Open Access Article
Open Access ArticleA high-throughput N-glycan analysis strategy with targeted mass spectrometry (HTnGQs-target) for liver disease diagnosis
Xuejiao
Liu†
ab,
Jierong
Chen†
c,
Bin
Fu†
b,
Sanfeng
Han
d,
Dongdong
Zheng
e,
Ying
Zhang
 *ab and
Haojie
Lu
*ab and
Haojie
Lu
 *ab
*ab
aLiver Cancer Institute of Zhongshan Hospital, Department of Chemistry, Fudan University, Shanghai 200032, China. E-mail: ying@fudan.edu.cn; luhaojie@fudan.edu.cn
bInstitutes of Biomedical Sciences, NHC Key Laboratory of Glycoconjugates Research, Fudan University, Shanghai 200032, China
cLaboratory Medicine of Guangdong Provincial People's Hospital, Guangdong Academy of Medical Sciences, Guangzhou, Guangdong 510000, China
dMinhang Hospital, Fudan University, Shanghai 200032, China
eDepartment of Ultrasound, Fudan University Shanghai Cancer Center, Shanghai, 200032, PR China
First published on 4th November 2025
Abstract
Hepatocellular carcinoma (HCC), a global leading cause of cancer-related mortality, is critically hindered by delayed diagnosis due to the lack of sensitive early biomarkers. Alterations in N-glycan composition and structure, which are closely associated with HCC pathogenesis, hold great promise for early detection; however, conventional mass spectrometry-based glycomic methods are limited by low sample throughput. To address this, we developed HTnGQs-target—a high-throughput targeted mass spectrometry approach integrating methylamine derivatization and 6-plex aminoxy TMT labeling. This approach enables robust quantitative analysis of up to 144 serum samples per day with high sensitivity and reproducibility. Applied to a cohort of 320 serum samples encompassing the full spectrum of liver disease—from healthy controls to chronic hepatitis B (CHB), liver cirrhosis, and HCC—our targeted MS method identified a novel panel of N-glycan biomarkers that effectively discriminates between benign and malignant stages. Furthermore, isomer-specific analysis revealed 12 sialylated N-glycan isomers with significant differential expression, further enhancing diagnostic specificity and underscoring the potential for clinical application in early HCC detection.
Introduction
Hepatocellular carcinoma (HCC) is one of the deadliest malignancies, ranking fourth in cancer mortality rates world-wide.1 A major factor contributing to this high mortality is the low curative rate (20–30%), primarily due to the difficulty in early detection.2 Only 30% of patients are diagnosed at an early stage, achieving a five-year survival rate of 70%, whereas those diagnosed at a later stage have a two-year survival rate of less than 16%.3 Glycosylation plays significant roles in the progression and prognosis of various cancers, including HCC.4–6 Altered glycosylation patterns, such as changes in glycan branching, sialylation, and fucosylation, are commonly observed in HCC and are closely associated with tumor growth, metastasis, and immune evasion.7 For instance, fucosylation and sialylation significantly increase during HCC progression and metastasis.8–10 These glycan structure changes can reflect subtle physiological or pathological state differences, offering new possibilities for early disease diagnosis.11 For instance, α2-3 sialylation promotes cell adhesion and metastatic potential, while α2-6 sialylation modulates immune checkpoint interactions. Consequently, N-glycans are increasingly recognized as valuable biomarkers, particularly for the early diagnosis and monitoring of liver disease and cancer.12,13Despite these insights, conventional N-glycome analysis techniques remain limited by challenges that impede their clinical translation, including complex and time-consuming workflows, as well as insufficient throughput for large-scale studies.14 Hydrophilic interaction liquid chromatography with fluorescence detection (HILIC-FLD), matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS), and liquid chromatography-mass spectrometry (LC-MS) have been developed to address some of these issues. However, these approaches are primarily designed for global profiling of N-glycan differences rather than targeted analysis. Although targeted mass spectrometry approaches, such as multiple reaction monitoring (MRM) or parallel reaction monitoring (PRM), offer superior sensitivity, specificity, and quantitative precision, which are essential for the rigorous verification of candidate biomarkers, their current implementations in N-glycan biomarker analysis are still low-throughput and not suitable for large-scale clinical validation. Consequently, a major unmet need in the field is the development of robust, high-throughput targeted mass spectrometry methods to enable the efficient validation of N-glycan biomarkers across extensive sample cohorts.13,15,16
To address the challenges of validating N-glycan biomarkers, we developed HTnGQs-target, a high-throughput targeted N-glycomics platform designed for simultaneous quantification and isomer-specific analysis of serum N-glycans. Building on our prior works for studying glycoproteins,17,18 while protein-specific glycopeptide analyses, such as IgG-focused studies, can capture fine-scale glycosylation dynamics during disease progression, our total N-glycan-based approach emphasizes global serum alterations, which are more suitable for high-throughput clinical screening but inherently less specific to protein origin. We here developed a dual labeling method that combines methylamine derivatization for sialic acid stabilization with 6-plex aminoxy TMT labeling at the reducing end of N-glycans, enabling multiplexed parallel reaction monitoring-MS (PRM-MS) of six samples per run. The dual labeling strategy with isotopic reagent (d0- and d3-methylamine) further introduces mass shifts to discriminate α2-3 and α2-6 linkages. Compared with conventional targeted-MS approaches, this strategy increases detection channels six-fold, achieving a throughput of up to 144 samples per day, with isomeric resolution.
