Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Peptide ligand isomerism drives divergent stability and guest binding in Pd3L4 metal-peptidic cages
Ben E.
Barber†
ab,
Ellen M. G.
Jamieson†
ab,
Leah E. M.
White†
ab and
Charlie T.
McTernan
 *ab
*ab
aArtificial Molecular Machinery Laboratory, The Francis Crick Institute, 1 Midland Road, London NW1 1AT, UK. E-mail: charlie.mcternan@crick.ac.uk
bDepartment of Chemistry, King's College London, Britannia House, 7 Trinity Street, London SE1 1DB, UK. E-mail: charlie.mcternan@kcl.ac.uk
First published on 9th December 2025
Abstract
The self-assembly of metal–organic cages enables the rapid creation of atomically defined, three-dimensional, nanoscale architectures from easily accessible building blocks. Rigid and flat aromatic panels are typically used as ligands, but limit the diversity and aqueous solubility of cages thus formed. Building on our recent success using oligoprolines to create defined metal-peptidic Pd2L4 cages with emergent head-to-tail isomer control, we now show that installation of an additional metal-binding motif enables formation of a new family of Pd3L4 dual-cavity anisotropic ‘peanut’ cages. Using automated solid-phase peptide synthesis enables generation of a ligand series by varying sequence isomer and/or the stereochemistry of the 4R/S-hydroxyproline. Small differences in ligand isomerism generate four distinct self-assembly outcomes, forming: the Pd3L4cis CCNN cage isomer, the Pd3L4 ‘All Up’ CCCC cage isomer, a mixture of all possible isomers of Pd3L4 cages, or an interpenetrated Pd6L8 cage. Finally, these subtle alterations in cage structure led to differing host–guest interactions and strikingly divergent stability profiles for the metal-peptidic cages when exposed to a range of stimuli. Certain isomers remain stable to base for more than six days, while others fully degrade within an hour. This work underscores the advantages of using biological building blocks in supramolecular chemistry to create systems with tuneable properties.
Introduction
Metal–organic cages are discrete, three-dimensional species formed from the self-assembly of metal ions with rigid, organic ligands.1–3 A variety of polyhedra with defined internal cavities can be readily formed, and the function of these systems often stems from the dramatically different properties of the internal cavity from the surrounding solution.4,5 Metal–organic cages have been shown to perform challenging separations,6 catalyse reactions at rates comparable to enzymes,7–9 act as contrast agents or transport cargoes in vivo,10–12 and sequester contaminants.13,14 Flat, aromatic panels are often used to provide the structural rigidity required to favour the self-assembly of defined, discrete species.15 This leads to two key problems; firstly, water solubility can be challenging and stability limited, due to the fundamental propensity of building blocks to precipitate from solution.16 Secondly, functionalisation of the internal cavity, where the most interesting properties of the cages lie, is challenging and few examples have been reported.17–19Creating anisotropic cavities has been an area of increasing interest, as researchers seek to move away from the pseudospherical cavities of the current generation of metal–organic cages, towards systems better able to mimic the selectivity and potency of biological systems.20 The use of less symmetric ligands, and heteroleptic systems, have provided routes to lower symmetry cages with augmented properties.21 However, they are still bounded by the limitations of aromatic and conjugated building blocks. Creating functionalised, and particularly chiral, cage cavities remains challenging.22,23
One way to functionalise the interior of metal–organic cages is to use tritopic, linear, ligands to create differentiated cavities in a single assembly in ‘peanut’ cages.24,25 However, the assembly of ‘peanut’ cages from low symmetry ligands is rare, limiting their anisotropy. Lewis and co-workers have reported a pseudo-heteroleptic Pd3L4 cage formed from a single asymmetric ligand, with geometric complementarity leading to self-assembly of a single cage isomer.26 The anisotropy of such cages is limited, however, by the planarity and lack of chirality of the planar building blocks.
