Open Access Article
Open Access ArticleChemically modified DNA aptamers and DNAzymes for expanded functional capabilities
Canyu
Zhang
a,
Ke-siyi
Ma
a,
Zifan
Zhang
a,
Wenyong
lou
 *a,
Hongzhou
Gu
*b and
Yufei
Cao
*a,
Hongzhou
Gu
*b and
Yufei
Cao
 *a
*a
aLab of Applied Biocatalysis, School of Food Science and Engineering, South China University of Technology, Guangzhou 510640, Guangdong, China. E-mail: yufeicao@scut.edu.cn
bDepartment of Chemical Biology, School of Chemistry and Chemical Engineering, Frontiers Science Center for Transformative Molecules, National Center for Translational Medicine, Shanghai Jiao Tong University, Shanghai, 200240, Shanghai, China. E-mail: hongzhou.gu@sjtu.edu.cn
First published on 19th December 2025
Abstract
DNA aptamers and DNAzymes, typically single-stranded DNA (ssDNA), are often selected through systematic evolution of ligands by exponential enrichment (SELEX), and have demonstrated great potential across various applications. However, the inherent properties and limited chemical space of ssDNA pose challenges in terms of stability and functionality. Chemical modifications have emerged as a powerful strategy to enhance the stability and extend the functionality of these molecules. In this review, we summarize the types of chemical modifications applied to DNA aptamers and DNAzymes, and highlight representative examples from the past five years that demonstrate how these modifications expand their functional capabilities. We also discuss the current challenges and future directions in advancing the methodologies and toolsets necessary to accelerate the development of chemically modified functional DNAs, aiming to further broaden their structural diversity, functional complexity, and application potential.
1 Introduction
DNA, traditionally viewed as a carrier of genetic information, has been redefined as a programmable and versatile functional molecule. Through in vitro systematic evolution of ligands by exponential enrichment (SELEX),1,2 single-stranded DNAs (ssDNAs) are evolved to fold into specific three-dimensional structures, including aptamers and deoxyribozymes (DNAzymes). These functional DNAs leverage their unique conformations to achieve high-affinity molecular recognition of target ligands or catalyze specific chemical reactions.3–9 These functional DNAs offer a unique combination of biocompatibility, modularity, and chemical addressability, enabling their applications across therapeutics, biosensing, gene regulation, and nanotechnology.10–18However, unmodified ssDNAs face significant challenges under physiological conditions. Although chemically more stable than RNA due to the absence of the 2′-hydroxyl group, ssDNAs remain vulnerable to nuclease degradation. In parallel, their limited chemical diversity constrains conformational complexity, which can compromise binding affinity, target specificity, and catalytic efficiency. To improve biostability, alternative scaffolds such as L-DNA and circular ssDNA (circ-ssDNA) have been developed. L-DNA, with its mirror-image configuration, resists nuclease degradation but suffers from high synthesis cost and lack of enzymatic amplification.19 Circ-ssDNA provides enhanced nuclease resistance and thermal stability, yet faces low synthesis yields and inefficient selection.20–23 These limitations underscore the need for more generalizable and tunable strategies to enhance functional DNA performance.
Chemical modification has thus emerged as a powerful strategy to expand the structural and functional landscape of DNA. By altering the nucleobase, sugar, or phosphate backbone, modifications expand the structural and functional landscape of ssDNAs—enhancing stability, enabling diverse molecular interactions, and unlocking new catalytic modes. These benefits are particularly evident in aptamers and DNAzymes. For aptamers, chemical modifications can stabilize folded structures, improve binding kinetics, and extend biological half-life.24–26 In DNAzymes, strategically placed groups enable light27 or enzyme-responsive activation,28–30 enhanced substrate recognition, and tunable catalytic properties—supporting applications from smart therapeutics to synthetic gene circuits.31–33
This review summarizes recent advances in the chemical modification of DNA aptamers and DNAzymes, with an emphasis on strategies that enhance stability, affinity, and responsiveness (Fig. 1). We examine various modification approaches and their underlying mechanisms for expanding DNA functionality. Key developments in biomedical and biotechnological applications are highlighted, illustrating how chemical modifications have enabled novel capabilities. While previous reviews have primarily addressed chemical modifications in either aptamers or DNAzymes within specific applications such as biosensing, diagnostics, and therapeutics,34–37 this work is the first to comprehensively explore how these modifications enhance the functional capabilities and performance of both systems. We also discuss current challenges and outline future opportunities for the rational design and implementation of chemically modified DNA systems, advancing the field toward broader biomedical and synthetic biology applications.
2 Types of chemical modifications
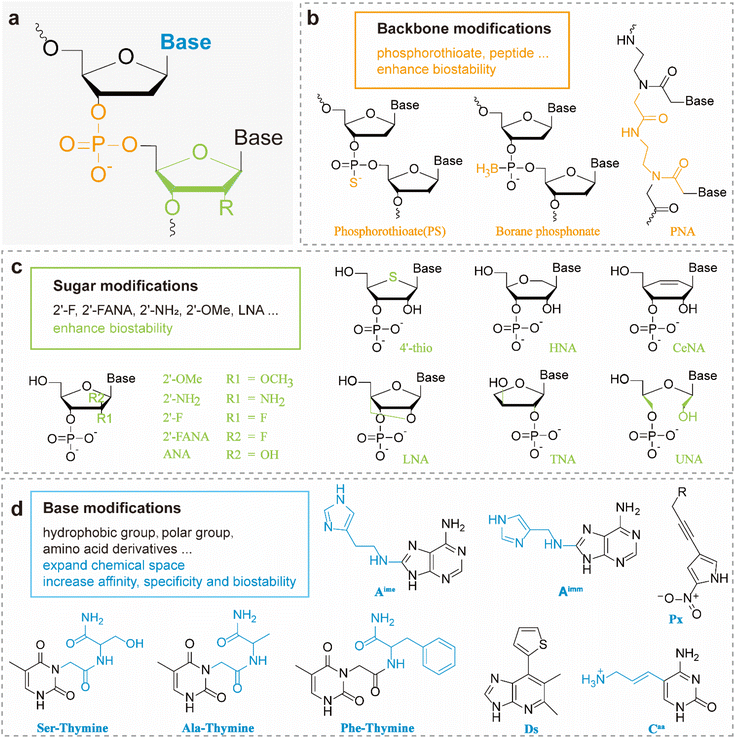
A variety of functional groups, including hydrophobic, hydrophilic and charged moieties, can be incorporated via well-established chemistries and many of those building blocks are commercially available today. The chemical modifications may be broadly classified according to the component of the nucleotide structure affected: the nucleobase, the sugar moiety, or the phosphate backbone. Each strategy offers unique advantages and has been tailored to different types of functional nucleic acids (Fig. 2).2.1 Sugar modifications
Chemical modifications of the sugar moieties in DNAs can profoundly influence physicochemical and biological properties of DNAs, including their stability, binding affinity, bioavailability, immunogenicity, and susceptibility to nuclease degradation. These effects are achieved by altering the molecular structure, conformational flexibility, and charge distribution of the nucleic acid backbone, thereby expanding their potential in biomedical applications. The most common modifications are the substitutions at the 2′ position of ribose or deoxyribose, including 2′-methoxy (2′-OMe),38 2′-fluoro (2′-F),39 and 2′-fluoroarabino (2′-FANA).40 Besides that, a diverse array of synthetic nucleic acid analogs have yielded, such as threose nucleic acid (TNA),41 hexitol nucleic acid (HNA),42 locked nucleic acid (LNA),43 unlocked nucleic acid (UNA),44 and cyclohexene nucleic acid (CeNA).45–47 These analogs exhibit distinct structural features that confer enhanced biochemical stability and target recognition capabilities.2′-FANA is a chemically modified xeno-nucleic acid (XNA) characterized by a unique 2′-fluoro substitution and an arabinose sugar backbone. This structural modification significantly enhances nuclease resistance, thermal stability, and conformational rigidity. As a result, FANA serves as an ideal scaffold for functional nucleic acids and has been widely employed in gene regulation, RNA-targeted therapeutics, and synthetic biology. Notably, FANA exhibits distinct advantages in improving molecular stability, target-binding affinity, and transmembrane delivery efficiency.40,48 TNA, in particular, replaces the natural deoxyribose with an unnatural four-carbon α-L-threofuranose sugar. This substitution imparts TNA with remarkable resistance to nuclease degradation and high binding affinity toward target molecules.24,45,49 HNA, in which the natural furanose ring is replaced with a six-membered hexitol ring, features a unique backbone geometry that supports the formation of non-canonical tertiary structures such as G-quadruplexes. This structural flexibility enhances its binding affinity toward target proteins, with dissociation constants reaching the nanomolar range.46,50 Owing to these properties, TNA, HNA and 2′-FANA serve as promising scaffold for the de novo selection of highly stable and high-affinity DNA aptamers, particularly in biosensing and therapeutic applications.51
2.2 Backbone modifications
Backbone modifications of DNA play a critical role in enhancing resistance to nuclease degradation, thereby significantly extending the circulation time of nucleic acids in vivo.52,53 In recent years, various strategies have been developed to substitute the non-bridging oxygen atoms in the phosphate backbone, leading to the creation of diverse DNA analogs.54,55 Chemical groups such as quinones can be introduced into the phosphate backbone of DNA to endow DNAzymes with responsiveness to light or enzymatic stimuli, enabling their integration with CRISPR-Cas9 systems for regulated gene editing.30Phosphorothioate (PS) is the most widely used modification which involves replacing a non-bridging oxygen atom in the phosphate group with a sulfur atom. This substitution not only enhances the nuclease resistance of functional DNAs but also introduces a chiral center at the phosphorus atom. The resulting Rp and Sp diastereomers may exhibit distinct biochemical behaviors, an aspect that warrants further investigation. Studies have shown that PS modifications can enhance the binding affinity of nucleic acids to proteins, particularly arginine-rich proteins through strengthened electrostatic and hydrophobic interactions, and also can change the metal binding preference from hard metals such as Mg2+ to softer metals such as Mn2+ and Cd2+.56–58 Despite its advantages, the stereochemical complexity and synthetic cost of PS modifications still pose challenges for broader mechanistic and functional exploration.