Applied to 320 serum samples from liver disease patients, spanning the full spectrum of liver disease progression from healthy controls to chronic hepatitis B (CHB), liver cirrhosis (LC), and hepatocellular carcinoma (HCC), HTnGQs-target revealed significant differences in N-glycan profiles between different status and identified a novel biomarker panel with good diagnostic performance (AUC = 0.93). By bridging high-throughput quantification with structural precision, HTnGQs-target unlocks the potential for isomer-resolved biomarker discovery, offering transformative capabilities for early HCC detection and personalized therapeutic monitoring.
Results and discussion
Workflow of HTnGQs-target
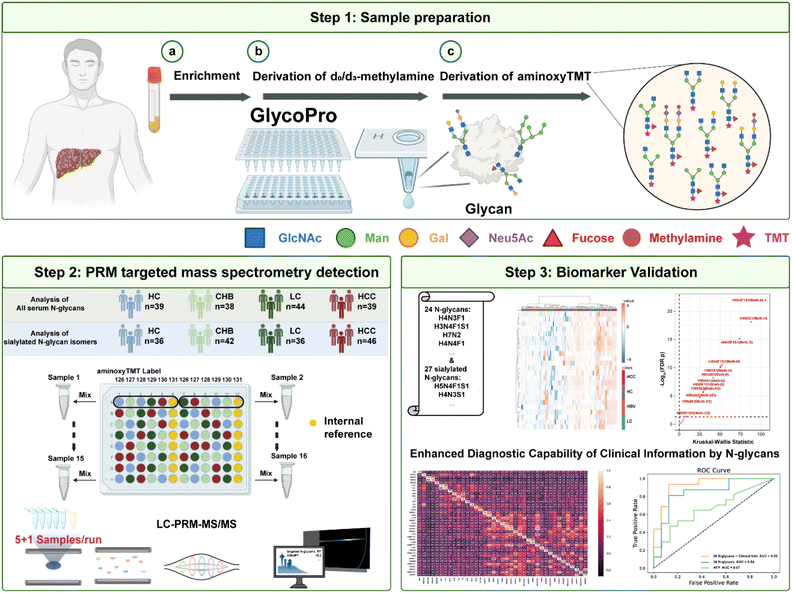
In this study, we developed a high-throughput N-glycan targeted analysis method (HTnGQs-target) to explore and vali-date N-glycan biomarkers. The experimental workflow is shown in Fig. 1. We collected serum samples from patients with liver diseases (chronic hepatitis B, liver cirrhosis and hepatocellular carcinoma) and healthy controls. The N-glycans were released from serum glycoproteins by overnight digestion with PNGase F, followed by a two-step labeling. The two-step labeling process involved methylamine labeling of sialic acid-containing N-glycans to protect sialic acids, and aminoxy-TMT 6-Plex labeling of all N-glycans at their reducing end, enabling multiplex detection in a single mass spectrometry run (Fig. S1). Before labeling, samples from patients with various liver diseases and healthy controls were randomly distributed into a 96-well plate to reduce systematic bias. Specifically, each set of samples (5 clinical samples) was evenly distributed across different aminoxy TMT labels. Samples from the three liver disease groups and healthy controls used different TMT labels in various mass spectrometry runs. This randomization strategy ensured that any technical variation or systematic error was not concentrated on specific labels across different runs, thereby improving the reliability and comparability of the results. Additionally, we included internal standards (mixed serum samples) in each run on the 131 channel to correct for technical variations between different runs. These internal standards analyzed throughout the experiment allowed us to calibrate variations between runs by comparing sample signals to the internal standard signals. Then, we used PRM-MS mode for targeted analysis of N-glycans, and N-glycans identified as significantly different between benign (HC & CHB) and malignant (LC & HCC) liver disease samples were further subjected to biomarker-screening. Specifically, QC-normalized, log-transformed reporter intensities were filtered using Mann–Whitney U tests with Benjamini–Hochberg correction (FDR < 0.05), and the shortlisted glycans were evaluated by cross-validated multivariable modeling to define a minimal diagnostic panel and assess diagnostic power via ROC–AUC.Qualitative and quantitative capability of HTnGQs-target
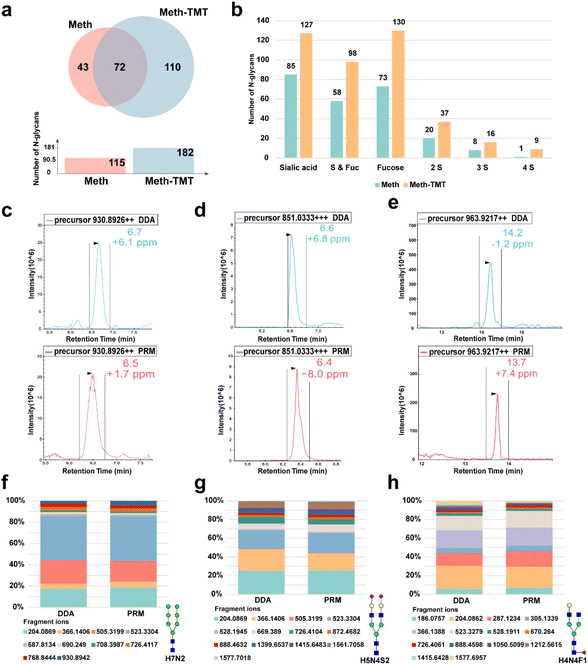