To address these challenges with solubility, biocompatibility, and anisotropy, we recently reported a new class of metal–organic cages with a defined and open structure – metal-peptidic cages – formed from oligoproline ligands, whose defined folding in solution provides the requisite rigidity for the formation of discrete species.27 Oligoprolines reliably form a left-handed polyproline II (PPII) helix in aqueous solutions, which contains all trans-amide bonds.28,29 This secondary structure has a rigid helical structure with a repeat length of 9 Å, with every third residue aligned on the same face of the helix, and tolerates substitution.30–33 This creates a platform for the design of intrinsically water-soluble cages whose interior can be functionalised.34 We have previously demonstrated that a family of Pd2L4 cages of different sizes could be readily formed, and that the richly chiral and helical surfaces of these cages led to unusual host–guest behaviour. More recently, Palma and co-workers have reported a modified system using alternative pyridine linkers – highlighting the robustness of the oligoproline platform.35 Others have investigated the use of metallo-peptidic systems in discrete multi-nuclear complexes and knots, and extended frameworks, achieving high levels of stereocontrol.36–44
Oligoproline ligands are intrinsically directional (the C-terminus is distinct from the N-terminus), and so there are four different head-to-tail cage isomers that can form, even though our cages are homoleptic (Fig. 1B).45–47 The C-termini can all be aligned at one end and bind one palladium(II) ion of the cage (the ‘All Up’ CCCC); three C-termini and one N-terminus can lie at one end (the CCCN); or there are two cases where two C-termini and two N-termini are at each end, either with C termini cis or trans to each other across the palladium(II) centre (cis CCNN and trans CNCN, respectively). Our previous research found the use of complex, chiral, and helical building blocks led to the unexpected emergence of isomer control, with the cis CCNN head-to-tail isomer of cage formed exclusively (Fig. 1A).27
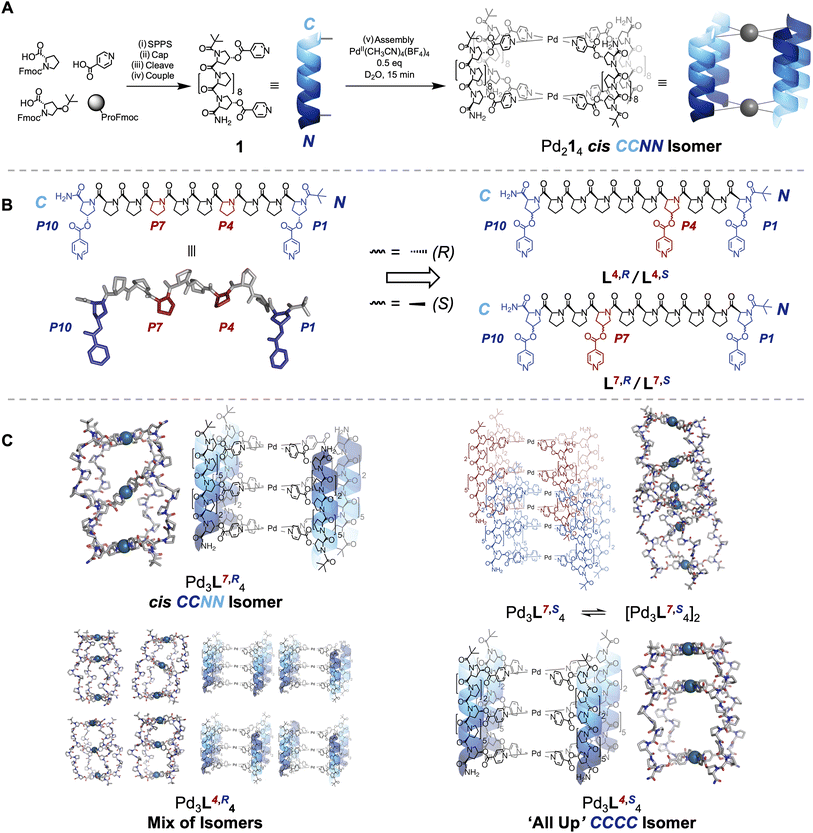 | ||
Fig. 1 (A) Synthesis of 10mer oligoproline ligand 1 and its self-assembly into Pd214 metal-peptide cage in our previous work.27 (i) Solid-phase peptide synthesis (see SI for protocol). (ii) Pivalic anhydride![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CH2Cl2 CH2Cl2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMF 1 DMF 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4.5 4.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4.5, r.t., 45 min. (iii) Trifluoroacetic acid 4.5, r.t., 45 min. (iii) Trifluoroacetic acid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) triisopropyl silane triisopropyl silane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O 38 H2O 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1:1, r.t, 2 h (iv) EDCI (6 equiv.), DMAP (3 equiv.), isonicotinic acid (6 equiv.), CH2Cl2, r.t., 16 h. (v) Pd(CH3CN)4(BF4)2 (0.5 equiv)., D2O, r.t., 15 min. (B) Chemical structure and molecular model of 1, with internally-aligned proline sidechains highlighted in red (left). Additional metal coordinating residues were installed at these positions to give oligoproline ligands L7,R, L4,R, L7,S and L4,S used in this paper (right, see SI for synthesis). (C) Self-assembly outcomes. (i) Pd(CH3CN)4(BF4)2 (0.75 equiv.), D2O, r.t., 4 days. 1:1, r.t, 2 h (iv) EDCI (6 equiv.), DMAP (3 equiv.), isonicotinic acid (6 equiv.), CH2Cl2, r.t., 16 h. (v) Pd(CH3CN)4(BF4)2 (0.5 equiv)., D2O, r.t., 15 min. (B) Chemical structure and molecular model of 1, with internally-aligned proline sidechains highlighted in red (left). Additional metal coordinating residues were installed at these positions to give oligoproline ligands L7,R, L4,R, L7,S and L4,S used in this paper (right, see SI for synthesis). (C) Self-assembly outcomes. (i) Pd(CH3CN)4(BF4)2 (0.75 equiv.), D2O, r.t., 4 days. | ||
Herein, the helical nature and modular synthesis of oligoprolines enables the design of cages with multiple internal cavities, and controlled head-to-tail isomerism, by installing additional metal coordinating residues. Using intrinsically tuneable subcomponents, and automated peptide synthesis, provides a unique platform for the synthesis of tritopic building blocks for Pd3L4 cages.48 Fine adjustments to the structure can be made by changing the peptide sequence and/or the point chirality of amino acid building blocks. This allows us to precisely adjust the relative location and geometry of metal-binding motifs along the peptidic backbone. We hypothesised that such adjustments would enable us to control the outcome, and head-to-tail isomerism, of metal-peptidic cage assembly. Differentiated, highly anisotropic, cavities form and highly complex behaviour can be generated from simple changes to peptide sequence isomers and/or point stereochemistry (Fig. 1). Four isomers of a single ligand deliver four unique outcomes – a cis CCNN Pd3L4 cage, an ‘All Up’ CCCC Pd3L4 cage, a mixture of all possible Pd3L4 cage isomers, and an interpenetrated Pd6L8 cage – demonstrating the flexibility and power of using complex chiral building blocks in supramolecular chemistry. Furthermore, each outcome of cage self-assembly shows significant differences in stability to a range of stimuli, and differentiated host–guest chemistry, informing future applications.