Peptide nucleic acids (PNAs) are nucleic acid analogs in which the sugar-phosphate backbone is replaced by repeating units of N-(2-aminoethyl)glycine, with the polyamide chain covalently linked to nucleobases via a carboxymethyl spacer. The absence of charged phosphate groups in PNAs confers increased stability against enzymatic and chemical degradation. Furthermore, the lack of phosphate groups facilitates the formation of stronger PNA/DNA duplexes, partly due to Hoogsteen-like base pairing interactions.59
These variants exhibit unique properties such as increased thermal stability, nuclease resistance, and the ability to form stable duplexes with complementary nucleic acids, making them valuable in biotechnology and therapeutic applications.
2.3 Base modifications
Natural DNA, composed of only four canonical bases (A, G, C, and T), possesses a limited chemical repertoire that constrains its ability to mimic the diverse side-chain functionalities of proteins. This restriction hinders the capacity of functional DNA to achieve high-affinity binding or catalytic specificity. Since nucleobases play a central role in molecular recognition and catalysis, modifying them provides a powerful strategy to expand the chemical functionality of nucleic acids and to fine-tune their interactions with target molecules.One of the most common and simple base modifications is N6-methyladenine (m6A) modification,60 which can be related to the m6A demethylase in vivo to apply in gene regulation or intracellular imaging. Chemical groups, such as amino, carboxyl, alkyl, and alkynyl groups, can also be introduced to the side chains of purine and pyrimidine to improve the properties of DNA aptamers.61,62 Base-appended base such as Ugu and Uad, are another kind of base modification that displays high affinity for small molecules and proteins.63,64 Apart from nucleobase analogs, unnatural bases (UBs) are also significant, such as Ds (7-(2-thienyl)imidazo[4,5-b]pyridine), Px (a diol-modified 2-nitro-4-propynylpyrrole) and 7-phenylbutyl-7-deazaadenine.65 By incorporating the hydrophobic base modification 7-phenylbutyl-7-deazaadenine, the study successfully developed a DNA aptamer with significantly enhanced binding affinity for Heat Shock Protein 70 (Hsp70), achieving a Kd value as low as 14.8 ± 0.3 nM, which is remarkably higher than that of its natural counterparts (>5 µM).62 At the meanwhile, Ichiro Hirao's group generated a series of high-affinity aptamers by genetic alphabet expansion for SELEX (ExSELEX).67,68
Modifying the nucleotide bases can broaden the structural and informational capacity of DNAs as well as effectively bridging the chemical gap between DNAs and proteins. These advances have been crucial for evolving aptamers that rival antibodies in binding performance and for pushing nucleic acid catalysts into new reaction spaces. Base modifications thus complement sugar modifications: while sugar/backbone tweaks mainly improve biostability and binding thermodynamics, base modifications can qualitatively expand the functions and targets accessible to nucleic acids.
3 Effects of chemical modifications on DNA aptamers
DNA aptamers, often regarded as the chemical equivalents of antibodies, have emerged as important molecular recognition elements in medical diagnostics, therapeutics, and biosensing applications due to their unique advantages, including low immunogenicity, scalable production capabilities, minimal batch-to-batch variability, chemical modifiability, and high programmability. Notwithstanding these benefits, their clinical and practical utility remains constrained by suboptimal performance in critical performance metrics, particularly structural stability, affinity maturation and target specificity under physiological conditions. Chemical modifications provide a way to enhance the reliability and functional efficacy of DNA aptamers for real-world applications.3.1 Enhancing stability
Despite DNA exhibiting greater chemical stability than RNA from a structural chemistry perspective, it remains challenging for DNA to maintain stability in biological environments due to exposure to nucleases. Researchers have undertaken extensive investigations to enhance the in vivo stability of DNA aptamers. The synthesis of mirror-image DNA aptamers, which are not recognized by nucleases, has demonstrated considerable potential.19,65,69 However, considering factors such as commercial availability and ease of application, chemical modification strategies to improve DNA aptamer stability are generally more advantageous. Phosphorothioate modification at the 3′ terminus of DNA aptamers substantially improves their resistance to nuclease degradation, enabling them to remain stable in serum for up to 8 hours.70Nucleic acid analogs represent an effective approach to resist nuclease degradation, which are also not recognized by nucleases. Aptamers selected from heteronucleic acid systems based on α-L-threofuranosyl nucleic acid (TNA) have demonstrated superior thermal stability compared to monoclonal antibodies, retaining full activity even after 48 hours of incubation at 75 °C (Fig. 3a).24 Recently, Ji's group proposed a universal strategy to improve the stability of DNA aptamers through glycosylation modification (Fig. 3b).26 This approach enhances aptamer stability by reducing the interaction between nucleases and the aptamer, resulting in superior stabilization compared to currently used chemical modification methods. Combining terminal and backbone chemical modifications represents an effective approach to improving the nuclease resistance and overall stability of DNA aptamers, particularly in biological environments.70
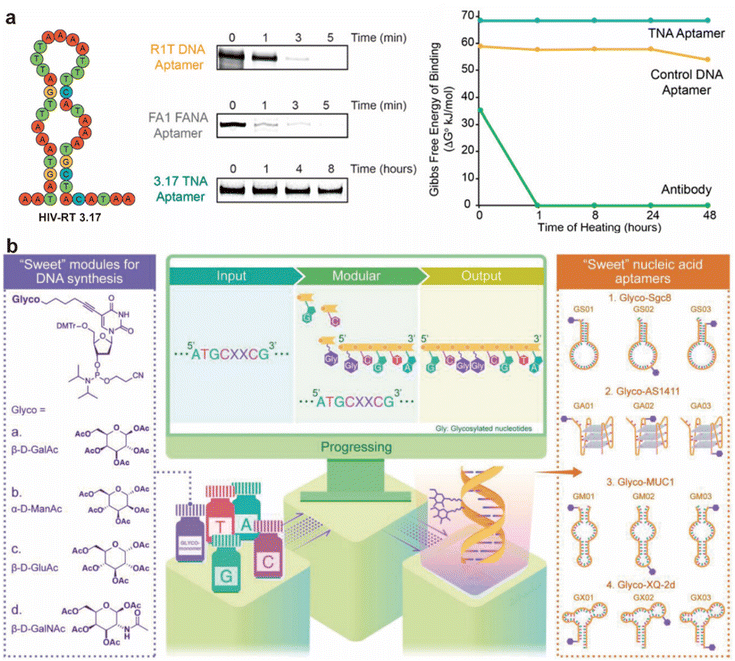 | ||
| Fig. 3 Chemical modification of aptamers to enhance stability. (a) TNA aptamer exhibited exceptional stability during HIV RT binding assays even in the presence of snake venom phosphodiesterase (SVPE), a highly active 3′ exonuclease.24 Adapted with permission from ref. 24. Copyright 2020 American Chemical Society. (b) The synthesis of glycol nucleic acid aptamers (GNAAs), highlighting the development of a novel method for their preparation using precise and programmable DNA solid-phase synthesis techniques.26 Reproduced with permission from ref. 26. Copyright 2025 Wiley-VCH. | ||
3.2 Improving affinity
Compared to conventional antibodies, DNA aptamers exhibit advantages such as low immunogenicity and facile chemical modifications.71 However, the limited chemical diversity of natural nucleic acids can restrict their binding interactions with target proteins. Chemical modifications-including nucleobase, sugar, and phosphate backbone alterations-expand aptamer diversity and enable additional non-covalent interactions, thereby enhancing binding affinity and specificity.Aptamers derived from chemically modified libraries are termed “SOMAmers” (Slow Off-rate Modified Aptamers) due to their notably low dissociation rate constants (koff) with target molecules.73 Recent advances demonstrate that pre-SELEX modifications, such as TNA,24,74 unnatural bases (UBs),68 and base-appended base,64,75 effectively expand the chemical diversity of the library, enabling the selection of high-affinity aptamers.
Thrombin-binding aptamer (TBA) is a well-defined G-quadruplex DNA aptamer widely used in biomedical research, including anticoagulant therapy. Previous studies have focused on modifying the sugar-phosphate backbone and nucleobases to improve TBA's stability and other properties. Timofeev et al. demonstrated that the recognition interface of the TBA can be expanded by introducing amino, carboxyl, and alkynyl groups to the side chains of residues T3.61 Yum et al. employed a modular strategy to investigate the effects of amino acid modifications on aptamers (Fig. 4a). Their results demonstrated that site-specific incorporation of amino acid-nucleic acid hybrids (ANHs) significantly enhanced the binding affinity, achieving a dissociation constant (Kd) of 2.94 nM, which represents a 2.5-fold improvement compared to the unmodified aptamer.72 Therefore, targeted site-specific modification is an effective approach to improving aptamer affinity.