Our previous work demonstrated the high efficiency of methylamine derivatization for labeling sialylated N-glycans.19 In this study, MALDI-MS results further confirmed that TMT labeling after methylamine derivatization maintains high labeling efficiency, as evidenced by the disappearance of native N-glycan peaks and the concomitant appearance of the expected TMT-derivatized N-glycans (Fig. S2). Three technical replicates consistently showed that the number of N-glycans identified with dual methylamine and TMT labeling increased by approximately 65% compared to methylamine labeling alone (Fig. 2a and S3). While 43 low-abundance, predominantly fucosylated N-glycans were uniquely detected in the Meth-only condition, likely due to minor analyte losses during the additional TMT step, which may lead to the dropout of some low-abundance glycan chains without introducing type-specific bias.Next, we analyzed potential biases in the types of N-glycans detected with Meth and Meth–TMT labeling. Specifically, we quantified the proportions of N-glycans containing only sialic acid, only fucose, both sialic acid and fucose, and multiple sialic acids. The results showed that the labeling methods had no bias on the types of N-glycans identified (Fig. 2b). The dual labeling method identified more N-glycans containing multiple sialic acids (Fig. 2b and S4). This could be attributed to the enhanced ionization efficiency and stability provided by the methylamine and TMT labeling, thereby improving detection sensitivity for more complex glycan structures, such as those with multiple sialic acids.
After confirming the high labeling efficiency of this dual labeling method and finding that it enhances the N-glycan identification, we further evaluated the qualitative performance of HTnGQs-target. We analyzed the chromatographic retention times and fragment ion ratios of the same N-glycan in both DDA and PRM detection modes. Three representative N-glycan types including sialylated, fucosylated, and high-mannose were displayed. The retention time deviation of chromatography peaks for N-glycans identified by DDA and PRM modes was within 0.5 minutes (Fig. 2c–e). Additionally, the relative abundances of tandem mass spectrometry fragment ions were highly similar between DDA and PRM modes (Fig. 2f–h and S5–S15). These results further demonstrate the robust qualitative capability of HTnGQs-target in N-glycan PRM-MS analysis. Overall, these results indicate that the dual labeling method with methylamine and TMT significantly increases the number of identified N-glycans and demonstrates high reliability in qualitative analysis.
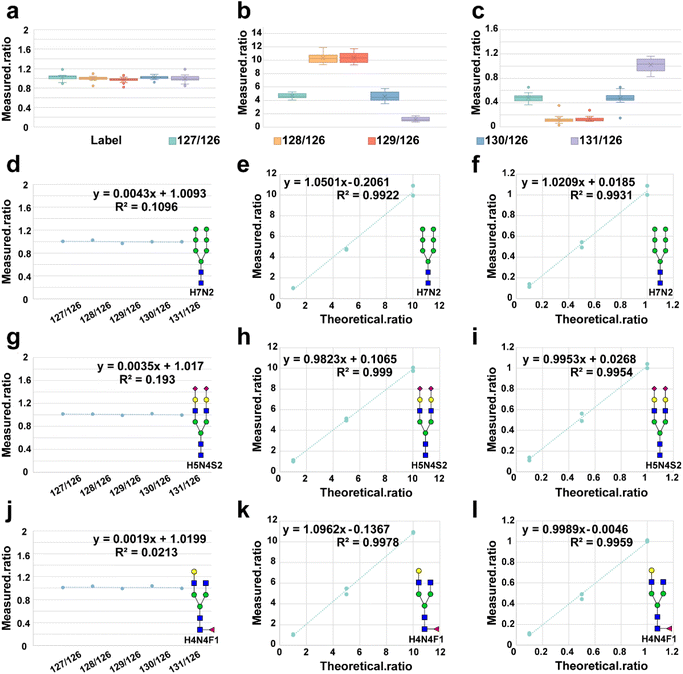
Next, we investigated the quantitative capability of HTnGQs-target. We divided the methylamine-labeled N-glycans into six aliquots and labeled them with 6-plex TMT reagents (126–131), respectively. The 6-plex TMT labeled N-glycans were then pooled in ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (group 1–1), 1
1 (group 1–1), 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (group 1–10), and 10
1 (group 1–10), and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (group 10–1). The results from tandem mass spectrometry quantification using HTnGQs-target were analyzed. The box plots demonstrated that the observed ratios closely matched the expected ratios (Fig. 3a–c).