Results and discussion
We initially targeted the installation of an additional metal binding site within the cavity in an attempt to override the intrinsic preference of our Pd2L4 systems to form cis CCNN cages (Fig. 1A),27 reasoning that an asymmetrically aligned additional binding site would favour the ‘All Up’ CCCC isomer, as the only way to achieve coordinative saturation. The alignment of every third residue on the same face of the helix in an idealised PPII structure provides a direct route to Pd3L4 cages (Fig. 1B).28 Ligands L7,R, L4,R, L7,S and L4,S were synthesised, consisting of seven proline and three hydroxyproline residues, with isonicotinic acids coupled to the Hyp positions, by solid phase peptide synthesis (SPPS), and purified by high performance liquid chromatography (HPLC). Due to the directionality of the PPII helix, in a 10mer helix the additional metal binding site can be closer to the N-terminus (L4,R, L4,S) or the C-terminus (L7,R, L7,S). The stereochemistry of the C–O bond was also varied, as this might further perturb isomer preference. L4,R and L7,R contained 4R stereocentres on Hyp sidechains, which occur naturally in biology, whilst L4,S and L7,S contained the unnatural 4S stereocentre. A tert-butyl carbonyl group was installed to provide additional PPII stability and a distinct NMR handle for cage assembly and assignment of isomer formation.49 Folding to a PPII conformation in all ligands (L7,R, L4,R, L4,S and L7,S) in H2O was confirmed by CD spectroscopy, showing characteristic minima and maxima at c. 205 and 225 nm (Fig. 2E and 3D, SI Section 9).50With the ligands in hand, L7,R and L4,R, both containing natural 4R Hyp stereocentres, were assembled to cages by addition of Pd(CH3CN)4(BF4)2 in a precisely 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 ligand
3 ligand![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
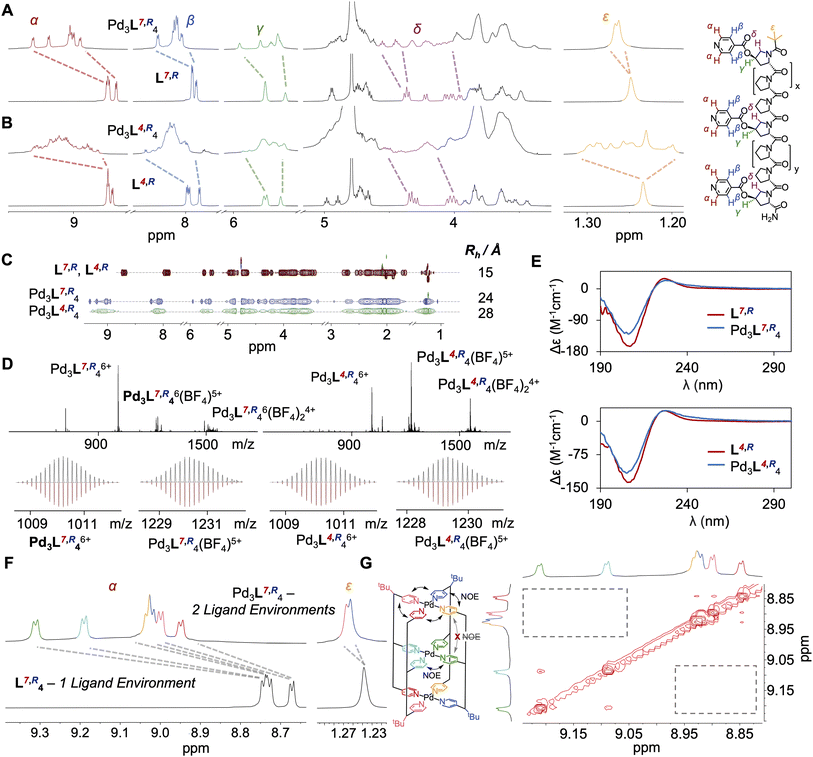
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) metal ratio. Discrete species Pd3L7,R4 and Pd3L4,R4 were formed in each case (Fig. 2A and B), with downfield shifts in the pyridyl protons indicative of palladium(II) co-ordination, and desymmetrisation of ligand signals along the oligoproline backbone, indicative of cage assembly. The 1H NMR of both assemblies showed changes over time, with initially broad signals sharpening gradually over 24 hours (Fig. S107–S110). This indicates that the self-assembly process faces a higher energetic barrier to equilibration than our Pd2L4 cages, which were equilibrated within five minutes. We attribute this to the costs of breaking additional coordination bonds (vide infra).
metal ratio. Discrete species Pd3L7,R4 and Pd3L4,R4 were formed in each case (Fig. 2A and B), with downfield shifts in the pyridyl protons indicative of palladium(II) co-ordination, and desymmetrisation of ligand signals along the oligoproline backbone, indicative of cage assembly. The 1H NMR of both assemblies showed changes over time, with initially broad signals sharpening gradually over 24 hours (Fig. S107–S110). This indicates that the self-assembly process faces a higher energetic barrier to equilibration than our Pd2L4 cages, which were equilibrated within five minutes. We attribute this to the costs of breaking additional coordination bonds (vide infra).
1H NMR, 13C NMR, Correlated Spectroscopy (COSY), Heteronuclear Single Quantum Coherence Spectroscopy (HSQC), Heteronuclear Multiple Bond Correlation Spectroscopy (HMBC), Diffusion-Ordered Spectroscopy (DOSY), High Resolution Electrospray Mass Spectroscopy (ESI-HRMS), Ion Mobility Mass Spectrometry (IMMS) and Circular Dichroism (CD) data were all consistent with the formation of metal-peptidic cages (Fig. 2, S41–S63, S163 and S164). DOSY spectra showed a single diffusion band for Pd3L7,R4 and Pd3L4,R4, with a hydrodynamic radius approximately twice that of the free ligand (Fig. 2C), consistent with cage assembly.51 ESI-HRMS showed clean formation of cage, and isotopic distributions matching simulations (Fig. 2D). IMMS showed peaks at a collision cross-section of 1046 Å2 and 1076 Å2 for Pd3L7,R4 and Pd3L4,R4, consistent with previous results for the Pd214 species (Fig. S163 and S164).27,52 CD confirmed retention of the PPII structure of ligands L7,R and L4,R on cage assembly (Fig. 2E, S166 and S167).
Interestingly, whilst HRMS, CD and DOSY suggested formation of identical species, interrogation of the 1H NMR told a different story. Ligand L7,R, where the additional binding site is close to the C-terminus, assembled into a sharply resolved single species Pd3L7,R4, with two-fold desymmetrisation throughout the ligand strand. Further analysis showed unexpected formation of cis CCNN, rather than the expected ‘All Up’.27 The assignment of a single isomer, rather than an equal mixture of ‘All Up’ CCCC and trans CNCN isomers, is further supported by NOESY analysis, with Hα pyridine environments corresponding to the internal co-ordinating motifs isolated from the N- and C-terminal Hα pyridine environments (Fig. 2G, S182 and S183, see SI Section 10 for detailed reasoning).