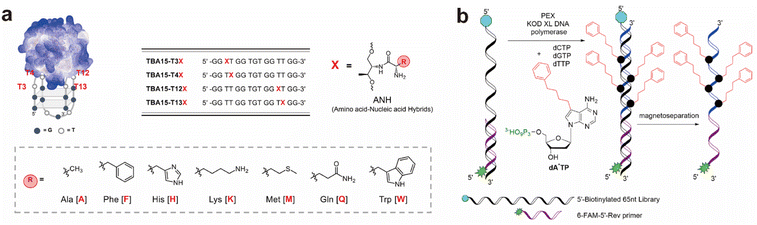 | ||
| Fig. 4 Chemical modification of aptamers to enhance binding affinity. (a) Thrombin-binding aptamer (TBA) sequence functionalized with amino-nucleic acid hybrids (ANHs) to improve target interaction.72 Adapted with permission from ref. 72. Copyright 2021 American Chemical Society. (b) Incorporation of hydrophobic 7-phenylbutyl-7 deazaadenine into the DNA library enables the selection of high binding affinity aptamers targeting Heat Shock Protein 70(HSP70).66 Reproduced with permission from ref. 66. Copyright 2023 Springer Nature. | ||
The binding affinity of aptamers of Heat Shock Protein 70 was enhanced by over 100-fold by introducing hydrophobic 7-phenylbutyl-7-deazaadenine modification (Fig. 4b). The most optimized aptamer, HSc-9.1, exhibited a dissociation constant (Kd) of 14.8 ± 0.3 nM.66 The indole modification is an important way enabling DNA aptamers to highly specifically recognize the glycan epitopes, either directly or by forming ligand-binding structural motifs that are stabilized by hydrophobic cores.76
Molecular dynamics (MD) simulations have provided preliminary evidence that the size, number, and position of chemical modifications can influence aptamer–target interactions by altering or inducing conformational changes in the target, thereby facilitating aptamer binding.77 Although chemical modifications have improved the properties of DNA aptamers, most current research has focused on aptamers targeting protein molecules,66,76,78 and there is still no universally effective strategy for optimizing the properties of aptamers against diverse targets through chemical modification.
As structural information on aptamers and their binding modes becomes increasingly detailed,79 rational design strategies—incorporating rational chemical modifications to regulate aptamer–target interactions collectively—are expected to emerge. Such approaches will enable the precise tuning of aptamer properties for a broader range of targets, thereby fully realizing the potential of functional nucleic acids in biomedicine.
4 Effects of chemical modifications on DNAzymes
RNA-cleaving DNAzymes 10–23 and 8–17 (ref. 80) are the most outstanding examples of DNA catalysts to date, which can be used as therapeutics to downregulate various RNAs (mRNA, miRNA). Due to their low efficacy in clinical trials, RNA-cleaving DNA enzymes (DNAzymes) as gene-silencing agents in therapeutic applications have stalled.77 As the catalytic cleavage ability of DNAzymes is critical for the therapeutic efficiency and detection limit of biosensors, much effort has been made to enhance the biological stability and the catalytic activity of DNAzymes in intracellular conditions.4.1 Enhancing stability
In the past five years, researchers have primarily employed xeno-nucleic acids (XNAs)-synthetic nucleic acid analogues that are chemically modified at the ribose moiety with natural nucleobases to enhance the biological stability of DNAzymes, especially conferring resistance to nucleases. Although XNAs can moderately improve the nuclease resistance and biostability of DNAzymes, these modifications alone offer only limited stability gains.82 Additional chemical modifications at the 5′ and 3′ termini have been shown to confer more pronounced improvements in both biological and thermal stability.Studies have demonstrated that incorporating phosphorothioate (PS) modifications at the terminal positions of DNAzyme 10–23, in addition to 2′-fluoroarabino nucleic acid (FANA) modifications, can extend its half-life in serum to approximately 16 hours, representing more than a tenfold increase in stability.83 Consequently, current approaches often combine multiple chemical modification strategies, targeting the binding arms, catalytic core, and the terminal ends of DNAzymes, achieving optimal stability and functional performance in biological environments.
4.2 Boosting catalytic activity
Foundational work demonstrated that the incorporation of Locked Nucleic Acid (LNA) monomers into the binding arms of DNAzymes markedly increases RNA cleavage efficiency.84 Specifically, LNAzymes achieved efficient cleavage under stoichiometric conditions, dramatically outperforming the unmodified enzyme, which required a significant molar excess. This established the principle that subtle sugar modifications could be a powerful tool for enhancing catalytic function. Building on this success, researchers began systematically investigating the impact of chemical alterations at other strategic positions, including the nucleobase and backbone, to further optimize DNAzyme properties. He's group has systematically explored the effects of various chemical modifications on deoxyadenosine residues within DNAzymes, particularly targeting the 10–23 and 8–17 motifs. Incorporation of functional groups such as amino, guanidino, or imidazole moieties into less conserved regions can enhance catalytic efficiency and alter metal ion preferences, presumably by modulating hydrogen bonding interactions and the spatial conformation of the catalytic site.85–88 These findings underscore the importance of structural and mechanistic insights into DNAzyme catalysis, providing valuable guidance for the rational design and chemical evolution of DNAzymes with improved properties.By systematically replacing each DNA residue in the catalytic core of 10–23 with the corresponding FANA nucleotide, researcher found that G2 and T8 are highly tolerant to substitution with the FANA residues, a designed version of 10–23 that carried both the G2 and U8 FANA mutations was nearly 50% more active than the parental enzyme (Fig. 5a).81 Based on a structure-guided chemical evolution strategy, the catalytic efficiency of DNAzyme 10–23 was further optimized by Nguyen et al., leveraging its acid-base catalytic mechanism driven by proton transfer at the catalytic core dG14 and synergistic metal ion coordination.32 The incorporation of a 2′-O-methoxyethyl (MOE) modification at dG14 could stabilize the A-form sugar pucker (3′-endo conformation) and enhance the catalytic microenvironment. Subsequent combinatorial modifications, including 2′-O-methyl (OMe) substitutions at C7/C8 and phosphorothioate (PS) backbone alterations in the binding arms, further constrained conformational dynamics, yielding mutant Dz 46 (Fig. 5c). This variant achieved exceptional catalytic performance under physiological conditions, exhibiting a turnover number of ∼65 min−1 within 30 minutes, over threefold greater than the unmodified parent enzyme. Structural analyses revealed that MOE and OMe modifications restricted DNAzyme flexibility, stabilizing critical interactions to enforce an active conformation. This study establishes a framework for rational design of high-performance DNAzymes with therapeutic potential. Recently, Yu's group reported CaBn, a DNA catalyst modified with both carboxyl and benzyl groups, exhibits a kobs of 3.76 min−1 in the presence of 1 mM Mg2+ (Fig. 5b), yield an approximately 700-fold increase in activity during an RNA cleavage reaction on a DNA–RNA chimeric substrate.89 This remarkable increase underscores the significant potential of chemical modifications in enhancing the catalytic efficiency of DNAzymes, highlighting their critical role in optimizing DNA-based catalytic systems for a wide range of biotechnological applications.
 | ||
| Fig. 5 Chemical modification on DNAzymes to enhance catalytic ability. (a) DNAzyme 10–23 with FANA modification has an observed pseudo first-order rate constant (kobs) of 0.57 min−1 under single-turnover conditions in a buffer that contained 10 mM MgCl2, which is nearly twofold faster than that of the unmodified parent enzyme.81 Adapted with permission from ref. 81. Copyright 2021 Springer Nature. (b) Schematic representation of the CaBn deoxyribozyme variant, featuring dual modifications: a carboxyl group at position 8 and a benzyl group at position 10. These modifications result in a 700-fold enhancement in the catalytic activity of deoxyribozyme 10–23.89 Reproduced with permission from ref. 89. Copyright 2024 American Chemical Society. (c) The synergistic effect of various chemical modifications, including LNA, OMe, MOE, and PS, leads to increased catalytic activity in 10–23 DNAzyme.32 Adapted with permission from ref. 32. Copyright 2023 Springer Nature. | ||
Peroxidase DNAzymes are an important class of DNA enzymes, typically featuring a G-quadruplex (G4) structure, that have emerged alongside RNA-cleaving DNAzymes. While less extensively studied, these DNAzymes are gaining attention for their potential in biosensing, environmental monitoring, and chemical catalysis. Recent studies have shown that chemical modifications can improve both catalytic activity and stability of peroxidase DNAzymes. For example, Shin et al. demonstrated that phosphorothioate (PS) backbone modifications significantly enhance the peroxidase-like activity of non-G-quadruplex DNAzymes by modulating electron density along the DNA backbone, thereby enhancing their biological detection efficiency for specific nucleic acid sequences.31
Beyond post-selection modifications, de novo selection from chemically modified nucleic acid libraries has emerged as a powerful strategy for discovering novel DNAzymes. The development of FANA and TNA enzymes highlights how such modifications can expand the functional capabilities and diversity of catalytic nucleic acids, driving progress in synthetic biology and molecular evolution.90,91
It is important to note, not all chemical modifications enhance DNAzyme activity. In fact, activity-blocking modifications can also play a crucial role in DNAzyme research. For example, to investigate the structural features of DNAzymes, researchers often introduce chemical groups at the cleavage site to inhibit catalysis, allowing the capture and analysis of DNAzymes in their pre-catalytic state.87,88 Moreover, such modifications can be strategically designed to impart responsiveness to specific stimuli, enabling the development of conditionally activatable DNAzymes.