10 (group 10–1). The results from tandem mass spectrometry quantification using HTnGQs-target were analyzed. The box plots demonstrated that the observed ratios closely matched the expected ratios (Fig. 3a–c).
We analyzed three representative N-glycans in detail: high-mannose N-glycan (H7N2) (Fig. 3d–f), disialylated N-glycan (H5N4S2) (Fig. 3g–i), and fucosylated N-glycan (H4N4F1) (Fig. 3j–l). The results demonstrated a strong linear correlation between the detected and expected ratios, indicating high reproducibility and good linearity (R2 > 0.99) in quantitation within one order of magnitude. These findings underscore the robust quantification capability of the HTnGQs-target method.
Application of HTnGQs-target for enhanced diagnosis of liver diseases
After validating the qualitative and quantitative capabilities of HTnGQs-target, we applied it to detect serum N-glycans in 192 samples (2 sets of 96-well plates) from patients with liver diseases and healthy controls. The total sample set included 32 internal reference samples and 160 clinical samples (Table S1). First, based on previous findings of significant differential serum N-glycans between liver disease patients and healthy controls (unpublished data, Table S2), we selected 24 serum N-glycans for subsequent PRM detection (Table S3).In the PRM-MS detection, we used aminoxy TMT-131 in every six aminoxy TMT labels as an internal reference to correct for potential instrument state errors between different runs. To assess the stability of the mass spectrometer during the detection process and correct the retention time of targeted N-glycans, 8 DDA injections were interspersed among the 32 PRM-MS injections (one DDA sample injected before every four sets of PRM samples).
First, we analyzed TMT-131 reporter ion intensities from internal reference samples across 8 DDA and 32 PRM runs to evaluate the intra-day and inter-day variations (Fig. S16). Both PRM and DDA demonstrated robust stability, with TMT-131 signal intensities showing low variability and consistent distributions, but PRM showed better stability than DDA. Moreover, no batch effects were observed during clinical sample analysis (Fig. S17).
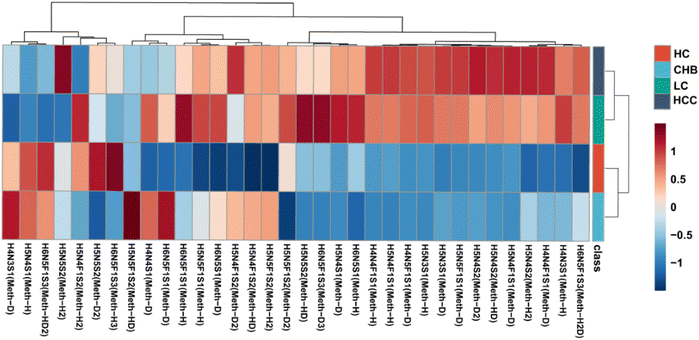
After confirming that the instrument was stable throughout the detection process and that no batch effects were present between samples, we analyzed the PRM-MS results of the 24 targeted N-glycans. The PRM-MS results for the relative expression levels and clustering of these 24 serum N-glycans in hepatocellular carcinoma (HCC), liver cirrhosis (LC), chronic hepatitis B (CHB) and healthy controls (HC) are shown in the heatmap (Fig. S18). The distinction between the healthy control group and the CHB patient group, as well as between the HCC and LC patient groups, was not obvious. Meanwhile, previous studies have reported overlapping biomarker profiles between healthy individuals and CHB patients without significant fibrosis progression, as well as between hepatocellular carcinoma and liver cirrhosis patients, due to shared glycosylation alterations.20,21 Certain N-glycan features, including hyperfucosylation, increased branching, and bisecting GlcNAc structures, recur in both LC and HCC, thereby complicating fine differentiation based solely on glycomic data. Therefore, we grouped the healthy control group and the CHB patient group as the benign control group, and the LC patient group and the HCC patient group as the malignant sample group. Subsequent analyses were conducted based on this two-group classification, which could better reflect the disease progression trends and aligns with glycosylation-based evidence observed in prior studies. Through Mann–Whitney U analysis of the differences between the benign controls and the malignant samples, we identified 24 significantly different N-glycans with FDR p-values < 0.05.