The 1H NMR spectrum for Pd3L4,R4 is more complex (Fig. 2B), but clearly resolves eight major tert-butyl signals, indicative of eight different ligand environments (Fig. S184). This is further supported by analysis of the aromatic pyridine Hα and Hβ protons (Fig. S181) showing a similar increase in identifiable environments. As no other major species were observed by ESI-HRMS (Fig. S60), and a single species was shown by DOSY, these ligand environments likely correspond to multiple cage isomers being present in solution. To observe eight different environments, all four cage isomers must be present, thus demonstrating a total lack of isomer selectivity, which was consistent across assembly temperatures (Fig. S64–S66). Our original Pd2L4 cages display an inherent energetic preference for the cis CCNN isomer, and our additional modifications are fighting this intrinsic bias (SI Section 12 for further discussion).
We sought to obtain crystallographic data to better understand the difference in cage isomer selectivity between Pd3L7,R4 and Pd3L4,R4. However, despite extensive attempts (>200 per sample), no crystals suitable for X-ray diffraction formed, consistent with previously reported difficulties in crystallising PPII structures.27,28 Molecular modelling studies were therefore undertaken. Our previous studies of Pd2L4 systems indicated that the cis CCNN cage isomer adopted a tilt, where the helical axis of the oligoproline rods was not perpendicular to the pyridine co-ordination vector (Fig. S197), which significantly decreased the energy of the cis CCNN relative to the other isomers.
Modelling of Pd3L7,R4 shows that the lowest energy isomer of the cage is likewise the tilted cis CCNN, with a distortion of the internal palladium co-ordination plane resulting in the formation of two symmetric cavities (Fig. 1C, S229 and S230). Computational models of Pd3L4,R4 gave the cis CCNN isomer as the highest energy structure (SI Section 12), suggesting it is strongly disfavoured. This supports the difference in behaviour between the two systems seen by 1H NMR.
Having noted the striking effect of moving the internal binding site between N- and C-terminal proximity, we next investigated the effect of epimerising the C–O bond in Hyp, hypothesising that this would enable further control. Ligands L7,S4 and L4,S4 (Fig. 1) are diastereoisomers of L7,R4 and L4,R4 respectively, with 4S-hydroxyprolines in place of 4R. Additional peaks were observed in the 1H NMR spectra of both ligands (Fig. S185 and S186). As both L7,S4 and L4,S4 were >98% pure by liquid chromatography mass spectrometry (Fig. S28 and S38), these peaks likely correspond to a subpopulation of ligand containing cis amide bonds, estimated by NMR to be <10%. This was confirmed by CD spectroscopy, with a reduction in the characteristic PPII peaks for both L7,S4 and L4,S4 relative to L7,R4 and L4,R4 (Fig. 3D, S171, S173 and S175). 4S-Hyp electron withdrawing substituents are known to destabilise PPII helices,53–55 inducing an endo-ring pucker that destabilises trans amide bonds relative to 4R-Hyp explaining this reduction.30
Ligands L7,S4 and L4,S4 were assembled with Pd(CH3CN)4(BF4)2 in a precisely 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 ligand
3 ligand![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) metal ratio, and fully characterised (Fig. 3, S67–S92, S165 and S166). Results were consistent with metal-peptidic cage assembly. For L4,S4 the 1H NMR yielded a sharp and discrete major species Pd3L4,S4 (c. 70% of the ligand converts to major isomer) after 48 h, in contrast to the mixture of cage isomers observed for Pd3L4,R4 (Fig. 3A and S113). DOSY showed a single diffusion band, indicating formation of a single species with a hydrodynamic radius of 28 Å (Fig. S85). ESI-HRMS confirmed successful self-assembly of cage Pd3L4,S4, and isotopic distributions matching simulations (Fig. 3B and S90–S92). IMMS showed a peak for Pd3L4,S4 at a collision cross-section of 1023 Å2, consistent with previous results (Fig. S166).
metal ratio, and fully characterised (Fig. 3, S67–S92, S165 and S166). Results were consistent with metal-peptidic cage assembly. For L4,S4 the 1H NMR yielded a sharp and discrete major species Pd3L4,S4 (c. 70% of the ligand converts to major isomer) after 48 h, in contrast to the mixture of cage isomers observed for Pd3L4,R4 (Fig. 3A and S113). DOSY showed a single diffusion band, indicating formation of a single species with a hydrodynamic radius of 28 Å (Fig. S85). ESI-HRMS confirmed successful self-assembly of cage Pd3L4,S4, and isotopic distributions matching simulations (Fig. 3B and S90–S92). IMMS showed a peak for Pd3L4,S4 at a collision cross-section of 1023 Å2, consistent with previous results (Fig. S166).
The major species of Pd3L4,S4 has a single ligand environment, which is not consistent with a cis CCNN metal-peptidic cage. The absence of desymmetrisation indicates formation of either an ‘All Up’ CCCC or trans CNCN arrangement as the major isomer (see SI Section 10). Geometric considerations, and molecular modelling, suggest the formation of the trans CNCN isomer would be unfavourable (see SI Section 12) without complete loss of PPII folding, which is not observed by CD. We therefore assign Pd3L4,S4 as the ‘All Up’ CCCC isomer.
Analysis of the NOESY correlations further supports this assignment (Fig. 3E and F). The NOESY spectrum of Pd3L4,S4 shows strong correlations and shielding between protons of two of the three pyridines on each ligand and proline sidechains (Fig. 3F and S190), which we have not observed in other systems. Significant upfield shifts of the α, β and γ protons of Pro2/Pro3 are consistent with close proximity to the pyridine's aromatic ring current (Fig. 3E and S191). The most likely mechanism involves compression of the smaller cavity of the ‘All Up’ CCCCPd3L4,S4 cage. This hypothesis is supported by CD changes of Pd3L4,S4 with respect to free ligand L4,S (Fig. 3D and S170), and the appearance of new absorbances (the characteristic PPII peak at 230 nm shifts and broadens to 238 nm) which do not correlate with PPII, PPI, or unstructured peptide,53,56 and could be due to a twisting and compression of the smaller cavity of Pd3L4,S4. Given the decrease in PPII helical character (1H NMR + CD; Fig. S175 and S186) in ligand L4,S compared to ligand L4,R, this process is likely mediated by trans to cis isomerism of some amide bonds in the smaller cavity of the cage, which contains a higher local concentration of PPII-destabilising 4S-Hyp. Minor species in the 1H NMR likely correspond to small amounts of the three other cage isomers, as no other major species were observed by ESI-HRMS (Fig. S90).