4.3 Modulating metal dependence
Most DNAzymes rely on divalent metal ions (M2+) for catalysis, which limits their activity under the low-metal conditions commonly found in biological environments. Pioneering work by Perrin and co-workers demonstrated that introducing protein-like functional groups such as imidazole, amine, and guanidinium into modified deoxyribonucleoside triphosphates during in vitro selection can yield M2+-independent RNA-cleaving DNAzymes.94,95 One representative catalyst, Dz7-38-32 exhibited a catalytic efficiency (kcat/KM) of approximately 106 M−1 min−1 at 37 °C with only 0.5 mM Mg2+, representing the highest efficiency reported for any nucleic acid catalyst under low-metal conditions at that time. These studies provided a conceptual breakthrough, showing that chemically introduced imidazole, amine, and guanidinium groups can mimic protein-like acid–base and electrostatic catalysis, effectively replacing divalent metal cofactors and overcoming the intrinsic metal dependence of nucleic acid catalysis.96–98Recent studies have shown that chemical modifications can not only eliminate but also tune metal-ion dependence.85,88 In the 8–17 DNAzyme family, targeted functionalization at specific adenine residues with amino, imidazolyl, or guanidinium groups modulated both catalytic efficiency and metal selectivity. These site-specific modifications enhanced cleavage rates and shifted metal preference, with certain variants showing higher activity under Ca2+-promoted conditions relative to Mg2+. Structural and kinetic analyses revealed that such modifications alter local folding and coordination environments, reshaping the catalytic core to favor distinct metal-binding geometries. Collectively, these findings laid the groundwork for subsequent efforts to eliminate or modulate the metal dependence of DNAzymes through rational chemical design.
4.4 Enabling external stimuli-responsiveness
Beyond improving biostability and catalytic efficiency, chemical modifications also expand the functional versatility of DNAzymes. When applied to selectively suppress DNAzyme activity, these modifications can be integrated into stimulus-responsive systems that restore function upon exposure to defined external triggers.30,99 Such systems hold great potential for applications in biosensing, gene regulation, and targeted disease diagnostics and therapeutics.Photoresponsive chemical modifications represent a powerful strategy to confer spatiotemporal control over the activity of functional DNAs, especially combined with upconversion nanoparticles. By incorporating light-sensitive moieties—such as azobenzene or photocleavable (PC) linkers100—into critical regions of DNAzymes or aptamers, the biological function of DNA can be reversibly masked and subsequently restored upon light irradiation.27,101 This dynamic modulation enables precise regulation of nucleic acid behavior in complex biological environments.
Recent advances have demonstrated the utility of such modifications across a range of applications. In the field of metal ion sensing, photocaged DNAzymes conjugated to upconversion nanoparticles have enabled NIR light-activated detection of Zn2+ in both living cells and zebrafish embryos, offering high specificity and spatial resolution (Fig. 6a).102 For protein regulation, DNA aptamer-functionalized spherical nucleic acids have been designed to inhibit thrombin activity upon NIR irradiation, facilitating deep-tissue modulation of enzyme functions (Fig. 6c).103 Furthermore, a modular nanoplatform based on photocleavable aptamers allows light-controlled manipulation of protein subcellular localization in live cells, providing a non-genetic strategy for dynamic control of intracellular signaling (Fig. 6b).104 More recently, azobenzene-modified aptamer agonists have been developed to reversibly modulate receptor tyrosine kinase activity and downstream ERK signaling in response to alternating light stimuli, enabling fine-tuned regulation of cell fate decisions with high temporal resolution (Fig. 6d).105 Collectively, these systems demonstrate the broad applicability of photo-responsive nucleic acid designs for programmable, reversible, and minimally invasive regulation in both basic research and biomedical contexts.
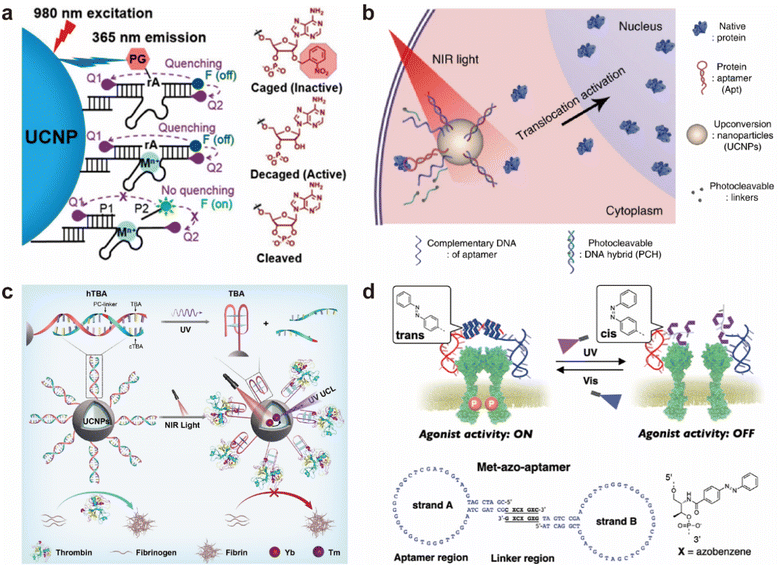 | ||
| Fig. 6 Photo-responsive systems based on modified DNA aptamers and DNAzymes. (a) Photocaged DNAzymes conjugated to lanthanide-doped upconversion nanoparticles (UCNPs), enabling the conversion of deeply penetrating near-infrared (NIR) light at 980 nm into UV emission at 365 nm.102 Reproduced with permission from ref. 102. Copyright 2018 American Chemical Society. (b) Upon NIR excitation, UCNPs emit UV light to activate a photocleavable DNA hybrid (PHC2), which releases the protein from the Apt/protein complex, thereby enabling spatiotemporal control of protein localization in living cells. Reproduced from ref. 104 with permission from, Copyright 2020 Springer Nature. (c) Covalent conjugation of an activatable hybridized thrombin-binding aptamer (hTBA) onto UCNPs allows remote control of thrombin activity.103 Reproduced with permission from ref. 103. Copyright 2022 Wiley-VCH. (d) Reversible optical regulation of receptor tyrosine kinase (RTK) activation using an azobenzene-modified DNA aptamer. The Met-azo-aptamer consists of two complementary strands, A and B, whose sequences are shown below.105 Reproduced with permission from ref. 105. Copyright 2025 American Chemical Society. | ||
In addition to photo stimuli-responsive systems, epigenetic enzyme-responsive platforms have recently gained considerable attention. Fat mass and obesity-associated protein (FTO), the first identified m6A demethylase, has emerged as a key biomarker due to its dysregulation in various cancers and metabolic disorders. Wang et al. pioneered the incorporated of m6A modifications into DNAzymes to construct an epigenetically responsive system. The resulting m6A-caged DNAzyme is specifically activated by FTO, enabling intracellular detection of FTO activity (Fig. 7a).60 To further enhance sensitivity, this system can be coupled with signal amplification strategies such as rolling circle amplification (RCA), CRISPR/Cas12a-based cascade amplification, hybridization chain reaction (HCR), and electrochemical sensing approaches,106–112 achieving detection limits as low as 0.596 fM.113 When combined with engineered nanopipette technology, these modified DNAzymes have also shown promise in single-cell epigenetic profiling (Fig. 7b).114
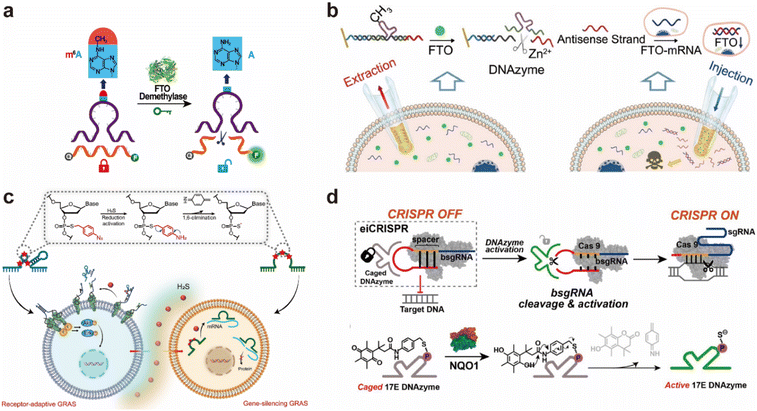 | ||
| Fig. 7 Other responsive systems based on modified DNAzymes. (a) An epigenetically responsive system constructed using an m6A-caged DNAzyme enables ultrasensitive detection of FTO demethylase expression levels.60 Adapted with permission from ref. 60. Copyright 2021 American Chemical Society. (b) A nanopipette sensor engineered with an m6A-bearing DNAzyme allows for profiling of the m6A-modifying enzyme FTO in single living cells.114 Adapted with permission from ref. 114. Copyright 2023 Wiley-VCH. (c) Gas-responsive artificial DNAzyme-based switches (GRAS) that specifically respond to hydrogen sulfide (H2S) signaling molecules.115 Adapted with permission from ref. 115. Copyright 2024 Wiley-VCH. (d) A phenyl quinone (QP)-conjugated 17EQP DNAzyme is selectively activated by quinone oxidoreductase (NQO1), enabling eiCRISPR-mediated genome editing in tumor cells.30 Adapted with permission from ref. 30. Copyright 2022 American Chemical Society. | ||
The concept of enzyme-responsive DNAzymes can be extended by introducing specific chemical groups at key catalytic or binding sites to sense disease-related enzymatic activity. For instance, a methyl-modified “repaired and activated” DNAzyme system has been developed to monitor the repair activity of methyltransferase in the context of drug treatment.28 Another example involves NAD(P)H:quinone oxidoreductase 1 (NQO1), a flavoprotein enzyme overexpressed in many tumor cells, which can orthogonally activate phenylquinone (QP)-conjugated 17EQP DNAzymes (Fig. 7d). This strategy enables tumor-selective genome editing through enzyme-inducible CRISPR (eiCRISPR). Recently, Xie et al. developed gas signaling molecule-responsive artificial DNAzyme-based Switches (GRAS) using chemically modified DNAzymes (Fig. 7c),115 offering a versatile platform for regulating diverse cellular processes across a range of physiological contexts.