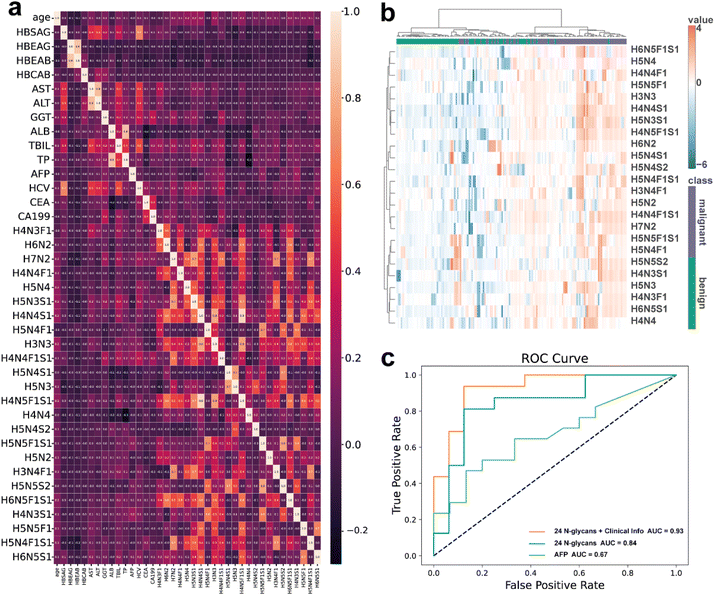
To assess the relationship between N-glycan features and clinical information, we plotted a correlation heatmap (Fig. 4a) showing the correlations between the 24 N-glycans and various clinical parameters. The clinical parameters included both virological markers (HBsAg, HBeAg, HBeAb, HBcAb, HCV), liver function indices (AST, ALT, GGT, ALB, TBIL, TP), and tumor markers (AFP, CEA, CA19-9). These parameters reflect viral infection status, hepatocellular injury, liver synthetic capacity, cholestasis, and tumor burden. Including these indices allows us to evaluate whether N-glycan alterations provide complementary diagnostic value beyond standard clinical markers. The results indicated a general lack of strong correlations between N-glycans and clinical parameters, suggesting that these N-glycan features provide unique and complementary information beyond traditional clinical markers. This independence enhances their diagnostic utility, significantly improving diagnostic accuracy when combined with traditional clinical data. The heatmap (Fig. 4b) indicated that with increasing severity of liver diseases, the abundance of all 24 N-glycans was upregulated. This clustering pattern was distinct between the benign and malignant groups. Although protein-origin is not resolved, the consistent upregulation in total serum N-glycans (Fig. 4b) suggests global glycosylation shifts associated with disease progression. Overall, our analysis demonstrated that these N-glycans have relatively consistent expression patterns across different validation sets. These findings strongly support the potential role of these N-glycans as biomarkers in the malignant progression of liver diseases. To comprehensively evaluate the diagnostic capability of these 24 N-glycans, we additionally assessed the performance of AFP alone as a traditional biomarker, as illustrated by the green curve in Fig. 4c. Using AFP alone to distinguish benign from malignant liver disease provided limited diagnostic performance (AUC = 0.67). In contrast, the independent diagnostic capability of the 24 targeted N-glycans, without incorporating clinical parameters (Fig. 4c, blue line), showed a significantly superior performance (AUC = 0.84) compared to AFP. Moreover, combining these glycan biomarkers with traditional clinical parameters markedly enhanced diagnostic capability, raising the AUC from 0.88 (Fig. 4c, red line) to 0.93 (glycans combined with clinical information, orange line). These findings emphasize the added value and clinical advantage of the 24-glycan panel, particularly when integrated into a multifactorial diagnostic model. We further analyzed the glycan characteristics, focusing on high-mannose, fucosylated, sialylated, and combined sialylated and fucosylated N-glycans.
The detailed comparison of the differential expression of these glycan types revealed that N-glycans containing sialic acid showed more significant differences, with FDR p-values < 0.001 for all four types of glycans (Fig. S19).
Application of HTnGQs-target for differential profiling of α2-3/α2-6 sialylated N-glycan isomers
To further explore the structural heterogeneity and biological significance of sialylation in liver disease, we collected an additional 160 serum samples from patients specifically to perform sialic acid linkage isomer analysis. Internal standards and detailed quality control data for this independent dataset are provided in the Fig. S20, confirming the robustness and reliability of our method when incorporating this modified derivatization strategy. Based on our previously established workflow, we employed a dual-labeling strategy utilizing light (d0-) and heavy (d3-)methylamine reagents to selectively distinguish α2-3 from α2-6 sialic acid linkages. This derivatization introduces a mass difference between linkage-specific sialylated glycan isomers, enabling linkage-level differentiation and quantification through PRM-MS detection.Using this approach, we analyzed a total of 27 sialylated N-glycans, comprising all 11 sialylated N-glycans previously validated in our primary glycan panel and an additional 16 sialic acid-containing glycans. The sialylated isomers precursors were derived from DDA on a subset of samples (Table S4), and quantification occurred via PRM with isotopic mass shifts for linkage resolution. The isomer-specific resolution provided by dual labeling allowed us to accurately quantify the individual α2-3 and α2-6 linkage signals for each glycan (Fig. 5). Differential expression analysis identified 12 glycan isomers exhibiting statistically significant differences (p < 0.05) across the four disease categories (HC, CHB, LC, and HCC), underscoring their diagnostic potential (Fig. S21).