Whilst the change in isomer selectivity surprised us, flipping the 4R-Hyp stereocentres of L4,R to 4S-Hyp does indeed lead to self-assembly of a single Pd3L4 cage isomer. This novel ‘All Up’ CCCC isomer has two separate and highly distinctive cavities, achieving one the initial aims of this project.
Self-assembly of L7,S with Pd(CH3CN)4(BF4)2 gave a broad species in the 1H NMR spectra which did not resolve over seven days (Fig. S111). ESI-HRMS showed the unexpected formation of both Pd3L4 and Pd6L8 species (Fig. S75–S81 and S187). DOSY NMR showed a single diffusion band with a hydrodynamic radius of 33.3 Å (Fig. S187), larger than the hydrodynamic radii seen for the three Pd3L4 cages Pd3L7,R4, Pd3L4,R4 and Pd3L4,S4 (24.0–28.3 Å), consistent with the presence of a larger metal-peptidic structure.
We assigned this to an interpenetrated Pd6L8 cage (Pd3L7,S4)2 formed in equilibrium by the interlocking of two Pd3L7,S4 cage complexes. Such structures have been reported in the literature, often driven by exclusion of solvent from cavities.57–59 The broadness in the 1H NMR is due to increased molecular weight leading to decreased tumbling, but also to the presence of isomers, along with the equilibrium between Pd3L7,S4 and (Pd3L7,S4)2. Dilution of the Pd6L8 cage (Pd3L7,S4)2 led to the disappearance of peaks corresponding to the interpenetrated species, leaving the Pd3L46+ peaks unchanged, supporting this assignment (Fig. S75, S76 and S187).59,60 This was supported by DOSY studies, in which a decrease in hydrodynamic radii was observed upon dilution (Fig. S121). Assembly studies (see SI Section 5) showed that an intermediate Pd2L4 species first forms, before assembly of (Pd3L7,S4)2. CD showed significant loss of PPII character, consistent with distortion on formation of the assembled cage. As such, the cavity compression seen in Pd3L4,S4 and the partial catenation seen in (Pd3L7,S4)2 are different responses to the same stressor – increasing disruption of PPII by incorporation of 4S-Hyp.
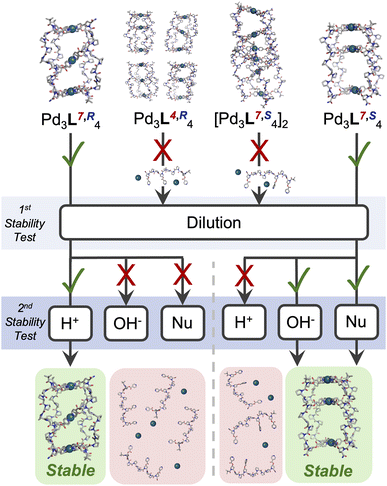
Finally, we sought to probe whether the different cage structures formed had distinct chemical properties. In the longer term, these cages hold promise for applications in biological systems, which requires tolerance to different stimuli. Firstly, the assemblies were diluted, and monitored by 1H NMR (Fig. 4, SI Section 7). Cages Pd3L7,R4 and Pd3L4,S4, the single isomer cages, were stable to a concentration of 50 µM in D2O (Fig. S115, S116, S122 and S123). This represents a fourfold increase from the stability of previously reported Pd214, likely due to the additional co-ordination bonds within Pd3L7,R4 and Pd3L4,S4.27 In contrast, cages Pd3L4,R4 and (Pd3L7,S4)2 only showed stability to concentrations of 500 µM in D2O (Fig. S117–S120). The most stable cages were then challenged with base, acid, and competitive metal binding ligands – glutathione and pyridine (Fig. 4). Pyridine was chosen due to the ubiquity of nitrogen heterocycles in drug molecules, and glutathione due to its high concentrations in cells and plasma.
Cages Pd3L7,R4 and Pd3L4,S4 were first tested for base stability by addition of 4 eq. NaOD, and showed distinct differences in behaviour. Whilst cage Pd3L7,R4 was almost immediately (<1 h) disassembled and its ester bonds hydrolysed (Fig. S133 and S134), cage Pd3L4,S4 was only partially disassembled after 120 h (<25% by 1H NMR; Fig. S140 and S141).
A similar trend in resilience was observed when Pd3L7,R4 and Pd3L4,S4 were challenged with competing ligands – glutathione and pyridine. Addition of 4 eq. of glutathione caused complete disassembly of Pd3L7,R4 within 5 min, but Pd3L4,S4 was not completely disassembled until 48 h post addition (Fig. S155, S156, S161 and S162). Both Pd3L7,R4 and Pd3L4,S4 showed significantly enhanced stability to pyridine addition, with 4 eq. failing to cause full disassembly even after 48 h (Fig. S143, S144, S151 and S152). However, after addition of 12 eq. pyridine, Pd3L4,S4 remained detectable in solution for c. 1 h, whilst Pd3L7,R4 was almost immediately (<10 min) disassembled (Fig. S145, S146, S153 and S154). The partially collapsed smaller cavity of the ‘All Up’ CCCCPd3L4,S4 cage may provide a more condensed cage structure, in which Pd metal centres and ester bonds are sterically protected from external attack, explaining this difference. The cis CCNN cage Pd3L7,R4 has two symmetric cavities with a more open structure, and could be rapidly attacked by −OD, glutathione, and pyridine. This is supported by the observation of complete hydrolysis of ligand esters upon treatment of cage Pd3L7,R4 with −OD, whilst Pd3L4,S4 showed significant residual cage (Fig. S135 and S142).