Collectively, both light-responsive and enzyme-responsive DNAzyme systems exemplify the transformative potential of chemical modifications in creating spatiotemporally controllable functional nucleic acids. By strategically introducing photolabile or enzymatically cleavable groups, researchers have developed DNAzymes that can be precisely activated by external stimuli such as light or disease-associated enzymes, enabling conditional control over catalytic function. These smart systems not only facilitate fundamental studies of enzyme dynamics and epigenetic regulation at the single-cell level, but also offer promising platforms for targeted diagnostics, precision therapeutics, and real-time cellular modulation. The convergence of chemical design, signal amplification strategies, and biological responsiveness underscores the growing versatility of modified DNAzymes and sets the stage for their broader application in complex biomedical environments.
5 Challenges and future directions
Current research on chemically modified DNA aptamers and DNAzymes primarily advances along two major trajectories: (1) the post-selection modification of these functional DNAs to enhance their stability, binding affinity, or catalytic activity, and (2) the de novo selection of functional sequences from DNA libraries incorporating chemically modified nucleotides.90,91 While these strategies have yielded significant functional improvements (Table 1), the field continues to face substantial challenges that limit both systematic development and broader translational application.| DNAzyme/DNA aptamer | Type of modification | Property improved | Quantitative improvement (fold change) | Ref. |
|---|---|---|---|---|
| HIV-RT 3.17 | TNA | Stability | >1400-fold increase in t1/2 (from 1–3 min to 72 h) | 24 |
| Sgc8 | Glycosylation | Stability | ∼14-fold increase in t1/2 | 26 |
| Aptamer BC15-31 | Phosphorothioate | Affinity; stability | ∼2-fold (Kd reduced from 1.49 to 0.73 µM; N/A | 70 |
| HD1 | N3-modified residues | Affinity | ∼2-fold (reduced KD from 20.2 ± 1.3 nM to 9.8 ± 0.6 nM) | 61 |
| TBA | Amino acid-nucleic acid hybrids | Affinity | 2.5-Fold (Kd reduced from 7.28 nM to 2.94 nM) | 72 |
| HSc-9 | 7-Phenylbutyl-7-deazaadenine | Affinity | N/A (Kd = 14.8 ± 0.3 nM) | 66 |
| SIgA binding aptamer | Base-appended bases | Affinity | N/A (Kd = 3.7 nM) | 64 |
| SARS-CoV-2 aptamer | Base-appended bases | Affinity | N/A (Kd = 1.2 nM) | 75 |
| DEN-NS1 aptamer | Unnatural bases (UBs) | Affinity | N/A (Kd = 27–182 pM) | 68 |
| Protein glycan-binding aptamer | Indole-modified bases | Affinity | N/A (Kd = 6–30 µM) | 76 |
| 8–17 DNAzyme | Nucleoside analogues | Catalytic activity | ∼1.6–3.6-fold increase | 85 and 88 |
| 10–23 DNAzyme | Nucleoside analogues | Catalytic activity | ∼2.2–4.2-fold increase | 86 and 87 |
| 10–23 DNAzyme | FANA, XNA | Catalytic activity | ∼50-fold increase (multiple-turnover) | 81 |
| 10–23 DNAzyme | LNA, OMe, MOE, phosphorothioate | Catalytic activity | N/A (kobs of ∼0.65 min−1) | 32 |
| 10–23 DNAzyme | Carboxyl and benzyl groups | Catalytic activity | ∼700-fold increase (multiple-turnover) | 89 |
One major constraint lies in the limited commercial availability and high cost of chemically modified nucleotides. Many non-canonical monomers, especially those with unconventional backbones or hydrophobic side chains, remain economically inaccessible or synthetically challenging. As a result, most studies rely on a narrow set of commercially available modifications, and only a few research groups with synthetic chemistry expertise are capable of exploring novel chemical spaces.26,72,86,89 The development of cost-efficient, modular, and scalable synthesis routes, along with expanded commercial offerings, is essential to democratize access and facilitate more comprehensive exploration of chemically modified nucleic acids.
Another critical bottleneck lies in the technical limitations of de novo selection from chemically diverse libraries. In addition to expanding the range of chemical modifications, there is an urgent need to enhance polymerase compatibility with unnatural or non-native substrates, keeping pace with the increasing chemical diversity of modified nucleotides.116 While some modifications, such as FANA,117 do not significantly impact polymerase amplification, most modified nucleotides, especially those inducing steric or electronic perturbations, reduce amplification efficiency. This poor substrate compatibility diminishes amplification efficiency and limits library diversity.118 Furthermore, standard next-generation sequencing platforms are frequently incompatible with heavily modified strands, complicating accurate sequence recovery and downstream analysis. Advances in polymerase engineering to improve substrate promiscuity,119 coupled with the development of modification-tolerant sequencing technologies, are essential for expanding the structural and chemical diversity that can be harnessed through in vitro evolution.
A third key challenge is the lack of high-resolution structural information on DNA aptamers and DNAzymes, with only a limited number of three-dimensional structures reported to date.92,93,120 In the absence of atomic-level insights into DNA folding architectures, binding interfaces, and catalytic cores, chemical modifications are often introduced through empirical screening89 rather than structure-guided design.61 The integration of structural techniques such as X-ray crystallography, NMR spectroscopy, and cryo-electron microscopy into functional nucleic acid research will be crucial for elucidating structure–function relationships. Recently, Tan's group utilized NMR spectroscopy to analyze the binding sites of the aptamer sgc8c, providing valuable guidance for the introduction of chemical modifications. This study achieved simultaneous improvements in both binding affinity and stability, underscoring the potential of structure-guided modification strategies.79
Additionally, computational studies predicting the three-dimensional structures of functional DNAs, along with molecular docking simulations to explore interactions with target molecules, will provide valuable insights for the integration of unnatural nucleotides and the selection of modification sites.121,122 Such approaches will enable precise site-directed chemical modifications, fine-tuning biomolecular performance and facilitating predictive control for future applications in functional DNA design.
As the field continues to evolve, the growing diversity of chemically modified functional DNAs highlights the urgent need for standardized nomenclature, annotation frameworks, and centralized databases. A curated resource documenting modification types, sequence contexts, structural motifs, and experimentally validated activities would facilitate comparative analysis, promote reproducibility, and accelerate the design of next-generation sequences using data-driven or machine learning–assisted approaches.
Most current studies focus on a small number of well-characterized aptamers and DNAzymes, such as the thrombin-binding aptamer and the 8–17 DNAzyme, mainly serving as proof-of-concept models. With the recent emergence of high-affinity and biostable aptamers generated through advanced SELEX technologies and machine learning-assisted discovery,123–127 new opportunities are arising for functional enhancement. Incorporating chemical modifications into these newly identified aptamers is expected to bridge the gap between conceptual studies and real-world applications in diagnostics, therapeutics, and biosensing.
Despite notable progress in enhancing stability and functionality through chemical modification, two persistent challenges continue to limit the practical translation of functional DNAs.
First, the performance gap between DNA aptamers and antibodies in complex biological matrices remains a major obstacle. DNA aptamers selected under idealized SELEX conditions often exhibit reduced binding specificity and affinity in physiological environments due to off–target interactions, glycan shielding, local ionic strength variations, and nuclease degradation. To bridge this gap, future research should prioritize physiologically relevant SELEX protocols that better mimic native biochemical conditions,128–130 combined with stabilization strategies such as 2′-O-methyl, phosphorothioate, or L-DNA modifications.52 Moreover, the integration of machine learning-assisted prediction and in silico affinity evaluation can accelerate the identification of aptamers with robust performance in complex biological systems.
Second, DNAzymes frequently display diminished catalytic turnover under intracellular or in vivo conditions, complicating the distinction between genuine catalysis and antisense-like hybridization effects. To clarify catalytic mechanisms, rigorous validation strategies are essential, including multiple-turnover assays under substrate-excess conditions, single-turnover kinetics to identify rate-limiting steps, and catalytic inhibition (poisoning) tests that confirm metal dependence and active-site integrity. Introducing inhibitory point mutations or chemical modifications at catalytic residues can further serve as negative controls. Meanwhile, site-specific 2′-O-methyl or phosphorothioate substitution patterns can suppress RNase H activation while preserving catalytic activity. The development of intracellular fluorescent reporters and real-time imaging platforms will facilitate direct observation of cleavage reactions, advancing the mechanistic understanding and optimization of DNAzyme catalysis in biological environments.
Looking forward, the convergence of synthetic nucleotide chemistry, polymerase engineering, structural biology, and high-throughput screening technologies is poised to reshape the development of functional DNAs. As interdisciplinary barriers continue to be broken, chemically modified aptamers and DNAzymes are expected to transition from experimental tools to programmable molecular platforms for precision diagnostics, targeted therapeutics, and other biology applications.
6 Conclusions
Chemically modified DNA aptamers and DNAzymes have opened new frontiers in the design of responsive, biostable, and highly functional nucleic acid-based tools. By enabling precise control over molecular interactions and resistance to biological degradation, chemical modifications expand the functional repertoire of DNA beyond its natural roles. As advances in synthetic chemistry, polymerase engineering, structural biology, and computational tools converge, the rational design and high-throughput selection of modified aptamers and DNAzymes will become increasingly feasible. Together, these developments hold promise for the creation of programmable nucleic acid systems with transformative applications in biosensing, gene regulation, nanotechnology, and biocatalysis.Author contributions
C. Z. wrote the original draft of the manuscript. K. M. and Z. Z. helped collect literature and drew the diagrams. Y. C., H. G., and W. L. contributed to the conception and revision of the manuscript.Conflicts of interest
The authors declare no conflict of interest.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Acknowledgements
This work was supported by the Key R&D Program of Shandong Province (2024CXGC010914), the National Natural Science Foundation of China (22408113, 22578144), the Fundamental Research Funds for the Central Universities (2024ZYGXZR078), and Science and Technology Projects in Guangzhou (2025A04J3891).References
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS PubMed.