We further conducted detailed pairwise comparisons among the four disease states (HC, CHB, LC, and HCC) for these 12 significantly altered sialylated N-glycan isomers. The objective was to identify specific disease stage transitions associated with these glycan changes. Fig. S22 provides a summary of the statistically significant pairwise differences observed. As a representative example, the glycan H5N4F1S1 exhibited pronounced differences specifically in its α2-3-linked sialic acid isomer, whereas its α2-6-linked isomer showed no significant change. Additionally, among these 12 glycans, 11 were sialylated N-glycans derived from our previously selected set of 24 target N-glycans. Beyond this, we identified a significant alteration in the α2-6-linked isomer of H5N5F1S2, which also demonstrated distinct changes during the development and progression of liver diseases. This finding not only reinforces the rationality of our initial target selection but also underscores the capability of this method to uncover novel, disease-specific glycan alterations.
Conclusions
In this work, to address the low throughput of conventional N-glycomic methods, we developed HTnGQs-target, a novel PRM-MS-based approach for targeted analysis of N-glycans. It significantly increases targeted-MS capacity for analyzing N-glycans, supporting the multiplexed analysis of up to 144 samples per day, which is six times greater than existing methods. In addition, the incorporation of dual isotopic labeling enhances structural specificity, permitting robust discrimination between α2-3 and α2-6 sialic acid linkages and enabling isomer-resolved quantification. Application of HTnGQs-target to a large clinical cohort encompassing the full spectrum of liver diseases revealed N-glycan biomarkers with strong diagnostic potential.Materials and methods
Clinical sample collection
Blood samples were collected and stored in plain tubes at −80 °C until analysis. The use of human serum samples was approved by the Guangdong Provincial People's Hospital (Guangdong Academy of Medical Sciences). For the differential analysis of serum N-glycans, a total of 160 samples were included, comprising 39 samples from HC, 38 samples from patients with CHB, 44 samples from patients with LC, and 39 samples from patients with HCC. For the linkage-specific analysis of sialylated N-glycan isomers, an additional cohort of 160 serum samples was analyzed, including 36 samples from HC, 42 samples from patients with CHB, 36 samples from patients with LC, and 46 samples from patients with HCC.Release of N-glycans
Serum proteins (2 µL) were diluted in 38 µL of 25 mM ABC buffer (pH 7.8), denatured at 100 °C for 5 min, cooled, and treated with 0.5 µL (500 U) PNGase F (New England Biolabs) for N-glycan release via over-night incubation at 37 °C.Methylamidation and aminoxy TMT labeling of N-glycans
The methylamine labeling of sialylated N-glycans was per-formed as previously described.19 Briefly, 10 µL each of 5 M methylamine hydrochloride (in DMSO) and 1 M PyAOP (in 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 DMSO
3 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4-NMM) were added and incubated at room temperature for 1 hour. The reaction was quenched with 180 µL of 80% acetonitrile (0.1% TFA), and excess reagents were removed via cotton wool enrichment. Aminoxy TMT labeling was performed per the manufacturer's protocol. The methylamine–aminoxy TMT-labeled N-glycans were then desalted, lyophilized, and normalized with aminoxy TMT-131 as an internal standard to mitigate batch effects.
4-NMM) were added and incubated at room temperature for 1 hour. The reaction was quenched with 180 µL of 80% acetonitrile (0.1% TFA), and excess reagents were removed via cotton wool enrichment. Aminoxy TMT labeling was performed per the manufacturer's protocol. The methylamine–aminoxy TMT-labeled N-glycans were then desalted, lyophilized, and normalized with aminoxy TMT-131 as an internal standard to mitigate batch effects.
