Genomics, 2011, 5, 283 CrossRef CAS PubMed
.
- J.-M. Guo, A.-J. Liu, P. Zang, W.-Z. Dong, L. Ying, W. Wang, P. Xu, X.-R. Song, J. Cai, S.-Q. Zhang, J.-L. Duan, J. L. Mehta and D.-F. Su, Cell Res., 2013, 23, 915–930 CrossRef CAS PubMed
.
- F. A. Dingler, M. Wang, A. Mu, C. L. Millington, N. Oberbeck, S. Watcham, L. B. Pontel, A. N. Kamimae-Lanning, F. Langevin, C. Nadler, R. L. Cordell, P. S. Monks, R. Yu, N. K. Wilson, A. Hira, K. Yoshida, M. Mori, Y. Okamoto, Y. Okuno, H. Muramatsu, Y. Shiraishi, M. Kobayashi, T. Moriguchi, T. Osumi, M. Kato, S. Miyano, E. Ito, S. Kojima, H. Yabe, M. Yabe, K. Matsuo, S. Ogawa, B. Göttgens, M. R. G. Hodskinson, M. Takata and K. J. Patel, Mol. Cell, 2020, 80, 996–1012 CrossRef CAS PubMed
.
- H. Reingruber and L. B. Pontel, Curr. Opin. Toxicol., 2018, 9, 28–34 CrossRef
.
- S. W. M. Crossley, L. Tenney, V. N. Pham, X. Xie, M. W. Zhao and C. J. Chang, J. Am. Chem. Soc., 2024, 146, 8865–8876 CrossRef CAS PubMed
.
- H. Li, S. Borinskaya, K. Yoshimura, N. Kal’ina, A. Marusin, V. A. Stepanov, Z. Qin, S. Khaliq, M.-Y. Lee, Y. Yang, A. Mohyuddin, D. Gurwitz, S. Q. Mehdi, E. Rogaev, L. Jin, N. K. Yankovsky, J. R. Kidd and K. K. Kidd, Ann. Hum. Genet., 2009, 73, 335–345 CrossRef CAS PubMed
.
- Y.-C. Chang, H.-L. Lee, W. Yang, M.-L. Hsieh, C.-C. Liu, T.-Y. Lee, J.-Y. Huang, J.-Y. Nong, F.-A. Li, H.-L. Chuang, Z.-Z. Ding, W.-L. Su, L.-Y. Chueh, Y.-T. Tsai, C.-H. Chen, D. Mochly-Rosen and L.-M. Chuang, Nat. Commun., 2023, 14, 5971 CrossRef CAS PubMed
.
- L. B. Pontel, I. V. Rosado, G. Burgos-Barragan, J. I. Garaycoechea, R. Yu, M. J. Arends, G. Chandrasekaran, V. Broecker, W. Wei, L. Liu, J. A. Swenberg, G. P. Crossan and K. J. Patel, Mol. Cell, 2015, 60, 177–188 CrossRef CAS PubMed
.
- J. I. Garaycoechea, G. P. Crossan, F. Langevin, L. Mulderrig, S. Louzada, F. Yang, G. Guilbaud, N. Park, S. Roerink, S. Nik-Zainal, M. R. Stratton and K. J. Patel, Nature, 2018, 553, 171–177 CrossRef CAS PubMed
.
- V. N. Pham, K. J. Bruemmer, J. D. W. Toh, E. J. Ge, L. Tenney, C. C. Ward, F. A. Dingler, C. L. Millington, C. A. Garcia-Prieto, M. C. Pulos-Holmes, N. T. Ingolia, L. B. Pontel, M. Esteller, K. J. Patel, D. K. Nomura and C. J. Chang, Science, 2023, 382, eabp9201 CrossRef CAS PubMed
.
- L. Tenney, V. N. Pham and C. J. Chang, ACS Chem. Biol., 2024, 19, 798–801 CrossRef CAS PubMed
.
- L. Tenney, V. N. Pham, T. F. Brewer and C. J. Chang, Chem. Sci., 2024, 15, 8080–8088 RSC
.