Remarkably, when cages Pd3L7,R4 and Pd3L4,S4 were challenged by addition of 4 eq. DCl, a reversal in the stability trend was seen. The ‘All Up’ Pd3L4,S4 cage was >80% disassembled after 48 h, whereas only c. 35% of the cis CCNNPd3L7,R4 cage had been disassembled (Fig. S124, S125, S131 and S132). The difference observed between acid and base addition is likely due to the differing mechanisms of cage destruction, with accessibility of the ester (base hydrolysis) and the pyridine nitrogen (protonation) varying between cages.
Having shown differential stability, varying host–guest properties between the differentially sized cavities were then explored (SI Section 11). Exemplar 1H NMR titrations of the cis CCNNPd3L7,R4 and ‘All Up’ CCCCPd3L4,S4 cages with negatively charged aromatic anions benzenesulfonate and pyrene-1-sulfonate were performed. Pleasingly, substantial differences in binding constants for sodium benzenesulfonate with Pd3L7,R4 and Pd3L4,S4 were found, with the cis CCNNPd3L7,R4 binding almost an order of magnitude more strongly than the ‘All Up’ CCCCPd3L4,S4 cage (Ka = 911 M−1 ± 6% for Pd3L7,R4vs. Ka = 120 M−1 ± 4% for Pd3L4,S4, Fig. S192–S204). A similar, but less pronounced, trend of interaction was found with sodium pyrene-1-sulfonate between the two cages (Ka = 1557 M−1 ± 7% for Pd3L7,R4vs. Ka = 2192 M−1 ± 6% for Pd3L4,S4, Fig. S205–S210). Further, evidence of differing binding modes between the two cages was seen by 1H NMR (Fig. 5). For Pd3L7,R4, 1H resonances of external and internal pyridine Hα residues (Fig. S206) showed significant Δppm shifts (up to c. 0.25 ppm), suggesting a cavity bound species interacting with the central pyridines. In Pd3L4,S4, only the external Hyp Hγ and pyridine residues showed significant Δppm shifts (up to 0.35 ppm for Hyp, Fig. S209), suggesting guest binding on the external aromatic Pd-pyridine panels of the cage, and so evidencing differential binding modes in cages with different cavities (Fig. 5).
Finally, guest binding with Irinotecan, an anticancer agent, was investigated. As Irinotecan is administered clinically as the HCl salt, this provides a bridge between the differential acid stabilities seen and guest binding. Binding constants were similar (Ka = 2188 M−1 ± 6% for Pd3L7,R4vs. Ka = 2629 M−1 ± 9% for Pd3L4,S4, Fig. S211–S216), but Pd3L4,S4, and not Pd3L7,R4, disassembled at higher Irinotecan. HCl equivalencies (>2 eq.) (Fig. S212 and S215). The interplay of differential guest binding and cage stability, therefore, provides a novel route to control guest binding.
Conclusions
Herein, we have shown that a series of four isomeric oligoproline ligands can be used to synthesise Pd3L4 metal-peptidic ‘peanut’ cages. Small changes in ligands, varying structural isomers and point chiral centres, can lead to dramatic changes in self-assembly and stability. Each of the four tritopic ligands produced a different cage upon addition of Pd(II): L7,R formed the cis CCNN Pd3L4 isomer, L4,R formed all possible Pd3L4 isomers simultaneously, L7,S formed an interpenetrated Pd6L8 cage, and L4,S formed the ‘All Up’ CCCC Pd3L4 isomer, generating a diverse range of anisotropic cavities. The cis and ‘All Up’ cages were significantly more stable, and showed contrasting differences in stability to various stimuli. They showed differences in host–guest behaviour, and binding site. Future work will explore how the asymmetric cavities generated can be leveraged for drug delivery and release, catalysis and sensing.Author contributions
Authorship is alphabetical between B. E. B., E. M. G. J., and L. E. M. W. Conceptualisation, B. E. B., E. M. G. J., L. E. M. W., and C. T. M.; formal analysis, B. E. B., E. M. G. J., and L. E. M. W.; investigation, B. E. B., E. M. G. J., and L. E. M. W.; resources, C. T. M.; writing – original draft, B. E. B., E. M. G. J., L. E. M. W.; writing – review & editing; B. E. B., E. M. G. J., L. E. M. W., and C. T. M.; visualisation, B. E. B., E. M. G. J., L. E. M. W., and C. T. M; supervision; C. T. M.; project administration, C. T. M.; funding acquisition, C. T. M.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5sc06441d.Acknowledgements
We thank Dr Meng Li, Warwick University, for recording IMMS data, Dr Peter Simpson, Dr Alain Oregioni, and Dr Geoff Kelly for their advice and support with NMR spectroscopy and Dr Laura Masino for assistance with CD spectroscopy. This work was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (CC2213), the UK Medical Research Council (CC2213), and the Wellcome Trust (CC2213) and by the UKRI under Future Leaders Fellowship MR/W00657X/1. This work was supported by the Francis Crick Institute through provision of access to the MRC Biomedical NMR Centre and the Structural Biology Science and Technology Platform. The Francis Crick Institute receives its core funding from Cancer Research UK (CC1078), the UK Medical Research Council (CC1078), and the Wellcome Trust (CC1078).References
- S. R. Seidel and P. J. Stang, Acc. Chem. Res., 2002, 35(11), 972–983, DOI:10.1021/ar010142d.
- M. Han, D. M. Engelhard and G. H. Clever, Chem. Soc. Rev., 2014, 43(6), 1848–1860, 10.1039/C3CS60473J.
- M. Fujita and K. Ogura, Bull. Chem. Soc. Jpn., 1996, 69(6), 1471–1482, DOI:10.1246/bcsj.69.1471.
- F. J. Rizzuto, L. K. S. Von Krbek and J. R. Nitschke, Nat. Rev. Chem., 2019, 3(4), 204–222, DOI:10.1038/s41570-019-0085-3.