Enrichment of N-glycans
Cotton enrichment was performed as described in the literature.19 Tips containing 3.75–4.25 mg of cotton were pretreated with 80 µL of 0.1% TFA (twice), then equilibrated with 80 µL of 80% acetonitrile containing 0.1% TFA. Samples were prediluted 20-fold with 80% acetonitrile 0.1% TFA and loaded three times. Cotton tips were rinsed six times with 80 µL of 80% acetonitrile 0.1% TFA. For elution, 50 µL of water was applied three times, totaling 150 µL of solution. A one-minute soaking period was used between sample addition and centrifugation to ensure complete saturation of the cotton.LC-MS/MS and LC-MS/MS-PRM analysis
Labeled N-glycans from serum samples were resuspended in 0.1% formic acid (FA) and separated by nano-liquid chromatography, followed by online electrospray tandem mass spectrometry using a Thermo Scientific™ Orbitrap Eclipse™ Tribrid™ mass spectrometer. Chromatographic separation was performed on an EASY-nLC 1200 system (Thermo Fisher Scientific) equipped with a C18 capillary column (Acclaim Pep-Map C18, 75 µm × 25 cm). The mobile phases consisted of solvent A (0.1% FA in water) and solvent B (80% acetonitrile with 0.1% FA). The gradient was 60 min in total for the samples: 2–8% from 0 to 1 min, 8–18% from 1 min to 31 min, 18–30% from 31 min to 35 min, 18–30% from 31 min to 35 min, 30–55% from 35 min to 55 min, and held for 95% for the last 5 min. The column flow rate was maintained at 300 nL min−1.In DDA mode, MS1 scans were acquired in the Orbitrap at a resolution of 120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, over an m/z range of 350–2000. The AGC target was set to 250%, with a maximum injection time of 50 ms. The charge state range was 1–5. The FAIMS compensation voltage was −55, and internal calibration using ETD was disabled. MS2 scans were acquired using stepped higher-energy collision dissociation (HCD) at 20%, 30%, and 40%, with an isolation window of 4m/z. MS2 detection was performed in the Orbitrap at a resolution of 30
000, over an m/z range of 350–2000. The AGC target was set to 250%, with a maximum injection time of 50 ms. The charge state range was 1–5. The FAIMS compensation voltage was −55, and internal calibration using ETD was disabled. MS2 scans were acquired using stepped higher-energy collision dissociation (HCD) at 20%, 30%, and 40%, with an isolation window of 4m/z. MS2 detection was performed in the Orbitrap at a resolution of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, with a scan range of 120–2000m/z and a maximum injection time of 200 ms. For PRM analysis, most settings remained consistent with those in DDA mode, except that the isolation window was narrowed to 1.2m/z, and targeted acquisition was performed using a retention time window of 3 minutes for each glycan precursor. GlycoNote software was used to analyze N-glycan MS/MS spectra from DDA and export the PRM precursor list (including m/z, charge state, and retention time).
000, with a scan range of 120–2000m/z and a maximum injection time of 200 ms. For PRM analysis, most settings remained consistent with those in DDA mode, except that the isolation window was narrowed to 1.2m/z, and targeted acquisition was performed using a retention time window of 3 minutes for each glycan precursor. GlycoNote software was used to analyze N-glycan MS/MS spectra from DDA and export the PRM precursor list (including m/z, charge state, and retention time).
In PRM mode, samples were reinjected using the PRM inclusion list, and raw files were analyzed using Skyline. TMT reporter ion intensities were extracted using a custom script (available at https://github.com/fubin1999/lab/blob/main/reporter_ions.py) to quantify labeled N-glycans across all samples.
Data analysis
Data preprocessing was conducted using the online platform https://www.metaboanalyst.ca/. Data visualization was performed using the R (version 4.3.3). Machine learning implementation in this study was carried out using Python (version 3.10.11), predominantly relying on the scikit-learn library (version 1.2.2).Author contributions
Xuejiao Liu: conceptualization, methodology, writing – review & editing. Jierong Chen, Bin Fu: methodology, investigation. Sanfeng Han: investigation, data analysis. Dongdong Zheng: visualization. Ying Zhang, Haojie Lu: supervision, project administration, writing – review & editing, funding acquisition.Conflicts of interest
There are no conflicts to declare.Data availability
The mass spectrometry data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the data set identifier PXD042528. For quantification of intact N-glycopeptides, the intensity of TMT reporter ion and the peak area of precursor ion were extracted directly. The code is available on https://github.com/FudanLuLab/useful-utilities/tree/main/tmt and https://github.com/FudanLuLab/useful-utilities/tree/main/ms1_quantify.Supplementary information is available. See DOI: https://doi.org/10.1039/d5sc06867c.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2025YFC3409504), National Natural Science Foundation of China (Grants 22174021, and 22434001) and Project supported by Shanghai Municipal Science and Technology Major Project (2023SHZDZX02).Notes and references
- J. D. Yang, P. Hainaut, G. J. Gores, A. Amadou, A. Plymoth and L. R. Roberts, A global view of hepatocellular carcinoma: trends, risk, prevention and management, Nat. Rev. Gastroenterol. Hepatol., 2019, 16, 589–604 CrossRef PubMed.
- A. G. Singal, F. Kanwal and J. M. Llovet, Global trends in hepatocellular carcinoma epidemiology: implications for screening, prevention and therapy, Nat. Rev. Clin. Oncol., 2023, 20, 864–884 CrossRef PubMed.
- K. D. Miller, L. Nogueira, T. Devasia, A. B. Mariotto, K. R. Yabroff, A. Jemal, J. Kramer and R. L. Siegel, Cancer treatment and survivorship statistics, Ca-Cancer J. Clin., 2022, 72, 409–436 CrossRef PubMed.
- L. Yang, Z. Sun, L. Zhang, Y. Cai, Y. Peng, T. Cao, Y. Zhang and H. Lu, Chemical labeling for fine mapping of IgG N-glycosylation by ETD-MS, Chem. Sci., 2019, 10, 9302–9307 RSC.
- M. R. S. Alvarez, X. A. Holmes, A. Oloumi, S. J. Grijaldo-Alvarez, R. Schindler, Q. Zhou, A. Yadlapati, A. Silsirivanit and C. B. Lebrilla, Integration of RNAseq transcriptomics and N-glycomics reveal biosynthetic pathways and predict structure-specific N-glycan expression, Chem. Sci., 2025, 16, 7155–7172 RSC.