- C. Tuerk and L. Gold, Science, 1990, 249, 505–510 CrossRef CAS PubMed.
- X. Chang, C. Zhang, C. Lv, Y. Sun, M. Zhang, Y. Zhao, L. Yang, D. Han and W. Tan, J. Am. Chem. Soc., 2019, 141, 12738–12743 CrossRef CAS PubMed.
- C. Shi, D. Yang, X. Ma, L. Pan, Y. Shao, G. Arya, Y. Ke, C. Zhang, F. Wang, X. Zuo, M. Li and P. Wang, Angew. Chem., Int. Ed., 2024, 63, e202320179 CrossRef CAS PubMed.
- H. Gu, K. Furukawa, Z. Weinberg, D. F. Berenson and R. R. Breaker, J. Am. Chem. Soc., 2013, 135, 9121–9129 CrossRef CAS PubMed.
- C. Zhang, Q. Li, T. Xu, W. Li, Y. He and H. Gu, Nucleic Acids Res., 2021, 49, 6364–6374 CrossRef CAS PubMed.
- M. M. Kennebeck, C. K. Kaminsky, M. A. Massa, P. K. Das, R. D. Boyd, M. Bishka, J. T. Tricarico and S. K. Silverman, Angew. Chem., Int. Ed., 2024, 63, e202317565 CrossRef CAS PubMed.
- M. Zandieh, X. Luo, Y. Zhao, C. Feng and J. Liu, Angew. Chem., Int. Ed., 2024, 64, e202421438 CrossRef PubMed.
- R. D. Boyd, M. M. Kennebeck, A. A. Miranda, Z. Liu and S. K. Silverman, Nucleic Acids Res., 2024, 52, 8702–8716 CrossRef CAS PubMed.
- H. Mann, S. Khan, A. Prasad, F. Bayat, J. Gu, K. Jackson, Y. Li, Z. Hosseinidoust, T. F. Didar and C. D. M. Filipe, Adv. Mater., 2025, 37, e2411173 Search PubMed.
- Q. Hu, Z. Tong, A. Yalikong, L. P. Ge, Q. Shi, X. Du, P. Wang, X. Y. Liu, W. Zhan, X. Gao, D. Sun, T. Fu, D. Ye, C. Fan, J. Liu, Y. S. Zhong, Y. Z. Jiang and H. Gu, Nat. Chem., 2024, 16, 122–131 Search PubMed.
- J. Yan, R. Bhadane, M. Ran, X. Ma, Y. Li, D. Zheng, O. M. H. Salo-Ahen and H. Zhang, Nat. Commun., 2024, 15, 3684 CrossRef CAS PubMed.
- Z. Zhou, Y. Liu, Y. Wang, H. Jiang, T. Chen, Y. Zhu, T. Fu and J. Li, Nat. Commun., 2025, 16, 3919 CrossRef CAS PubMed.
- V. Behnam, A. M. McManamen, H. G. Ballard, B. Aldana, M. Tamimi, N. Milosavic, M. N. Stojanovic, M. R. Rubin and S. K. Sia, Angew. Chem., Int. Ed., 2025, 64, e202414871 Search PubMed.
- M. Lyu, L. Kong, X. Shao and Y. Lu, Angew. Chem., Int. Ed., 2025, e202509127, DOI:10.1002/anie.202509127.
- R. A. Scheele, Y. Weber, F. E. H. Nintzel, M. Herger, T. S. Kaminski and F. Hollfelder, ACS Catal., 2024, 14, 6259–6271 Search PubMed.
- J. Wang, S. Yu, Q. Wu, X. Gong, S. He, J. Shang, X. Liu and F. Wang, Angew. Chem., Int. Ed., 2021, 60, 10766–10774 CrossRef CAS.
- H. Fan, Z. Zhao, G. Yan, X. Zhang, C. Yang, H. Meng, Z. Chen, H. Liu and W. Tan, Angew. Chem., Int. Ed., 2015, 54, 4801–4805 CrossRef CAS.
- J. Chen, M. Chen and T. F. Zhu, Nat. Biotechnol., 2022, 40, 1601–1609 CrossRef CAS.
- C. Riccardi, A. Meyer, J.-J. Vasseur, I. R. Krauss, L. Paduano, F. Morvan and D. Montesarchio, Bioorg. Chem., 2020, 94, 103379 CrossRef CAS PubMed.
- H. Kuai, Z. Zhao, L. Mo, H. Liu, X. Hu, T. Fu, X. Zhang and W. Tan, J. Am. Chem. Soc., 2017, 139, 9128–9131 Search PubMed.
- L. Yao, T. Liu, L. Sun, S. Gao, S. Liu, H. Qu, Y. Mao and L. Zheng, J. Agric. Food Chem., 2025, 73, 3222–3231 CrossRef CAS PubMed.
- Y. Yan, D. Chang, Y. Xu, Y. Chang, Q. Zhang, Q. Yuan, B. J. Salena, Y. Li and M. Liu, J. Am. Chem. Soc., 2023, 145, 2630–2637 CrossRef CAS PubMed.
- M. R. Dunn, C. M. McCloskey, P. Buckley, K. Rhea and J. C. Chaput, J. Am. Chem. Soc., 2020, 142, 7721–7724 CrossRef CAS.
- M. Kovacic, P. Podbevsek, H. Tateishi-Karimata, S. Takahashi, N. Sugimoto and J. Plavec, Nucleic Acids Res., 2020, 48, 3975–3986 CrossRef CAS.
- Y. Han, R. Zhang, H.-L. Bao, M. Yang, Y. Gao, X. Gao, R. Wang, W. Tan and D.-K. Ji, Adv. Sci., 2025, 12, 2408168 Search PubMed.
- Z. Wu, H. Fan, N. S. R. Satyavolu, W. Wang, R. Lake, J. H. Jiang and Y. Lu, Angew. Chem., Int. Ed., 2017, 56, 8721–8725 CrossRef CAS PubMed.
- X. Wang, X. Yi, Z. Huang, J. He, Z. Wu, X. Chu and J.-H. Jiang, Angew. Chem., Int. Ed., 2021, 60, 19889–19896 Search PubMed.
- X. Wang, S. Yu, Y. He, F. Wang and X. Liu, Chin. J. Chem., 2024, 42, 951–956 CrossRef CAS.
- W. Cai, J. Liu, X. Chen, L. Mao and M. Wang, J. Am. Chem. Soc., 2022, 144, 22272–22280 CrossRef CAS.
- H. Shin, J.-Y. Cho, B. Y. Park and C. Jung, Chem. Eng. J., 2024, 493, 152548 CrossRef CAS.
- K. Nguyen, T. N. Malik and J. C. Chaput, Nat. Commun., 2023, 14, 2413 CrossRef CAS.
- R. Wang, W. He, X. Yi, Z. Wu, X. Chu and J. H. Jiang, J. Am. Chem. Soc., 2023, 145, 17926–17935 CrossRef CAS.
- D. Ji, H. Feng, S. W. Liew and C. K. Kwok, Trends Biotechnol., 2023, 41, 1360–1384 CrossRef CAS.
- R. Oliveira, E. Pinho, A. L. Sousa, J. J. DeStefano, N. F. Azevedo and C. Almeida, Improving aptamer performance with nucleic acid mimics: de novo and post-SELEX approaches, Trends Biotechnol., 2022, 40, 549–563 CrossRef PubMed.
- Q. Wang, Z. Wang, Y. He, B. Xiong, Y. Li and F. Wang, TrAC, Trends Anal. Chem., 2023, 159, 116910 CrossRef CAS.
- J. P. Elskens, J. M. Elskens and A. Madder, Int. J. Mol. Sci., 2020, 21, 4522 CrossRef CAS.
- G. Maio, O. Enweronye, H. E. Zumrut, S. Batool, N. Van and P. Mallikaratchy, ChemistrySelect, 2017, 2, 2335–2340 CrossRef CAS PubMed.
- D. Thirunavukarasu, T. Chen, Z. Liu, N. Hongdilokkul and F. E. Romesberg, J. Am. Chem. Soc., 2017, 139, 2892–2895 CrossRef CAS PubMed.
- R. El-Khoury and M. J. Damha, Acc. Chem. Res., 2021, 54, 2287–2297 CrossRef CAS PubMed.
- L. Zhang and J. C. Chaput, Molecules, 2020, 25, 4194 CrossRef CAS.
- M. De Fenza, E. Eremeeva, R. Troisi, H. Yang, A. Esposito, F. Sica, P. Herdewijn, D. D'Alonzo and A. Guaragna, Chemistry, 2020, 26, 9589–9597 CrossRef CAS PubMed.
- G. Ying, X. Lu, J. Mei, Y. Zhang, J. Chen, X. Wang, Z. Ou and Y. Yi, Bioorg. Med. Chem., 2019, 27, 3201–3207 CrossRef CAS.
- W. Kotkowiak, J. Wengel, C. J. Scotton and A. Pasternak, J. Med. Chem., 2019, 62, 2499–2507 CrossRef CAS.
- H. Shi, X. He, W. Cui, K. Wang, K. Deng, D. Li and F. Xu, Anal. Chim. Acta, 2014, 812, 138–144 CrossRef CAS.