- M. Yoshizawa, T. Kusukawa, M. Kawano, T. Ohhara, I. Tanaka, K. Kurihara, N. Niimura and M. Fujita, J. Am. Chem. Soc., 2005, 127(9), 2798–2799, DOI:10.1021/ja043953w.
- D. Zhang, T. K. Ronson, Y. Zou and J. R. Nitschke, Nat. Rev. Chem., 2021, 5(3), 168–182, DOI:10.1038/s41570-020-00246-1.
- C. J. Hastings, M. D. Pluth, R. G. Bergman and K. N. Raymond, J. Am. Chem. Soc., 2010, 132(20), 6938–6940, DOI:10.1021/ja102633e.
- W. Cullen, M. C. Misuraca, C. A. Hunter, N. H. Williams and M. D. Ward, Nat. Chem., 2016, 8(3), 231–236, DOI:10.1038/nchem.2452.
- R. G. DiNardi, S. Rasheed, S. S. Capomolla, M. H. Chak, I. A. Middleton, L. K. Macreadie, J. P. Violi, W. A. Donald, P. J. Lusby and J. E. Beves, J. Am. Chem. Soc., 2024, 146(31), 21196–21202, DOI:10.1021/jacs.4c04846.
- R. Cosialls, C. Simó, S. Borrós, V. Gómez-Vallejo, C. Schmidt, J. Llop, A. B. Cuenca and A. Casini, Chem. Eur. J., 2023, 29(3), e202202604, DOI:10.1002/chem.202202604.
- B. P. Burke, W. Grantham, M. J. Burke, G. S. Nichol, D. Roberts, I. Renard, R. Hargreaves, C. Cawthorne, S. J. Archibald and P. J. Lusby, J. Am. Chem. Soc., 2018, 140(49), 16877–16881, DOI:10.1021/jacs.8b09582.
- G. E. Sokolow, M. R. Crawley, D. R. Morphet, D. Asik, J. A. Spernyak, A. J. R. McGray, T. R. Cook and J. R. Morrow, Inorg. Chem., 2022, 61(5), 2603–2611, DOI:10.1021/acs.inorgchem.1c03660.
- T. K. Ronson, J. P. Carpenter and J. R. Nitschke, Chem, 2022, 8(2), 557–568, DOI:10.1016/j.chempr.2021.12.017.
- E. G. Percástegui, Chem. Commun., 2022, 58(33), 5055–5071, 10.1039/D2CC00604A.
- S. Pullen and G. H. Clever, Acc. Chem. Res., 2018, 51(12), 3052–3064, DOI:10.1021/acs.accounts.8b00415.
- E. G. Percástegui, T. K. Ronson and J. R. Nitschke, Chem. Rev., 2020, 120(24), 13480–13544, DOI:10.1021/acs.chemrev.0c00672.
- M. Tominaga, K. Suzuki, T. Murase and M. Fujita, J. Am. Chem. Soc., 2005, 127(34), 11950–11951, DOI:10.1021/ja054069o.
- A. M. Johnson and R. J. Hooley, Inorg. Chem., 2011, 50(11), 4671–4673, DOI:10.1021/ic2001688.
- P. Howlader, P. Das, E. Zangrando and P. S. Mukherjee, J. Am. Chem. Soc., 2016, 138(5), 1668–1676, DOI:10.1021/jacs.5b12237.
- C. T. McTernan, J. A. Davies and J. R. Nitschke, Chem. Rev., 2022, 122(11), 10393–10437, DOI:10.1021/acs.chemrev.1c00763.
- S. Pullen, J. Tessarolo and G. H. Clever, Chem. Sci., 2021, 12(21), 7269–7293, 10.1039/D1SC01226F.
- K. Suzuki, M. Kawano, S. Sato and M. Fujita, J. Am. Chem. Soc., 2007, 129(35), 10652–10653, DOI:10.1021/ja073629b.
- X. Tang, H. Jiang, Y. Si, N. Rampal, W. Gong, C. Cheng, X. Kang, D. Fairen-Jimenez, Y. Cui and Y. Liu, Chem, 2021, 7(10), 2771–2786, DOI:10.1016/j.chempr.2021.07.017.
- K. Yazaki, M. Akita, S. Prusty, D. K. Chand, T. Kikuchi, H. Sato and M. Yoshizawa, Nat. Commun., 2017, 8(1), 15914, DOI:10.1038/ncomms15914.
- D. Preston, J. E. M. Lewis and J. D. Crowley, J. Am. Chem. Soc., 2017, 139(6), 2379–2386, DOI:10.1021/jacs.6b11982.
- J. E. M. Lewis, Angew. Chem., Int. Ed., 2022, 61(44), e202212392, DOI:10.1002/anie.202212392.
- B. E. Barber, E. M. G. Jamieson, L. E. M. White and C. T. McTernan, Chem, 2024, 10(9), 2792–2806, DOI:10.1016/j.chempr.2024.05.002.
- P. Wilhelm, B. Lewandowski, N. Trapp and H. Wennemers, J. Am. Chem. Soc., 2014, 136(45), 15829–15832, DOI:10.1021/ja507405j.
- T. J. El-Baba, D. R. Fuller, D. A. Hales, D. H. Russell and D. E. Clemmer, J. Am. Soc. Mass Spectrom., 2019, 30(1), 77–84, DOI:10.1007/s13361-018-2034-7.
- M. Kümin, L.-S. Sonntag and H. Wennemers, J. Am. Chem. Soc., 2007, 129(3), 466–467, DOI:10.1021/ja067148o.
- S. Dobitz, M. R. Aronoff and H. Wennemers, Acc. Chem. Res., 2017, 50(10), 2420–2428, DOI:10.1021/acs.accounts.7b00340.
- C.-L. Tsai, J.-W. Chang, K.-Y. Cheng, Y.-J. Lan, Y.-C. Hsu, Q.-D. Lin, T.-Y. Chen, O. Shih, C.-H. Lin, P.-H. Chiang, M. Simenas, V. Kalendra, Y.-W. Chiang, C.-H. Chen, U.-S. Jen and S.-K. Wang, Nanoscale Adv., 2024, 6, 947–959, 10.1039/D3NA00945A.