- Y. Shi, F. Lan, C. Matson, P. Mulligan, J. R. Whetstine, P. A. Cole, R. A. Casero and Y. Shi, Cell, 2004, 119, 941–953 CrossRef CAS PubMed
.
- K. J. Bruemmer, O. Green, T. A. Su, D. Shabat and C. J. Chang, Angew. Chem., Int. Ed., 2018, 57, 7508–7512 CrossRef CAS PubMed
.
- T. F. Brewer and C. J. Chang, J. Am. Chem. Soc., 2015, 137, 10886–10889 CrossRef CAS PubMed
.
- T. F. Brewer, G. Burgos-Barragan, N. Wit, K. J. Patel and C. J. Chang, Chem. Sci., 2017, 8, 4073–4081 RSC
.
- K. J. Bruemmer, R. R. Walvoord, T. F. Brewer, G. Burgos-Barragan, N. Wit, L. B. Pontel, K. J. Patel and C. J. Chang, J. Am. Chem. Soc., 2017, 139, 5338–5350 CrossRef CAS PubMed
.
- W. Liu, C. Truillet, R. R. Flavell, T. F. Brewer, M. J. Evans, D. M. Wilson and C. J. Chang, Chem. Sci., 2016, 7, 5503–5507 RSC
.
- J. Ohata, K. J. Bruemmer and C. J. Chang, Acc. Chem. Res., 2019, 52, 2841–2848 CrossRef CAS PubMed
.
- A. Roth, H. Li, C. Anorma and J. Chan, J. Am. Chem. Soc., 2015, 137, 10890–10893 CrossRef CAS PubMed
.
- Y. Du, Y. Zhang, M. Huang, S. Wang, J. Wang, K. Liao, X. Wu, Q. Zhou, X. Zhang, Y.-D. Wu and T. Peng, Chem. Sci., 2021, 12, 13857–13869 RSC
.
- L. He, X. Yang, Y. Liu, X. Kong and W. Lin, Chem. Commun., 2016, 52, 4029–4032 RSC
.
- J. Li, D. Ding, W. Song, J. Wang, W. Quan, L. Huang and W. Lin, Sens. Actuators, B, 2022, 368, 132136 CrossRef CAS
.
- K. J. Bruemmer, S. W. M. Crossley and C. J. Chang, Angew. Chem., Int. Ed., 2020, 59, 13734–13762 CrossRef CAS PubMed
.
- K. J. Bruemmer, T. F. Brewer and C. J. Chang, Curr. Opin. Chem. Biol., 2017, 39, 17–23 CrossRef CAS PubMed
.
- J. Chan, S. C. Dodani and C. J. Chang, Nat. Chem., 2012, 4, 973–984 CrossRef CAS PubMed
.
- P. Španěl, D. Smith, T. A. Holland, W. A. Singary and J. B. Elder, Rapid Commun. Mass Spectrom., 1999, 13, 1354–1359 CrossRef
.
- S. Kato, P. J. Burke, T. H. Koch and V. M. Bierbaum, Anal. Chem., 2001, 73, 2992–2997 CrossRef CAS PubMed
.
- S. E. Ebeler, A. J. Clifford and T. Shibamoto, J. Chromatogr., B, 1997, 702, 211–215 CrossRef CAS PubMed
.
- G. Lukinavičius, C. Blaukopf, E. Pershagen, A. Schena, L. Reymond, E. Derivery, M. Gonzalez-Gaitan, E. D'Este, S. W. Hell, D. Wolfram Gerlich and K. Johnsson, Nat. Commun., 2015, 6, 8497 CrossRef PubMed
.
- A. Nakamura, K. Takigawa, Y. Kurishita, K. Kuwata, M. Ishida, Y. Shimoda, I. Hamachi and S. Tsukiji, Chem. Commun., 2014, 50, 6149–6152 RSC
.
- B. Weisblum and E. Haenssler, Chromosoma, 1974, 46, 255–260 CrossRef CAS PubMed
.
- A. Adhikary, V. Buschmann, C. Müller and M. Sauer, Nucleic Acids Res., 2003, 31, 2178–2186 CrossRef CAS PubMed
.