- Y. Li, J. Wang, W. Chen, H. Lu and Y. Zhang, Comprehensive review of MS-based studies on N-glycoproteome and N-glycome of extracellular vesicles, Proteomics, 2024, 24, e2300065 CrossRef PubMed.
- X. Cao, Y. Shao, P. Meng, Z. Cao, G. Yan, J. Yao, X. Zhou, C. Liu, L. Zhang, H. Shu and H. Lu, Nascent Proteome and Glycoproteome Reveal the Inhibition Role of ALG1 in Hepatocellular Carcinoma Cell Migration, Phenomics, 2022, 2, 230–241 CrossRef CAS PubMed.
- Y. Wang and H. Chen, Protein glycosylation alterations in hepatocellular carcinoma: function and clinical implications, Oncogene, 2023, 42, 1970–1979 CrossRef CAS.
- T. Pongracz, O. A. Mayboroda and M. Wuhrer, The Human Blood N-Glycome: Unraveling Disease Glycosylation Patterns, JACS Au, 2024, 4, 1696–1708 CrossRef CAS PubMed.
- L. Cao, T. M. Lih, Y. Hu, M. Schnaubelt, S. Y. Chen, Y. Zhou, C. Guo, M. Dong, W. Yang, R. V. Eguez, L. Chen, D. J. Clark, A. Sodhi, Q. K. Li and H. Zhang, Characterization of core fucosylation via sequential enzymatic treatments of intact glycopeptides and mass spectrometry analysis, Nat. Commun., 2022, 13, 3910 CrossRef.
- L. Zhu, S. Zou, D. Yao, J. Li, Y. Xiong, Q. Wu, Y. Du, J. Wang, T. Wu and B. Wei, Profiling of aberrant sialylated N-glycans in hepatocellular carcinoma by liquid chromatography mass spectrometry, Clin. Chim. Acta, 2024, 555, 117827 CrossRef CAS.
- J. Han, X. Huang, H. Liu, J. Wang, C. Xiong and Z. Nie, Laser cleavable probes for in situ multiplexed glycan detection by single cell mass spectrometry, Chem. Sci., 2019, 10, 10958–10962 RSC.
- Y. Li, B. Fu, M. Wang, W. Chen, J. Fan, Y. Li, X. Liu, J. Wang, Z. Zhang, H. Lu and Y. Zhang, Urinary extracellular vesicle N-glycomics identifies diagnostic glycosignatures for bladder cancer, Nat. Commun., 2025, 16, 2292 CrossRef CAS.
- A. Shirakawa, Y. Manabe and K. Fukase, Recent Advances in the Chemical Biology of N-Glycans, Molecules, 2021, 26(4), 1040 CrossRef CAS PubMed.
- X. Liu, Z. Sun, Z. Li, Y. Zhang and H. Lu, Mass spectrometry-based analysis of IgG glycosylation and its applications, Int. J. Mass Spectrom., 2022, 474, 73–135 Search PubMed.
- Z. Sun, B. Fu, G. Wang, L. Zhang, R. Xu, Y. Zhang and H. Lu, High-throughput site-specific N-glycoproteomics reveals glyco-signatures for liver disease diagnosis, Natl. Sci. Rev., 2022, 10(1), nwac059 CrossRef.
- X. Liu, B. Fu, J. Chen, Z. Sun, D. Zheng, Z. Li, B. Gu, Y. Zhang and H. Lu, High-throughput intact Glycopeptide quantification strategy with targeted-MS (HTiGQs-target) reveals site-specific IgG N-glycopeptides as biomarkers for hepatic disorder diagnosis and staging, Carbohydr. Polym., 2024, 325, 121499 CrossRef CAS PubMed.
- G. Wang, S. Ma, H. Song, Y. Liang, X. Li, L. Zhang, H. Lu and Y. Zhang, A Chemoproteomic Approach for System-Wide and Site-Specific Uncovering of Functional Protein N-Glycosylation, J. Am. Chem. Soc., 2025, 147, 24127–24139 CrossRef CAS PubMed.
- Y. Peng, L. Wang, Y. Zhang, H. Bao and H. Lu, Stable Isotope Sequential Derivatization for Linkage-Specific Analysis of Sialylated N-Glycan Isomers by MS, Anal. Chem., 2019, 91, 15993–16001 CrossRef CAS PubMed.
- L. Guo, L. Wan, Y. Hu, H. Huang, B. He and Z. Wen, Serum N-glycan profiling as a diagnostic biomarker for the identification of hepatitis B virus-associated hepatocellular carcinoma, J. Gastrointest. Oncol., 2022, 13, 344–354 CrossRef.
- B. Blomme, C. Van Steenkiste, N. Callewaert and H. Van Vlierberghe, Alteration of protein glycosylation in liver diseases, J. Hepatol., 2009, 50, 592–603 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2026 |