- E. Eremeeva, A. Fikatas, L. Margamuljana, M. Abramov, D. Schols, E. Groaz and P. Herdewijn, Nucleic Acids Res., 2019, 47, 4927–4939 Search PubMed.
- K. Nauwelaerts, E. Lescrinier, G. Sclep and P. Herdewijn, Nucleic Acids Res., 2005, 33, 2452–2463 Search PubMed.
- I. Alves Ferreira-Bravo, C. Cozens, P. Holliger and J. J. DeStefano, Nucleic Acids Res., 2015, 43, 9587–9599 Search PubMed.
- M. C. Culbertson, K. W. Temburnikar, S. P. Sau, J. Y. Liao, S. Bala and J. C. Chaput, Bioorg. Med. Chem. Lett., 2016, 26, 2418–2421 CrossRef CAS.
- P. Schofield, A. I. Taylor, J. Rihon, C. D. Pena Martinez, S. Zinn, C. A. Mattelaer, J. Jackson, G. Dhaliwal, G. Schepers, P. Herdewijn, E. Lescrinier, D. Christ and P. Holliger, Nucleic Acids Res., 2023, 51, 7736–7748 CrossRef CAS.
- X. Li, Z. Zhang, F. Gao, Y. Ma, D. Wei, Z. Lu, S. Chen, M. Wang, Y. Wang, K. Xu, R. Wang, F. Xu, J. Y. Chen, C. Zhu, Z. Li, H. Yu and X. Guan, J. Am. Chem. Soc., 2023, 145, 9334–9342 CrossRef CAS PubMed.
- P. Amero, G. L. R. Lokesh, R. R. Chaudhari, R. Cardenas-Zuniga, T. Schubert, Y. M. Attia, E. Montalvo-Gonzalez, A. M. Elsayed, C. Ivan, Z. Wang, V. Cristini, V. Franciscis, S. Zhang, D. E. Volk, R. Mitra, C. Rodriguez-Aguayo, A. K. Sood and G. Lopez-Berestein, J. Am. Chem. Soc., 2021, 143, 7655–7670 CrossRef CAS.
- K. Yamada, V. N. Hariharan, J. Caiazzi, R. Miller, C. M. Ferguson, E. Sapp, H. H. Fakih, Q. Tang, N. Yamada, R. C. Furgal, J. D. Paquette, A. Biscans, B. M. Bramato, N. McHugh, A. Summers, C. Lochmann, B. Godinho, S. Hildebrand, S. O. Jackson, D. Echeverria, M. R. Hassler, J. F. Alterman, M. DiFiglia, N. Aronin and A. Khvorova, Nat. Biotechnol., 2025, 43, 904–913 CrossRef CAS.
- P. Kumar and M. H. Caruthers, Acc. Chem. Res., 2020, 53, 2152–2166 CrossRef CAS PubMed.
- S. Roy, S. Paul, M. Roy, R. Kundu, L. Monfregola and M. H. Caruthers, J. Org. Chem., 2017, 82, 1420–1427 CrossRef CAS.
- M. Hyjek-Skladanowska, T. A. Vickers, A. Napiorkowska, B. A. Anderson, M. Tanowitz, S. T. Crooke, X. H. Liang, P. P. Seth and M. Nowotny, J. Am. Chem. Soc., 2020, 142, 7456–7468 CrossRef CAS.
- R. Saran, Z. Huang and J. Liu, Coord. Chem. Rev., 2021, 428, 213624 Search PubMed.
- P. J. Huang and J. Liu, Nucleic Acids Res., 2015, 43, 6125–6133 CrossRef CAS.
- M. S. Islam, S. Aktar, N. Moetamedirad, N. Xie, C. T. Lu, V. Gopalan, A. K. Lam and M. J. A. Shiddiky, Biosens. Bioelectron., 2025, 267, 116813 CrossRef CAS.
- Q. Wang, K. Tan, H. Wang, J. Shang, Y. Wan, X. Liu, X. Weng and F. Wang, J. Am. Chem. Soc., 2021, 143, 6895–6904 CrossRef CAS.
- I. Smirnov, N. Kolganova, R. Troisi, F. Sica and E. Timofeev, Mol. Ther.--Nucleic Acids, 2021, 23, 863–871 CrossRef CAS PubMed.
- Z. Chen, P. A. Lichtor, A. P. Berliner, J. C. Chen and D. R. Liu, Nat. Chem., 2018, 10, 420–427 Search PubMed.
- H. Minagawa, K. Onodera, H. Fujita, T. Sakamoto, J. Akitomi, N. Kaneko, I. Shiratori, M. Kuwahara, K. Horii and I. Waga, Sci. Rep., 2017, 7, 42716 Search PubMed.
- H. Minagawa, A. Shimizu, Y. Kataoka, M. Kuwahara, S. Kato, K. Horii, I. Shiratori and I. Waga, Anal. Chem., 2020, 92, 1780–1787 CrossRef CAS.
- M. Kimoto, R. Yamashige, K. Matsunaga, S. Yokoyama and I. Hirao, Nat. Biotechnol., 2013, 31, 453–457 CrossRef CAS.
- C. Mulholland, I. Jestrabova, A. Sett, M. Ondrus, V. Sykorova, C. L. Manzanares, O. Simoncik, P. Muller and M. Hocek, Commun. Chem., 2023, 6, 65 CrossRef CAS.
- K. I. Matsunaga, M. Kimoto and I. Hirao, J. Am. Chem. Soc., 2017, 139, 324–334 CrossRef CAS.
- K. I. Matsunaga, M. Kimoto, V. W. Lim, H. P. Tan, Y. Q. Wong, W. Sun, S. Vasoo, Y. S. Leo and I. Hirao, Nucleic Acids Res., 2021, 49, 11407–11424 CrossRef CAS.
- Z. Lai, D. Jin, Y. Tian, X. Chen, D. Han, H. Chen, J. Wang and Y. Yang, Adv. Sci., 2024, 11, e2410642 Search PubMed.
- J. Wu, S. Wang, X. Li, Q. Zhang, J. Yang, Y. Ma, Z. Guan and Z. Yang, Front. Cell Dev. Biol., 2021, 9, 660233 Search PubMed.
- Z. Fu and J. Xiang, Int. J. Mol. Sci., 2020, 21, 2793 CrossRef CAS.
- J. H. Yum, T. Ishizuka, K. Fukumoto, D. Hori, H. L. Bao, Y. Xu, H. Sugiyama and S. Park, ACS Biomater. Sci. Eng., 2021, 7, 1338–1343 CrossRef CAS.
- D. R. Davies, A. D. Gelinas, C. Zhang, J. C. Rohloff, J. D. Carter, D. O'Connell, S. M. Waugh, S. K. Wolk, W. S. Mayfield, A. B. Burgin, T. E. Edwards, L. J. Stewart, L. Gold, N. Janjic and T. C. Jarvis, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 19971–19976 CrossRef CAS PubMed.
- C. M. McCloskey, Q. Li, E. J. Yik, N. Chim, A. K. Ngor, E. Medina, I. Grubisic, L. Co Ting Keh, R. Poplin and J. C. Chaput, ACS Synth. Biol., 2021, 10, 3190–3199 Search PubMed.
- H. Minagawa, H. Sawa, T. Fujita, S. Kato, A. Inaguma, M. Hirose, Y. Orba, M. Sasaki, K. Tabata, N. Nomura, M. Shingai, Y. Suzuki and K. Horii, Biochem. Biophys. Res. Commun., 2022, 614, 207–212 Search PubMed.
- A. M. Yoshikawa, A. Rangel, T. Feagin, E. M. Chun, L. G. Wan, A. P. Li, L. Moekl, D. A. Wu, M. Eisenstein, S. Pitteri and H. T. Soh, Nat. Commun., 2021, 12, 7106 CrossRef CAS PubMed.
- M. T. Murray and S. D. Wetmore, Nucleic Acids Res., 2024, 52, 10823–10835 CrossRef CAS PubMed.
- Z. Chen, H. Luo, A. Gubu, S. Yu, H. Zhang, H. Dai, Y. Zhang, B. Zhang, Y. Ma, A. Lu and G. Zhang, Front. Cell Dev. Biol., 2023, 11, 1091809 CrossRef PubMed.
- A. He, L. Wan, Y. Zhang, Z. Yan, P. Guo, D. Han and W. Tan, Proc. Natl. Acad. Sci. U. S. A., 2024, 121, e2404060121 CrossRef CAS.
- S. W. Santoro and G. F. Joyce, A general purpose RNA-cleaving DNA enzyme, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 4262–4266 CrossRef CAS.
- Y. Wang, K. Nguyen, R. C. Spitale and J. C. Chaput, Nat. Chem., 2021, 13, 319–326 CrossRef CAS.
- Y. Wang, Y. Wang, D. Song, X. Sun, Z. Li, J. Y. Chen and H. Yu, Nat. Chem., 2022, 14, 350–359 Search PubMed.
- A. I. Taylor, C. J. K. Wan, M. J. Donde, S. Y. Peak-Chew and P. Holliger, Nat. Chem., 2022, 14, 1295–1305 CrossRef CAS.
- B. Vester, L. B. Lundberg, M. D. Sorensen, B. R. Babu, S. Douthwaite and J. Wengel, J. Am. Chem. Soc., 2002, 124, 13682–13683 Search PubMed.
- S. Du, Y. Li, Z. Chai, W. Shi and J. He, Bioorg. Chem., 2020, 94, 103401 CrossRef CAS PubMed.
- S. Du, Y. Li, Z. Chai, W. Shi and J. He, RSC Adv., 2020, 10, 19067–19075 RSC.
- S. Du, Y. Li and J. He, Bioorg. Med. Chem. Lett., 2020, 30, 126961 Search PubMed.