- Y. Yasueda, T. Tamura and I. Hamachi, Chem. Lett., 2016, 45, 265–267 CrossRef CAS
.
- Y. Ohsaki, P. O'Connor, T. Mori, R. P. Ryan, B. C. Dickinson, C. J. Chang, Y. Lu, S. Ito and A. W. Cowley, Am. J. Physiol. Ren. Physiol., 2012, 302, F95–F102 CrossRef CAS PubMed
.
- H. J. Forman, H. Zhang and A. Rinna, Mol. Aspects Med., 2009, 30, 1–12 CrossRef CAS PubMed
.
- G. V. Los, L. P. Encell, M. G. McDougall, D. D. Hartzell, N. Karassina, C. Zimprich, M. G. Wood, R. Learish, R. F. Ohana, M. Urh, D. Simpson, J. Mendez, K. Zimmerman, P. Otto, G. Vidugiris, J. Zhu, A. Darzins, D. H. Klaubert, R. F. Bulleit and K. V. Wood, ACS Chem. Biol., 2008, 3, 373–382 CrossRef CAS PubMed
.
- A. Gautier, A. Juillerat, C. Heinis, I. R. Corrêa, M. Kindermann, F. Beaufils and K. Johnsson, Chem. Biol., 2008, 15, 128–136 CrossRef CAS PubMed
.
- L. D. Lavis, Annu. Rev. Biochem., 2017, 86, 825–843 CrossRef CAS PubMed
.
- Y. Tsukada, J. Fang, H. Erdjument-Bromage, M. E. Warren, C. H. Borchers, P. Tempst and Y. Zhang, Nature, 2006, 439, 811–816 CrossRef CAS PubMed
.
- X. Shen, R. Wang, M. J. Kim, Q. Hu, C.-C. Hsu, J. Yao, N. Klages-Mundt, Y. Tian, E. Lynn, T. F. Brewer, Y. Zhang, B. Arun, B. Gan, M. Andreeff, S. Takeda, J. Chen, J. Park, X. Shi, C. J. Chang, S. Y. Jung, J. Qin and L. Li, Mol. Cell, 2020, 80, 1013–1024 CrossRef CAS PubMed
.
- M. Wang, L. T. L. Brandt, X. Wang, H. Russell, E. Mitchell, A. N. Kamimae-Lanning, J. M. Brown, F. A. Dingler, J. I. Garaycoechea, T. Isobe, S. J. Kinston, M. Gu, G. S. Vassiliou, N. K. Wilson, B. Göttgens and K. J. Patel, Mol. Cell, 2023, 83, 2417–2433 CrossRef CAS PubMed
.
- M. D. Taylor, S. Sadhukhan, P. Kottangada, A. Ramgopal, K. Sarkar, S. D'Silva, A. Selvakumar, F. Candotti and Y. M. Vyas, Sci. Transl. Med., 2010, 2, 37ra44 Search PubMed
.
- S.-S. Han, K.-K. Wen, M. L. García-Rubio, M. S. Wold, A. Aguilera, W. Niedzwiedz and Y. M. Vyas, Nat. Commun., 2022, 13, 3743 CrossRef CAS PubMed
.
- J. Nieminuszczy, P. R. Martin, R. Broderick, J. Krwawicz, A. Kanellou, C. Mocanu, V. Bousgouni, C. Smith, K.-K. Wen, B. L. Woodward, C. Bakal, F. Shackley, A. Aguilera, G. S. Stewart, Y. M. Vyas and W. Niedzwiedz, Nucleic Acids Res., 2023, 51, 6337–6354 CrossRef CAS PubMed
.
- B. R. Schrank, T. Aparicio, Y. Li, W. Chang, B. T. Chait, G. G. Gundersen, M. E. Gottesman and J. Gautier, Nature, 2018, 559, 61–66 CrossRef CAS PubMed
.
- K. Sarkar, S.-S. Han, K.-K. Wen, H. D. Ochs, L. Dupré, M. M. Seidman and Y. M. Vyas, J. Allergy Clin. Immunol., 2018, 142, 219–234 CrossRef CAS PubMed
.
| This journal is © The Royal Society of Chemistry 2026 |