- D. F. Brightwell, G. Truccolo, K. Samanta, H. J. Shepherd and A. Palma, ACS Macro Lett., 2023, 12(7), 908–914, DOI:10.1021/acsmacrolett.3c00304.
- A. K. Pandey, D. Naduthambi, K. M. Thomas and N. J. Zondlo, J. Am. Chem. Soc., 2013, 135(11), 4333–4363, DOI:10.1021/ja3109664.
- D. F. Brightwell, K. Samanta, J. Muldoon, P. C. Fleming, Y. Ortin, L. Mardiana, P. G. Waddell, M. J. Hall, E. R. Clark, F. Fantuzzi and A. Palma, A. ChemistryEurope, 2024, e202400050, DOI:10.1002/ceur.202400050.
- T. Sawada, Y. Inomata, K. Shimokawa and M. Fujita, Nat. Commun., 2019, 10(1), 5687, DOI:10.1038/s41467-019-13594-4.
- T. Sawada, A. Matsumoto and M. Fujita, Angew. Chem., Int. Ed., 2014, 53(28), 7228–7232, DOI:10.1002/anie.201403506.
- M. Yamagami, T. Sawada and M. Fujita, J. Am. Chem. Soc., 2018, 140(28), 8644–8647, DOI:10.1021/jacs.8b04284.
- Y. Inomata, T. Sawada and M. Fujita, Chem, 2020, 6(1), 294–303, DOI:10.1016/j.chempr.2019.12.009.
- T. Schnitzer, E. Paenurk, N. Trapp, R. Gershoni-Poranne and H. Wennemers, J. Am. Chem. Soc., 2021, 143(2), 644–648, DOI:10.1021/jacs.0c11793.
- J. Dong, Y. Liu and Y. Cui, J. Am. Chem. Soc., 2021, 143(42), 17316–17336, DOI:10.1021/jacs.1c08487.
- P.-M. Cheng, T. Jia, C.-Y. Li, M.-Q. Qi, M.-H. Du, H.-F. Su, Q.-F. Sun, L.-S. Long, L.-S. Zheng and X.-J. Kong, Nat. Commun., 2024, 15(1), 9034, DOI:10.1038/s41467-024-53320-3.
- X. Kang, L. Wang, B. Liu, S. Zhou, Y. Li, S.-L. Yang, R. Yao, L. Qiao, X. Wang, W. Gong, Y. Liu, L. Shi, J. Dong, Y. Cui and A. P. Davis, Nat. Synth., 2024, 4(1), 43–52, DOI:10.1038/s44160-024-00640-3.
- C. T. McTernan, B. E. Barber, E. M. G. Jamieson and L. E. M. White, Angew. Chem., Int. Ed., 2025, 64, e202512014, DOI:10.1002/anie.202512014.
- J. E. M. Lewis, A. Tarzia, A. J. P. White and K. E. Jelfs, Chem. Sci., 2020, 11(3), 677–683, 10.1039/C9SC05534G.
- J. E. M. Lewis, Chem. Eur. J., 2021, 27(13), 4454–4460, DOI:10.1002/chem.202005363.
- A. Tarzia, J. E. M. Lewis and K. E. Jelfs, Angew. Chem., Int. Ed., 2021, 60(38), 20879–20887, DOI:10.1002/anie.202106721.
- R. Behrendt, P. White and J. Offer, J. Pept. Sci., 2016, 22(1), 4–27, DOI:10.1002/psc.2836.
- M. R. Bhatt and N. J. Zondlo, Chem. Eur. J., 2024, 30(41), e202401454, DOI:10.1002/chem.202401454.
- M. Kuemin, J. Engel and H. Wennemers, J. Pept. Sci., 2010, 16(10), 596–600, DOI:10.1002/psc.1245.
- J. E. M. Lewis, E. L. Gavey, S. A. Cameron and J. D. Crowley, Chem. Sci., 2012, 3(3), 778–784, 10.1039/C2SC00899H.
- J. Ujma, M. De Cecco, O. Chepelin, H. Levene, C. Moffat, S. J. Pike, P. J. Lusby and P. E. Barran, Chem. Commun., 2012, 48(37), 4423–4425, 10.1039/C2CC30778B.
- P. Ruzza, G. Siligardi, A. Donella-Deana, A. Calderan, R. Hussain, C. Rubini, L. Cesaro, A. Osler, A. Guiotto, L. A. Pinna and G. Borin, J. Pept. Sci., 2006, 12(7), 462–471, DOI:10.1002/psc.750.
- J. Horng and R. T. Raines, Protein Sci., 2006, 15(1), 74–83, DOI:10.1110/ps.051779806.
- N. V. Costantini, H. K. Ganguly, M. I. Martin, N. A. Wenzell, G. P. A. Yap and N. J. Zondlo, Chem. Eur. J., 2019, 25(48), 11356–11364, DOI:10.1002/chem.201902382.
- J. L. S. Lopes, A. J. Miles, L. Whitmore and B. A. Wallace, Protein Sci., 2014, 23(12), 1765–1772, DOI:10.1002/pro.2558.
- S. Löffler, J. Lübben, L. Krause, D. Stalke, B. Dittrich and G. H. Clever, J. Am. Chem. Soc., 2015, 137(3), 1060–1063, DOI:10.1021/ja5130379.
- R. Zhu, J. Lübben, B. Dittrich and G. H. Clever, Angew. Chem., Int. Ed., 2015, 54(9), 2796–2800, DOI:10.1002/anie.201408068.
- S. Freye, R. Michel, D. Stalke, M. Pawliczek, H. Frauendorf and G. H. Clever, J. Am. Chem. Soc., 2013, 135(23), 8476–8479, DOI:10.1021/ja403184a.
- T. Tateishi, Y. Yasutake, T. Kojima, S. Takahashi and S. Hiraoka, Commun. Chem., 2019, 2(1), 25, DOI:10.1038/s42004-019-0123-6.
Footnote |
| † Authorship is alphabetical. |
| This journal is © The Royal Society of Chemistry 2026 |