- W. Zhang, Y. Li, S. Du, Z. Chai and J. He, Bioorg. Med. Chem. Lett., 2021, 48, 128234 CrossRef CAS PubMed.
- Z. Zhang, W. Wei, S. Chen, J. Yang, D. Song, Y. Chen, Z. Zhao, J. Chen, F. Wang, J. Wang, Z. Li, Y. Liang and H. Yu, J. Am. Chem. Soc., 2024, 146, 7052–7062 CrossRef CAS PubMed.
- J. Wang, F. Han, Y. Zou, M. Wang, Y. Wang, J. Y. Chen and H. Yu, J. Am. Chem. Soc., 2025, 147, 18349–18358 CrossRef CAS.
- Y. Wang, A. K. Ngor, A. Nikoomanzar and J. C. Chaput, Nat. Commun., 2018, 9, 5067 CrossRef.
- H. Liu, X. Yu, Y. Chen, J. Zhang, B. Wu, L. Zheng, P. Haruehanroengra, R. Wang, S. Li, J. Lin, J. Li, J. Sheng, Z. Huang, J. Ma and J. Gan, Nat. Commun., 2017, 8, 2006 CrossRef.
- J. Borggrafe, J. Victor, H. Rosenbach, A. Viegas, C. G. W. Gertzen, C. Wuebben, H. Kovacs, M. Gopalswamy, D. Riesner, G. Steger, O. Schiemann, H. Gohlke, I. Span and M. Etzkorn, Nature, 2022, 601, 144–149 CrossRef PubMed.
- D. M. Perrin, T. Garestier and C. Helene, J. Am. Chem. Soc., 2001, 123, 1556–1563 CrossRef CAS.
- L. Lermer, Y. Roupioz, R. Ting and D. M. Perrin, J. Am. Chem. Soc., 2002, 124, 9960–9961 CrossRef CAS PubMed.
- M. Hollenstein, C. J. Hipolito, C. H. Lam and D. M. Perrin, ACS Comb. Sci., 2013, 15, 174–182 Search PubMed.
- Y. Wang, E. Liu, C. H. Lam and D. M. Perrin, Chem. Sci., 2018, 9, 1813–1821 RSC.
- M. Hollenstein, C. J. Hipolito, C. H. Lam and D. M. Perrin, Nucleic Acids Res., 2009, 37, 1638–1649 CrossRef CAS.
- Y. Liu, Y. Shi, L. Yu, Z. Wu and J. H. Jiang, Anal. Chem., 2023, 95, 6490–6495 CrossRef CAS PubMed.
- Y. Jia, B. Han, G. Zhou, Z. Li, M. Xue, B. Liang, P. Liu and Y. Cheng, Anal. Chim. Acta, 2025, 1346, 343786 CrossRef CAS PubMed.
- J. Zhan, Z. Liu, R. Liu, J. J. Zhu and J. Zhang, Anal. Chem., 2022, 94, 16622–16631 Search PubMed.
- Z. Yang, K. Y. Loh, Y. T. Chu, R. Feng, N. S. R. Satyavolu, M. Xiong, S. M. Nakamata Huynh, K. Hwang, L. Li, H. Xing, X. Zhang, Y. R. Chemla, M. Gruebele and Y. Lu, J. Am. Chem. Soc., 2018, 140, 17656–17665 CrossRef CAS.
- J. Zhang, P. Zhao, W. Li, L. Ye, L. Li, Z. Li and M. Li, Angew. Chem., Int. Ed., 2022, 61, e202117562 CrossRef CAS PubMed.
- S. Xie, Y. Du, Y. Zhang, Z. Wang, D. Zhang, L. He, L. Qiu, J. Jiang and W. Tan, Nat. Commun., 2020, 11, 1347 Search PubMed.
- M. Wakano, M. Tsunoda, K. Murayama, J. Morimoto, R. Ueki, S. Aoyama-Ishiwatari, Y. Hirabayashi, H. Asanuma and S. Sando, J. Am. Chem. Soc., 2025, 147, 11477–11484 CrossRef CAS.
- H. Gao, X. Song, Q. Chen, R. Yuan and Y. Xiang, Anal. Chim. Acta, 2023, 1247, 340902 CrossRef CAS PubMed.
- T. Shen, X. Huang, Y. Zhang, Y. Tong, Y. Shi, J. Guo, X. Zou, Y. Xu and Z. Dai, Anal. Chem., 2023, 96, 1686–1692 CrossRef.
- L. Shi, X. Ma, H. Xie, Y. Qin, Y. Huang, Y. Zhang, L. Sun, J. Yang and G. Li, Biosens. Bioelectron., 2023, 222, 115007 Search PubMed.
- N.-n. Zhao, Q. Wang, H. Liu, M. Liu, Q. Xu, H. Yuan and C. y. Zhang, Chem. Eng. J., 2024, 493, 152548 Search PubMed.
- L.-P. Fan, X.-M. Wang, J. Li, T. Shao, Y.-X. Wang and D.-M. Kong, Sens. Actuators, B, 2025, 431, 137431 CrossRef CAS.
- X. Yang, S. Qiao, W. Zhao, S. Li, Y. Qiao, Y. Jiang, Y. Zhou and Y. Li, Anal. Chem., 2023, 95, 11420–11428 CrossRef CAS PubMed.
- Q. Chen, J. Cao, Y. Zhao, R. Yuan and Y. Xiang, Sens. Actuators, B, 2024, 412, 135799 CrossRef CAS.
- N. Zhao, Y. Liu, L. Zhang, W. Liu, X. Zou, Q. Xu and C. Zhang, Anal. Chem., 2023, 95, 12974–12981 CrossRef CAS.
- X. Shi, Y. Xu, B. Wang, Z. Li, S. Yu, H. Dong, W. Zhao, D. Jiang, H. Chen and J. Xu, Angew. Chem., Int. Ed., 2023, 62, e202302930 CrossRef CAS.
- X. Xie, H. Nan, J. Peng, K. Zeng, H. H. Wang, Y. Huang and Z. Nie, Angew. Chem., Int. Ed., 2024, 63, e202410380 CrossRef CAS.
- A. Hottin and A. Marx, Acc. Chem. Res., 2016, 49, 418–427 CrossRef CAS.
- Y. Liu, J. Wang, Y. Wu and Y. Wang, Chem. Sci., 2024, 15, 12534–12542 RSC.
- S. A. Lapa, A. V. Chudinov and E. N. Timofeev, Mol. Biotechnol., 2016, 58, 79–92 Search PubMed.
- H. Hoshino, Y. Kasahara, M. Kuwahara and S. Obika, J. Am. Chem. Soc., 2020, 142, 21530–21537 CrossRef CAS.
- H. Liu, Y. Gao, J. Mathivanan, Z. Armour-Garb, Z. Shao, Y. Zhang, X. Zhao, Q. Shao, W. Zhang, J. Yang, C. Cao, H. Li, J. Sheng and J. Gan, Nucleic Acids Res., 2023, 51, 4625–4636 CrossRef CAS.
- N. A. Ahmad, R. Mohamed Zulkifli, H. Hussin and M. H. Nadri, J. Mol. Graphics, 2021, 105, 107872 CrossRef CAS PubMed.
- R. Oliveira, E. Pinho, A. L. Sousa, O. Dias, N. F. Azevedo and C. Almeida, PLoS One, 2022, 17, e0264701 CrossRef CAS PubMed.
- A. Bashir, Q. Yang, J. Wang, S. Hoyer, W. Chou, C. McLean, G. Davis, Q. Gong, Z. Armstrong, J. Jang, H. Kang, A. Pawlosky, A. Scott, G. E. Dahl, M. Berndl, M. Dimon and B. S. Ferguson, Nat. Commun., 2021, 12, 2366 CrossRef CAS.
- K. Yang, N. M. Mitchell, S. Banerjee, Z. Cheng, S. Taylor, A. M. Kostic, I. Wong, S. Sajjath, Y. Zhang, J. Stevens, S. Mohan, D. W. Landry, T. S. Worgall, A. M. Andrews and M. N. Stojanovic, Science, 2023, 380, 942–948 CrossRef CAS.
- N. K. Singh, Y. Wang, C. Wen, B. Davis, X. Wang, K. Lee and Y. Wang, Nat. Biotechnol., 2024, 42, 1224–1231 CrossRef CAS PubMed.
- L. Wang, O. Alkhamis, J. Canoura, H. Yu and Y. Xiao, J. Am. Chem. Soc., 2024, 146, 21296–21307 CrossRef PubMed.
- X. Yang, C. H. Chan, S. Yao, H. Y. Chu, M. Lyu, Z. Chen, H. Xiao, Y. Ma, S. Yu, F. Li, J. Liu, L. Wang, Z. Zhang, B. T. Zhang, L. Zhang, A. Lu, Y. Wang, G. Zhang and Y. Yu, Mol. Ther.--Nucleic Acids, 2025, 36, 102436 CrossRef CAS PubMed.
- M. Chen, P. Chang, Z. Zhang, D. Liu, R. Hou, M. Shi, J. Li and K. Xu, Adv. Sci., 2025, 12, e2506746 CrossRef.
- L. Zhang, L. Zhou, H. Zhang, Y. Zhang, L. Li, T. Xie, Y. Chen, X. Li, N. Ling, J. Dai, X. Sun, J. Liu, J. Zhao, T. Peng and M. Ye, ACS Appl. Mater. Interfaces, 2021, 13, 54656–54664 CrossRef CAS PubMed.
- P. Yadav, T. A. Asa, M. Cebeci, M. Choi and Y. J. Seo, J. Med. Chem., 2025, 68, 16227–16236 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2026 |